Abstract
The archaeal community in a sulfide- and sulfur-rich spring with a stream water salinity of 0.7 to 1.0% in southwestern Oklahoma was studied by cloning and sequencing of 16S rRNA genes. Two clone libraries were constructed from sediments obtained at the hydrocarbon-exposed source of the spring and the microbial mats underlying the water flowing from the spring source. Analysis of 113 clones from the source library and 65 clones from the mat library revealed that the majority of clones belonged to the kingdom Euryarchaeota, while Crenarchaeota represented less than 10% of clones. Euryarchaeotal clones belonged to the orders Methanomicrobiales, Methanosarcinales, and Halobacteriales, as well as several previously described lineages with no pure-culture representatives. Those within the Halobacteriales represented 36% of the mat library and 4% of the source library. All cultivated members of this order are obligately aerobic halophiles. The majority of halobacterial clones encountered were not affiliated with any of the currently described genera of the family Halobacteriaceae. Measurement of the salinity at various locations at the spring, as well as along vertical gradients, revealed that soils adjacent to spring mats have a much higher salinity (NaCl concentrations as high as 32%) and a lower moisture content than the spring water, presumably due to evaporation. By use of a high-salt-plus-antibiotic medium, several halobacterial isolates were obtained from the microbial mats. Analysis of 16S rRNA genes indicated that all the isolates were members of the genus Haloferax. All isolates obtained grew at a wide range of salt concentrations, ranging from 6% to saturation, and all were able to reduce elemental sulfur to sulfide. We reason that the unexpected abundance of halophilic Archaea in such a low-salt, highly reduced environment could be explained by their relatively low salt requirement, which could be satisfied in specific locations of the shallow spring via evaporation, and their ability to grow under the prevalent anaerobic conditions in the spring, utilizing zero-valent sulfur compounds as electron acceptors. This study demonstrates that members of the Halobacteriales are not restricted to their typical high-salt habitats, and we propose a role for the Halobacteriales in sulfur reduction in natural ecosystems.
Zodletone Spring in southwestern Oklahoma is a mesophilic spring characterized by high dissolved sulfide and sulfur concentrations throughout its course, an abundance of microbial mats that harbor a complex bacterial community, and high concentrations of short-chain gaseous alkanes (methane, ethane, and propane), especially at the source of the spring (19, 53). Activity studies and molecular characterization of the bacterial community at several locations along the spring have revealed a highly diverse bacterial population involved in phototrophically driven sulfur-cycling processes (19). Elshahed et al. also observed a diverse nonphototrophic community with novel division-level diversity, especially at the areas exposed to gaseous alkanes (19).
The presence of an extremely diverse bacterial community within the spring, coupled with the broad diversity currently recognized for the domain Archaea, led us to speculate on the composition of the archaeal community within Zodletone spring. Cultivated members of the domain Archaea are currently classified into 2 kingdoms, 8 orders, and 17 families that include methanogens, heterotrophic thermophiles, thermophilic sulfur and sulfate reducers, and obligate halophiles (23). However, culture-independent surveys of different habitats have indicated that the diversity and geographical distribution within the domain Archaea are much broader than previously inferred from culture-dependent estimates (for reviews, see references 12, 14, 15, and 55).
Although several studies have documented the presence of methanogenic Archaea at a variety of thermophilic (65), psychrophilic (40), and hypersaline (10, 11, 28, 58) microbial mats, few reports exist on detailed culture-independent analysis of the archaeal component in multispecies microbial mats. 16S rRNA gene analysis of the archaeal community in Solar Lake (Sinai, Egypt) hypersaline microbial mats revealed the presence of methanogens (of the genera Methanobacterium and Methanococcus), halobacteria (extremely halophilic Archaea), and members of two groups with no pure-culture representatives (11). A recently described mat system in the Black Sea appeared to be formed mainly of anaerobic methane-oxidizing group I (ANME-1) Archaea and sulfate reducers, both apparently involved in anaerobic, syntrophic methane oxidation (39).
In contrast to the scarcity of reports on the archaeal components of stratified multispecies microbial mats, archaeal diversity had been fairly well documented in a variety of hydrocarbon-impacted environments including locations with methane hydrates (32, 35, 44, 48, 57), hydrocarbon-contaminated sites (16), oil reservoirs (66), and hydrocarbon-degrading enrichments (21). These studies indicated the presence of an extremely diverse archaeal population at such sites, with a particular abundance of clones belonging to known families of methanogens, as well as members of several lineages that have no cultured representatives so far.
In this work, we surveyed the Zodletone Spring archaeal community within the microbial mat and at the spring source. Our original intent was to determine whether clones belonging to previously documented methane-oxidizing archaeal lineages (5, 27, 32, 39, 44, 45, 57) were present at the site and to examine archaeal diversity in an anoxic sulfur-cycling microbial mat. We encountered an unexpected abundance of members of the family Halobacteriaceae (extremely halophilic Archaea) in the microbial mats, in spite of the low concentration of salts in the spring water (0.7 to 1.0%). These observations led us to isolate several halophilic Archaea from the microbial mats. Based on physiological features of these organisms, we suggest a role for this group in anaerobic sulfur cycling at the spring. We also provide evidence that evaporation in the shallow banks of the stream creates a relatively high salinity niche, where halophilic Archaea can survive and grow in this seemingly nonhalophilic setting.
MATERIALS AND METHODS
Site description.
Zodletone Spring is located north of the Anadarko basin in southwestern Oklahoma. Water at the spring source emerges at a rate of 8 liters/min, and the spring flows approximately 20 m before discharging into a nearby creek. The hydrological and geochemical characteristics of Zodletone Spring (26, 51), as well as its bacterial diversity and sulfur transformations, have been described previously (19, 53). Spring water has a high dissolved-sulfide concentration (8 to 10 mM), providing ideal conditions for anaerobic, photosynthetic bacteria. As a result, phototrophic microbial mats are abundant, and they mediate sulfide oxidation and barium sulfate precipitation in the spring (53). Another important characteristic is the continuous bubbling of gaseous short-chain alkanes (methane, ethane, and propane) from the source of the spring, resulting in high hydrocarbon levels, especially in the source area.
Sampling.
Mat and source materials for DNA extractions were carefully removed from the spring with a sterile spatula and stored in sterile whirl pack bags. The samples were immediately frozen in dry ice and stored at −20°C in the laboratory. Samples used for microbial isolation and salinity measurements were stored on ice until transfer to the laboratory, where they were stored at 4°C and used within 24 h. For salinity measurements, 5-cm-deep cores were collected in 30-ml syringes as previously described (53) from locations at 5 and 30 cm from the bank of the spring.
DNA extraction, PCR amplification, cloning, and sequencing.
DNA extraction from sediment was carried out by an indirect Percoll separation of cells (19, 29), followed by a lysis bead-beating protocol (16), and the extracted DNA was visualized on an ethidium bromide-stained 2% agarose gel. Custom primers (Invitrogen Corp., Carlsbad, Calif.) used for amplifying the 16S rRNA gene were the archaeon-specific primer 25f (5′ CYGGTTGATCCTGCCRG 3′) (61) and primer 958r (5′ YCCGGCGTTGAMTCCAAT T 3′) (46). These two degenerate primers have been used to amplify archaeal 16S rRNA genes from a variety of environments (see, e.g., references 8, 13, 16, 30, 36, 61, and 62) and have not been shown to be biased toward a single group of microorganisms. For pure cultures, DNA was extracted from every isolate with no Percoll separation, and the primers used for amplification were Arch. 21F and Uni1492R (49). 16S rRNA genes of the archaeal community were amplified from the bulk community DNA in a 50-μl reaction mixture containing the following (final concentrations): 2 μl of a 1:100 dilution of extracted DNA, 1× PCR buffer (Invitrogen), 2.5 mM MgSO4, 0.2 mM deoxynucleoside triphosphate mixture, 1.5 U of high-fidelity Taq polymerase (Invitrogen), and 10 μM (each) forward and reverse primers. PCR amplification was carried out on a Gene Amp PCR system 9700 thermocycler. 16S rRNA amplification used a protocol involving initial denaturation for 5 min at 94°C; 39 cycles of 94°C for 1.5 min, 55°C for 1.5 min, and 72°C for 2 min; and a final extension at 72°C for 12 min. The PCR products obtained were either purified by using a gel purification kit (Qiagen Inc., Valencia, Calif.) or directly cloned into an Invitrogen TOPO-TA vector by using the cloning kit according to the manufacturer's instructions. Sequencing and assembly procedures were performed as previously described in detail in reference 19 and at http://www.genome.ou.edu/ds_seq_template_isol_hydra.html.
For phylogenetic placement, sequences were initially checked by using the Blast algorithm (1) to determine their rough phylogenetic affiliation. Sequences with more than 98% similarity were considered to belong to the same operational taxonomic unit (OTU). The occurrence of chimeras was checked by using the chimera-check program in the Ribosomal Database Project (34). Zodletone sequences and GenBank-downloaded sequences were aligned by using the ClustalX program (59), and the alignments were manually checked and corrected by using the seqApp program (D. Gilbert, Indiana University). Evolutionary distance trees (neighbor-joining algorithm with Jukes-Cantor corrections) were constructing by using PAUP (version 4.01b10; Sinauer Associates, Sunderland, Mass.). All phylogenetic trees show the frequencies of occurrence of specific OTUs in the source and mat clone libraries.
Isolation and partial characterization of halophilic Archaea.
Halophile medium (HM) modified from the formulation of Oren (42) and containing MgSO4 (20 g/liter), K2SO4 (5 g/liter), CaCl2 · 2H2O (0.1 g/liter), yeast extract (as the only carbon source) (5 g/liter), and NaCl was used for enrichment and isolation of halophilic microorganisms from the mat community. Three different NaCl concentrations (7, 12, or 18%) were utilized in the isolation procedure. To select against halophilic and halotolerant Bacteria, ampicillin and kanamycin were included in the medium at a final concentration of 75 μg/ml. Mat material was either serially diluted in liquid medium and directly plated onto HM agar plates (2% agar) or initially incubated for 3 weeks in liquid medium prior to streaking. Cultures were incubated at 37°C in the light. Colonies were picked from HM solid medium containing the antibiotics, restreaked twice, and microscopically checked for purity.
Tests for optimum salt concentrations were carried out in liquid medium with shaking at 150 rpm, except that the concentration of Mg2+ was lowered to 30 mM and the NaCl concentration was varied (between 0 and 37%). Aerobic heterotrophic growth of halophilic isolates was monitored by measuring the increase in optical density at 600 nm. Survival and viability of halophilic isolates at salt concentrations below 6% were tested by suspending washed cell pellets grown at 15% salt in saline solution with the desired salinity (1 to 5%). Salinity was further checked by using a hand-held refractometer (model S-10; Atago Co. Ltd., Tokyo, Japan) to ensure that no carryover of NaCl occurred with the cell pellets. Cell integrity was determined by counting the cells in a hemocytometer (Reichart Scientific, Buffalo, N.Y.). To test for the ability of the culture to retain viability, 1 ml of culture in saline was reinoculated into HM liquid, streaked onto solid medium (15% salt), and monitored for growth and the formation of characteristic red colonies. Haloferax volcanii (DSM 3757) was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany) and was used as a reference strain for survival and viability studies.
HM was also prepared anaerobically (3, 7) with 40 mM NaHCO3, using 20% CO2:80% N2 in the gas phase. Either Na2SO4, Na2S2O3 (30 mM), or elemental sulfur (0.5 g/liter) was used as an electron acceptor. Elemental sulfur was tested alone or with added ferrous ammonium sulfate (0.2 g/liter) to precipitate excess sulfide. This medium was used for attempts at isolation of anaerobic halophiles and to test the ability of halobacterial isolates to grow on elemental sulfur, sulfate, or thiosulfate as terminal electron acceptors. Sulfur reduction was measured by comparing sulfide production in inoculated media to that in uninoculated, non-sulfur-amended, or non-substrate (yeast extract)-amended controls. Anaerobic growth under sulfur-reducing conditions was evaluated by cell counts and protein measurements of washed cells (6) using the bicinchoninic acid reagents (Pierce Chemical Co., Rockford, Ill.).
Analytical methods.
Sulfate and thiosulfate were measured by ion chromatography (18). For sulfide analysis, 0.2-ml samples were withdrawn by using N2-flushed syringes, and sulfide was trapped in 0.2 ml of 10% zinc acetate solution. Sulfide was then quantified by the methylene blue assay (9). In situ salinity measurements at various locations in the spring were taken by using a hand-held refractometer as described above. Water for determination of microbial mat salinity was obtained by centrifugation of the collected wet mat materials. For salinity measurements along a vertical gradient from the soil top to a depth of 5 cm, cores were cut at 1-cm intervals and sliced into four quadrants. Moisture content was determined in two quadrants. The other two quadrants were suspended in distilled water, shaken (200 rpm for 16 h), and centrifuged, and the resulting supernatant was used for salinity measurements. X-ray diffraction of mineral precipitates encountered on the samples from the banks of the spring was determined with an automated Rigaku diffractometer (53).
Nucleotide sequence accession numbers.
Sequences obtained in this study were deposited in GenBank under accession numbers AY341267 to AY341334.
RESULTS
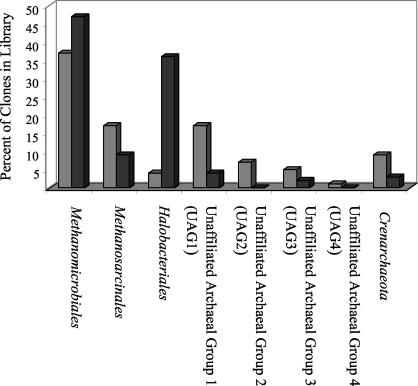
Composition of the mat and source clone libraries. A total of 178 clones (113 from the source and 65 from the mat) were sequenced. A breakdown of the group affiliations of the mat and source clone libraries is shown in Fig. 1. The majority (96% of the mat clones and 91% of the source clones) belonged to the kingdom Euryarchaeota. Clones belonging to euryarchaeotal lineages with pure cultured representatives belonged to the orders Methanomicrobiales, Methanosarcinales, and Halobacteriales. Some of the source (36%) and mat (6%) clones clustered within four euryarchaeotal lineages that have no pure-culture representatives and are designated UAG1 (unaffiliated archaeal group 1) to UAG4 (Fig. 1).
FIG. 1.
Distributions of source (shaded bars) and mat (solid bars) clones among various archaeal groups as percentages of the total population (y axis).
Phylogenetic analysis. (i) Methanomicrobiales and Methanosarcinales.
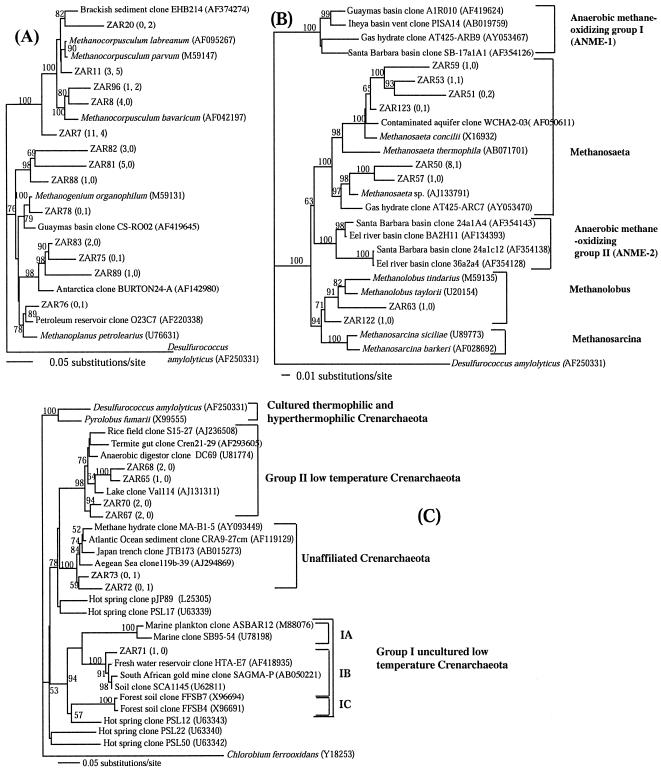
Zodletone clones belonging to the Methanomicrobiales could be clustered into five groups (Fig. 2A). The best-represented group was phylogenetically related to the family Methanocorpusculaceae, members of which use hydrogen as well as secondary alcohols as substrates for methane production (69). The majority of clones belonging to the Methanosarcinales belonged to the family Methanosaetaceae. Cultivated organisms within this group inhabit anaerobic, organic-rich environments and produce methane from acetate (67). (Fig. 2B).
FIG. 2.
Distance dendrograms of the Methanomicrobiales (A), Methanosarcinales (B), and Crenarchaeota (C) lineages within the domain Archaea. Zodletone clones are designated ZAR, and numbers in parentheses are the frequencies of occurrence of a specific OTU in the source and mat clone libraries, respectively. Bootstrap values (expressed as percentages) are based on 1,000 replicates and are shown for branches with more than 50% bootstrap support.
(ii) Unaffiliated clones.
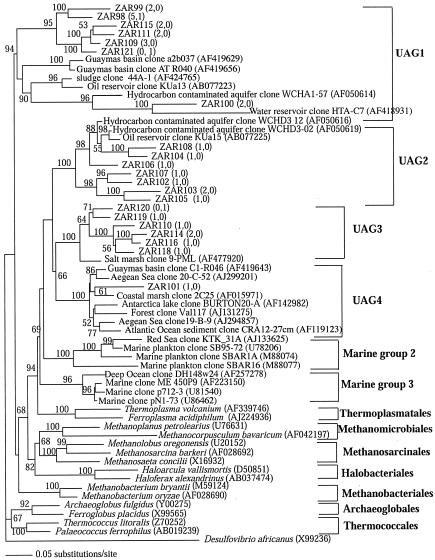
Clones that were not affiliated with any of the previously recognized lineages within the domain Archaea were especially abundant in the source library (36% of the total archaeal clones). These sequences, together with several previously published or database-deposited sequences, clustered into four different groups (UAG1 to UAG4) (Fig. 3). UAG1 contains a few clones retrieved from naturally or anthropogenically hydrocarbon impacted sites (16, 57, 66), anaerobic digestor sludge (52), or a water reservoir (54) (Fig. 3). This group is referred to as “unaffiliated Euryarchaeota” in reference 57 and as candidate division 5 in reference 66. UAG2, UAG3, and UAG4 formed a deep-branching cluster related to the order Thermoplasmatales but still phylogenetically distinct from the two previously recognized Thermoplasmatales-related marine groups II (13) and III (22). UAG2 was first reported by Dojka et al. (16) and subsequently detected in a variety of habitats. This group is referred to as candidate division II in reference 66 and as rice root cluster III in reference 25. UAG3 includes a salt marsh clone, 9PML, that was previously reported as a member of marine euryarchaeotal group III (4) and six Zodletone OTUs. UAG4 was originally described as “marine benthic group IV” (63). However, recent studies (31; this study) indicate that this group is not restricted to marine environments.
FIG. 3.
Distance dendrogram demonstrating the phylogenetic position of UAG clones within the domain Archaea. Numbers in parentheses represent the frequencies of occurrence of a specific OTU in the source and mat clone libraries, respectively. Bootstrap values (expressed as percentages) are based on 1,000 replicates and are shown for branches with more than 50% bootstrap support.
(iii) Crenarchaeota.
The crenarchaeotal populations in the source and the mat were each unique, as inferred from the 16S rRNA gene analysis. According to the phylogenetic nomenclature of DeLong and Pace (15) and Dawson et al. (12), crenarchaeotal clones from the source belonged to the 1B cluster of the low-temperature uncultured Crenarchaeota (1 clone) or to the group 2 Crenarchaeota (7 clones; 4 OTUs), members of which have been retrieved from a wide variety of anaerobic environments (Fig. 2C). On the other hand, members of the mat crenarchaeotal community belonged to a novel lineage of the Crenarchaeota that was recently recognized (48). This lineage has members from gas hydrate formations (referred to as NTAK2 in reference 48) and from Japanese deep-sea (33), Aegean Sea, and Atlantic Ocean (63) sediments. Their presence in Zodletone mats indicates that this group could be also retrieved from terrestrial habitats and is not only restricted to marine environments.
(iv) Halobacteriales.
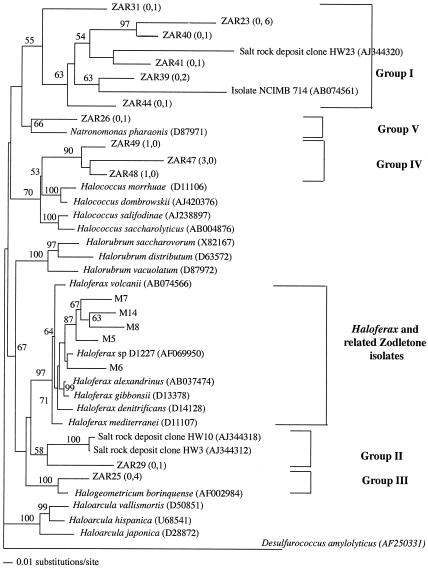
In spite of the low salt concentrations (<1.0%) in the emerging spring water, as well as the apparent anaerobic to microaerophilic conditions prevailing at Zodletone Spring, 36% of the mat clones and 4% of the source clones belonged to the order Halobacteriales, members of which are known to grow in aerobic, high-salt environments. A phylogenetic tree of the clones and closely related sequences, as well as sequences of other selected members of the family Halobacteriaceae, is shown in Fig. 4. Interestingly, halobacterial sequences were not confined to a specific lineage within the tree but rather were dispersed into five different lineages (designated groups I to V) in Fig. 4. The majority of clones (groups I and II) were not affiliated with any of the described genera of the family Halobacteriaceae but were closely related to clones retrieved from Permo-Triassic rock salt and yet uncharacterized isolates (Fig. 4). Groups III and V, represented by ZAR25 and ZAR26, respectively, were closely related to members of two previously described genera (Halogeometricum and Natronomonas). Group IV formed a deep branch within the genus Halococcus. A marked difference was observed between the halobacterial community of the source and that of the mat. All source clones belonged to a single group (Halococcus-related group IV), while mat clones belonged to the other four groups.
FIG. 4.
Distance dendrogram of the order Halobacteriales, including clones encountered in mat and source clone libraries. Zodletone clones are designated ZAR, and numbers in parentheses represent the frequencies of occurrence of a specific OTU in the source and mat clone libraries, respectively. Bootstrap values (expressed as percentages) are based on 1,000 replicates and are shown for branches with more than 50% bootstrap support.
Isolation of halophilic Archaea from the mat materials.
Mat material was diluted up to 10−7, inoculated into aerobic enrichment cultures (HM) and anaerobic enrichment cultures (with sulfur as the electron acceptor), and incubated in the presence of antibiotics at three different salt concentrations. In anaerobic enrichments, no growth or sulfur reduction was observed. Aerobic enrichment cultures showing turbidity were streaked out onto solid medium and developed white, pale-yellow, and red colonies, with a predominance of white and pale-yellow colonies in 7% NaCl plates and a predominance of red colonies in 12 and 18% NaCl plates. These colonies were picked into liquid medium, restreaked twice, and microscopically checked for purity. 16S rRNA gene analysis revealed that the white and yellow colonies belonged to the genera Halomonas and Marinobacter within the γ-Proteobacteria, while the red colonies belonged to the family Halobacteriaceae. All archaeal isolates sequenced (10 isolates; designations beginning with M in Fig. 4) were most closely related to members of the genus Haloferax (Fig. 4). The numerical abundance of the Halobacteriales was evident, since isolates were obtained from serial dilutions up to 10−5.
Partial characterization of halobacterial isolates.
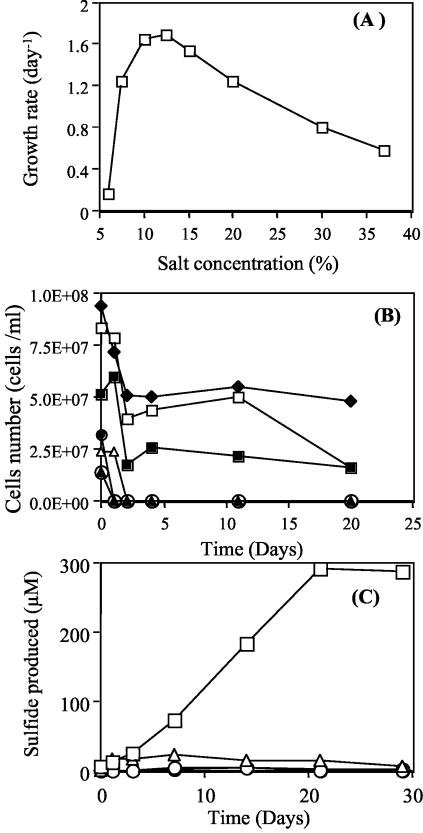
All archaeal isolates were able to grow at a similar range of NaCl concentrations (6 to 37%) and showed the highest growth rate at 12.5 to 15% NaCl. A representative graph for strain M6 is shown in Fig. 5A. Survival and viability studies indicated that cells of strain M6 lyse immediately at 0% salt, within 24 h at 1 and 2% salt, and within 4 days at 3% salt. The cells, however, survive and retain viability at 4 and 5% NaCl in incubations as long as 21 days (Fig. 5B). Control experiments using H. volcanii (DSM 3757) showed a similar ability to survive prolonged incubations at 4 and 5% NaCl.
FIG. 5.
(A) Relationship between growth rate and NaCl concentration in the medium for isolate M6. (B) Survival of isolate M6 at various salinities. Symbols: ▴, distilled water (0% salt); ○, 1% NaCl; •, 2% NaCl; ▵, 3% NaCl; ▪, 4% NaCl; □, 5% NaCl; ♦, 15% NaCl. (C) Sulfide production by isolate M6 when inoculated into an anaerobic medium (12% NaCl) either supplemented with elemental sulfur (□), containing elemental sulfur but no substrate (yeast extract) (▵), or without elemental sulfur (○). An uninoculated medium with elemental sulfur but no microorganisms produced no detectable sulfide. All values shown are averages from duplicate tubes.
No halobacterial strains were able to grow in an anaerobic medium supplemented with sulfate or thiosulfate as the electron acceptor. However, sulfur-amended anaerobic media showed a slow production of sulfide in active cultures. Non-yeast extract-amended, non-S0-amended, or uninoculated controls produced no detectable sulfide (Fig. 5C). In experiments where ferrous ammonium sulfate was included as a medium component, a black precipitate (presumably FeS) was observed within 3 days of the start of the experiment. No significant increase in cell numbers or protein concentrations was observed in any of these cultures. These data suggest that these isolates are capable of sulfur reduction but provide no conclusive evidence regarding their ability to couple sulfur reduction to growth and biomass production. Experiments testing the ability of H. volcanii DSM 3757 to reduce elemental sulfur indicated that this microorganism was also able to reduce sulfur, albeit at a much lower rate (sulfide production, 50 μM in 3 months, compared to 0.3 mM in 3 weeks for strain M6).
Salinity measurements at different areas at the spring.
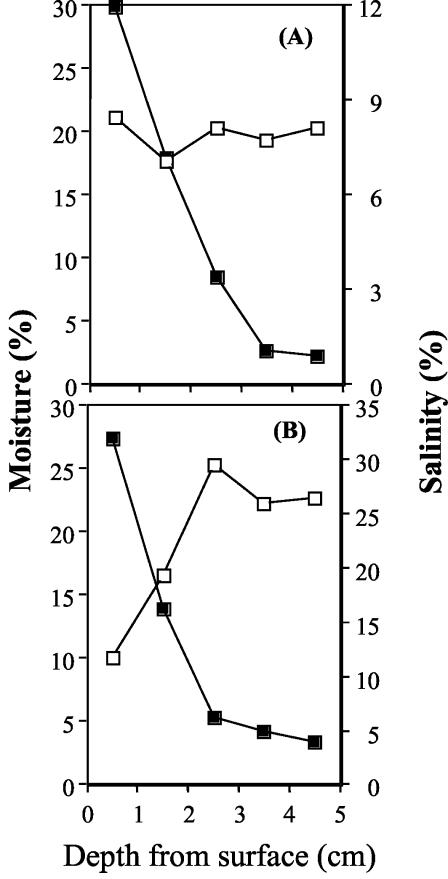
Salinity measurements were conducted on water samples at the spring source and at 1, 6, 10, 15, and 20 m along the spring. Results showed little variation in salinity (0.7 to 1%) at all locations along the spring. However a salinity and moisture depth profile conducted on two cores collected along the stream bank showed that salinity increases in the top 1 cm of soil and decreases with depth (Fig. 6). Salinity was as high as 12% in the top 1 cm of a core (core A) collected 5 cm from the stream bank (3 m from the source) and decreased with depth until it reached 1%, comparable to that of the spring water. Similarly, in another core (core B), collected 30 cm out from the stream (10 m from source), the increase in salinity and decrease in moisture content in the top layers were more apparent (salinity was as high as 32% in the top layer, and the moisture level was down to 10% at the top 1 cm of soil, compared to approximately 25% at a 3- to 5-cm depth). These results indicate that a higher salinity is attained, probably due to evaporation, in localized niches in the stream. White mineral precipitates collected on the stream banks were analyzed by X-ray diffraction. These minerals contained a mixture of 58% gypsum, 24% barite, 9.6% halite, and 5.2% calcite (percentages [by weight] of total evaporite minerals).
FIG. 6.
Salinity (solid squares) and moisture content (open squares) of two representative cores collected on the stream bank. Core A was collected 5 cm from the stream, and core B was collected approximately 30 cm from the stream.
DISCUSSION
In this study, a culture-independent survey of the archaeal community in an anaerobic sulfur spring was undertaken. 16S rRNA gene analysis confirmed the abundance of novel archaeal lineages and revealed the presence of a diverse halobacterial community in the spring, the viability of which was confirmed via isolation procedures. Therefore, the use of a sequence-guided approach in isolation procedures can prove useful in the isolation of novel microorganisms (2, 47) or to confirm the viability and study the ecological significance of a specific group of microorganisms detected by culture-independent approaches (this study).
The fact that clones belonging to methanogenic lineages (Methanomicrobiales and Methanosarcinales) are present in both clone libraries (54 and 46% of the source and mat libraries, respectively) is consistent with previous studies demonstrating that high rates of methanogenesis are usually associated with microbial mats (28, 58, 65). Moreover, methanogenic activity observed in enrichment studies might not be mediated solely by members of these two groups, since some microorganisms belonging to UAGs might be involved in the process. H2-based methanogenesis has been observed in predominantly phototrophic mats, even in the presence of excess terminal electron acceptors (28). We reason that members of the Methanomicrobiales are involved in the conversion of hydrogen to methane in the microbial mats. The presence of acetate-utilizing methanogens of the Methanosaetaceae is an indication of the importance of acetoclastic methanogenesis in the degradation of organic matter in Zodletone Spring, especially at the spring source. Members of the family Methanosaetaceae are common inhabitants of high-organic-content, electron acceptor-limited environments, such as anaerobic digestors (38, 52), sewage sludge, freshwater lake sediments (46), and hydrocarbon-contaminated sites (16, 66).
The abundance of UAG clones in the source compared to the mat is an important manifestation of the role of hydrocarbons in shaping the microbial community at Zodletone Spring. UAG1 and UAG2 clones had been detected from a variety of environments; however, they are most commonly obtained from hydrocarbon-impacted sediments (57). The physiology of microorganisms belonging to these two lineages is still unknown. However, based on the environments from which these clones were detected, it appears that UAG1 and UAG2 are anaerobes. UAG4 sequences (benthic group IV) have been retrieved primarily from marine or high-salt environments (4, 63) but have recently been detected in terrestrial ecosystems (31; this study). As with UAG1 and UAG2, no evidence regarding the physiological capabilities of this group of microorganisms is yet available.
Perhaps the most unexpected finding of this study is the presence of members of the family Halobacteriaceae at the spring, especially in the microbial mats. Halobacteria were readily isolated from dilutions of mat material up to 10−5, confirming their viability and numerical abundance in the microbial mats. The biogeochemical characteristics of Zodletone Spring are clearly not suggestive of the presence of a thriving halobacterial population. The low salinity (0.7 to 1.0%) provides an unsuitable ionic environment for members of the Halobacteriaceae, whose cells generally lyse at NaCl concentrations lower than 8 to 10% (42). Zodletone halobacterial isolates belonged to the genus Haloferax, members of which are known to have the lowest salt optimum and the minimum NaCl requirement for retaining cell wall integrity among the halophilic Archaea (17, 43). This is manifested by the ability of our isolates to readily grow at an NaCl concentration of 7.5%. It should be noted, however, that 16S rRNA gene analysis revealed a diverse halobacterial community in the spring including several halobacterial lineages other than Haloferax. We speculate that our ability to isolate only Haloferax species is due to their relative ease of isolation compared to that of other halobacterial species and is not necessarily indicative of their numerical abundance or relative ecological significance in the microbial mats. Extrapolation of the isolates' salt profiles to the entire halobacterial community is therefore not appropriate. A similar or lower salt requirement for other Halobacteriales in the spring is possible. Salinity and moisture depth profiles suggest that as spring water with low salinity diffuses to the banks of the stream, evaporation results in a decrease in moisture and an increase in salinity at the top soil layers, resulting in the creation of a suitable environment for the halophilic Archaea. Therefore, this work indicates that members of the Halobacteriales are not restricted to hypersaline ecosystems such as salt lakes, salterns, and the Dead Sea (42) but can also inhabit lower-salinity environments where localized NaCl concentrations are sufficient to prevent their lysis. Indeed, few culture-dependent or -independent surveys have either isolated Halobacteriales (37, 50) or encountered 16S rRNA Halobacteriales clones (41, 56) in low-salt environments such as low-salinity salterns, black smoker chimney structures, seawater, and coastal salt marshes.
Since the original soil salinity in the spring area is low, it is unlikely that the halophilic Archaea detected are native inhabitants of the spring or soil. However, previous studies have suggested that the emerging water in Zodletone Spring represents a mixture of deeper basinal brine (3%) and shallow groundwater (97%) (68). A few investigators (20, 64) have isolated halophilic Archaea from deep brine deposits and brine-saturated sediments. Therefore, we postulate that the halophilic Archaea isolated from the spring mats originated from deeper brine ejected together with shallow groundwater in minor quantities at the spring source. Regardless of the inoculum source, the abundance and viability of Halobacteriales in the mats indicates their ability to adapt and survive in Zodletone Spring.
Aside from the low salt concentration in the stream, the apparently anaerobic conditions prevailing in Zodletone Spring are in contrast to the aerobic mode of metabolism observed for all cultivated members of the Halobacteriales, as well as to the aerobic habitats from which halophilic Archaea have previously been isolated. The viability of these isolates in the anaerobic environment of the spring suggests that these microorganisms can use an alternative electron acceptor(s). Field studies showed a high concentration of zero-valent sulfur (53) in Zodletone Spring, and 16S rRNA gene analysis of the bacterial community in Zodletone Spring indicated the importance of sulfur in shaping the bacterial community at the site (19, 53). A large fraction of the bacterial clones recovered belonged to lineages that are known for their sulfur-reducing or sulfur-disproportionating abilities (19). Previous reports have previously speculated on the ability of halophilic Archaea to reduce elemental sulfur (24, 60). Tindall and Trüper (60) noted the ability of several halobacteria to slowly reduce elemental sulfur within several months, although the process was never thoroughly documented. The ability of Zodletone halobacterial isolates to slowly reduce sulfur (Fig. 5C) is consistent with this observation. Sulfur reduction by members of the Halobacteriaceae might be occurring at a higher rate under in situ conditions, and the slow pattern observed in laboratory experiments could be a function of our inability to provide suitable conditions for growth or to isolate dominant sulfur-reducing species from the stream. Recently, Takai et al. (56) reported on the presence of halobacterial clones on the inside of a black smoker chimney structure. Analysis of the elemental composition of the structure indicated that sulfur is one of its major components (56). Therefore, the presence of halobacteria in the black smoker chimney as well as in Zodletone Spring might indicate that they could play a role in sulfur metabolism in sulfur-rich anaerobic ecosystems.
Finally, since methane is continuously ejected from the spring source, the site could be regarded as an ideal environment for exploring the anaerobic oxidation of methane in freshwater ecosystems. In our survey of the archaeal community, we found no clones belonging to ANME-1 or ANME-2, both of which have been reported to be involved in anaerobic methane oxidation (AOM) in a variety of marine environments (5, 27, 32, 39, 44, 45, 57). Biogeochemical and enrichment studies to explore the AOM potential in Zodletone sediments were beyond the scope of this study. However, it should be noted that ANME-1 and ANME-2 have been shown to be associated with methane-rich marine ecosystems and not with terrestrial ecosystems. The low sulfate concentration in the upper part of the spring (60 to 150 μM) may be responsible for the absence of this group of microorganisms, since they mediate AOM in syntrophic association with sulfate-reducing bacteria (5). Indeed, studies from deeper, sulfate-limited, methane-rich hydrates indicated the absence of ANME-1 and ANME-2 clones from these structures (35, 48). Whether their absence indicates the absence of AOM activity and whether sulfate-dependent AOM mediated by ANME-1 and ANME-2 is the only mechanism for AOM remain to be answered.
Acknowledgments
This work was supported by a grant from the National Science Foundation microbial observatories program (grant MCB_0240683).
We thank Kristin Rorabaugh and Kristen Savage for excellent technical assistance.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antón, J., A. Oren, S. Benlloch, F. Rodríguez-Valera, R. Amann, and R. Rosselló-Mora. 2002. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int. J. Syst. Evol. Microbiol. 52:485-491. [DOI] [PubMed] [Google Scholar]
- 3.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benlloch, S., A. López-López, E. O. Casamayor, L. Øvreas, V. Goddard, F. L. Daae, G. Smerdon, R. Massana, I. Joint, F. Thingstad, C. Pedrós-Alió, and F. Rodríguez-Valera. 2002. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4:349-360. [DOI] [PubMed] [Google Scholar]
- 5.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jørgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Bryant, M. P. 1972. Commentary on the Hungate technique for culture of anaerobic bacteria. Am. J. Clin. Nutr. 25:1324-1328. [DOI] [PubMed] [Google Scholar]
- 8.Chapelle, F. H., K. O'Neill, P. M. Bradley, B. A. Methe, S. A. Ciufo, L. L. Knobel, and D. R. Lovley. 2002. A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415:312-315. [DOI] [PubMed] [Google Scholar]
- 9.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 10.Cohen, Y., B. B. Jørgensen, E. Padan, and M. Shilo. 1986. Adaptation to hydrogen sulfide of oxygenic and anoxygenic photosynthesis among cyanobacteria. Appl. Environ. Microbiol. 51:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cytryn, E., D. Minz, R. S. Oremland, and Y. Cohen. 2000. Distribution and diversity of archaea corresponding to the limnological cycle of a hypersaline stratified lake (Solar Lake, Sinai, Egypt). Appl. Environ. Microbiol. 66:3269-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson, S. C., E. F. DeLong, and N. R. Pace. 2000. Phylogenetic and ecological perspectives on uncultured Crenarchaeota and Korarchaeota. In M. Dworkin, N. Falkow, H. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The Prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.3. [Online.] Springer-Verlag, New York, N.Y. http://141.150.157.117:8080/prokPUB/index.htm.
- 13.DeLong, E. F. 1992. Archaea in coastal marine environment. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLong, E. F. 1998. Everything in moderation: archaea as “non-extremophiles.” Curr. Opin. Genet. Dev. 8:649-654. [DOI] [PubMed] [Google Scholar]
- 15.DeLong, E. F., and N. R. Pace. 2001. Environmental diversity of Bacteria and Archaea. Syst. Biol. 50:470-478. [PubMed] [Google Scholar]
- 16.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Souza, S. E., W. Altekar, and S. F. D'Souza. 1997. Adaptive response of Haloferax mediterranei to low concentrations of NaCl (<20%) in the growth medium. Arch. Microbiol. 168:68-71. [DOI] [PubMed] [Google Scholar]
- 18.Elshahed, M. S., L. M. Gieg, M. J. McInerney, and J. M. Suflita. 2001. Signature metabolites attesting to the in situ attenuation of alkylbenzenes in anaerobic environments. Environ. Sci. Technol. 35:682-689. [DOI] [PubMed] [Google Scholar]
- 19.Elshahed, M. S., J. M. Senko, F. Z. Najar, S. M. Kenton, B. A. Roe, T. A. Dewers, J. R. Spear, and L. R. Krumholz. 2003. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl. Environ. Microbiol. 69:5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emerson, D., C. Chauhan, P. Oriel, and J. A. Breznak. 1994. Haloferax sp. D1227, a halophilic archaeon capable of growth on aromatic compounds. Arch. Microbiol. 161:445-452. [Google Scholar]
- 21.Ficker, M., K. Krastel, S. Orlicky, and E. Edwards. 1999. Molecular characterization of a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 65:5576-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuhrman, J. A., and A. A. Davis. 1997. Widespread Archaea and novel Bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar. Ecol. Prog. Ser. 150:275-285. [Google Scholar]
- 23.Garrity, G. M., and J. G. Holt. 2001. The road map to the Manual, p. 119-141. In G. M. Garrity, D. R. Boone, and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y. [Google Scholar]
- 24.Grant, W. D., and H. N. M. Ross. 1986. The ecology and taxonomy of halobacteria. FEMS Microbiol. Rev. 39:9-15. [Google Scholar]
- 25.Großkopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Havens, J. S. 1983. Reconnaissance of ground water in the vicinity of the Wichita Mountains, southwestern Oklahoma. Oklahoma Geological Survey circular 85. Oklahoma Geological Survey, Norman, Okla.
- 27.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 28.Hoehler, T. M., D. B. Albert, M. J. Alperin, B. M. Bebout, C. S. Martens, and D. J. Des Marais. 2002. Comparative ecology of H2 cycling in sedimentary and phototrophic ecosystems. Antonie Leeuwenhoek 81:575-585. [DOI] [PubMed] [Google Scholar]
- 29.Holben, W. E. 1996. Isolation and purification of bacterial community DNA from environmental samples, p. 431-444. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 30.Huber, J. A., D. A. Butterfield, and J. A. Baross. 2002. Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol. 68:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurgens, G., F. Glockner, R. Amann, A. Saano, L. Montonen, M. Likolammi, and U. Munster. 2000. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34:45-56. [DOI] [PubMed] [Google Scholar]
- 32.Lanoil, B. D., R. Sassen, M. T. La Duc, S. T. Sweet, and K. H. Nealson. 2001. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, L., C. Kato, and K. Horikoshi. 1999. Microbial diversity in sediments collected from the deepest cold-seep area, the Japan trench. Mar. Biotechnol. 1:391-400. [DOI] [PubMed] [Google Scholar]
- 34.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchesi, J. R., A. J. Weightman, B. A. Cragg, R. J. Parkes, and J. C. Fry. 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. Ecol. 34:221-228. [DOI] [PubMed] [Google Scholar]
- 36.Massana, R., E. F. DeLong, and C. Pedrós-Alió. 2000. A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl. Environ. Microbiol. 66:1777-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGenity, T. J., R. T. Gemmell, and W. D. Grant. 1998. Proposal of a new halobacterial genus Natrinema gen. nov., with two species Natrinema pellirubrum nom. nov. and Natrinema pallidum nom. nov. Int. J. Syst. Bacteriol. 48:1187-1196. [DOI] [PubMed] [Google Scholar]
- 38.McInerney, M. J. 1999. Anaerobic metabolism and its regulation. Bio/Technology 11a:456-478.
- 39.Michaelis, W., R. Seifert, K. Nauhaus, T. Treude, V. Thiel, M. Blumenberg, K. Knittel, A. Gieseke, K. Peterknecht, T. Pape, A. Boetius, R. Amann, B. B. Jørgensen, F. Widdel, J. Peckmann, N. V. Pimenov, and M. B. Gulin. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013-1015. [DOI] [PubMed] [Google Scholar]
- 40.Mountfort, D. O., H. F. Kaspar, R. A. Asher, and D. Sutherland. 2003. Influences of pond geochemistry, temperature, and freeze-thaw on terminal anaerobic processes occurring in sediments of six ponds of the McMurdo Ice Shelf, near Bratina Island, Antarctica. Appl. Environ. Microbiol. 69:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munson, M. A., D. B. Nedwell, and T. M. Embley. 1997. Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh. Appl. Environ. Microbiol. 63:4729-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oren, A. 2000. The order Halobacteriales. In M. Dworkin, N. Falkow, H. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The Prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.2. [Online.] Springer-Verlag, New York, N.Y. www.prokaryotes.com.
- 43.Oren, A. 2002. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS. Microbiol. Ecol. 39:1-7. [DOI] [PubMed] [Google Scholar]
- 44.Orphan, V. J., K. U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. DeLong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pudry, K. J., M. A. Munson, D. B. Nedwell, and T. M. Embley. 2002. Comparison of the molecular diversity of the methanogenic community at the brackish and marine ends of a UK estuary. FEMS Microbiol. Ecol. 39:17-21. [DOI] [PubMed] [Google Scholar]
- 47.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 48.Reed, D. W., Y. Fujita, M. E. Delwiche, D. B. Blackwelder, P. P. Sheridan, T. Uchida, and F. S. Colwell. 2002. Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl. Environ. Microbiol. 68:3759-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reysenbach, A. L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez-Valera, F., F. Ruiz-Berraquero, and A. Ramos-Cormenzana. 1979. Isolation of extreme halophiles from seawater. Appl. Environ. Microbiol. 38:164-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanders, W. E. 1998. Rate and mechanism of barite mineralization at Zodletone mountain, sowthwestern Oklahoma. M.S. thesis. University of Oklahoma, Norman.
- 52.Sekiguchi, Y., Y. Kamagata, K. Syutsubo, A. Ohashi, H. Harada, and K. Nakamura. 1998. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144:2655-2665. [DOI] [PubMed] [Google Scholar]
- 53.Senko, J. M., B. S. Campbell, J. R. Henriksen, M. S. Elshahed, T. A. Dewers, and L. R. Krumholz. 2004. Barite deposition mediated by phototrophic sulfide oxidizing bacteria. Geochim. Cosmochim. Acta 68:773-780.
- 54.Stein, L. Y., G. Jones, B. Alexander, K. Elmund, C. Wright-Jones, and K. H. Nealson. 2002. Intriguing microbial diversity associated with metal-rich particles from a freshwater reservoir. FEMS Microbiol. Ecol. 42:431-440. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki, M. T., and E. F. DeLong,. 2002. Marine microbial diversity, p. 209-235. In J. T. Staley and A. L. Reysenbach (ed.), Biodiversity of microbial life. Wiley-Liss Press, New York, N.Y.
- 56.Takai, K., T. Komatsu, F. Inagaki, and K. Horikoshi. 2001. Distribution of archaea in a black smoker chimney structure. Appl. Environ. Microbiol. 67:3618-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teske, A., K.-U. Hinrichs, V. Edgcomb, A. V. Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teske, A., and D. A. Stahl. 2002. Microbial mats and biofilms: evolution, structure, and function of fixed microbial communities, p. 49-101. In J. T. Staley and A. L. Reysenbach (ed.), Biodiversity of microbial life. Wiley-Liss Press, New York, N.Y.
- 59.Thompson, J. D., T. J. Gibson, F. Pleioniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tindall, B. J., and H. G. Trüper. 1986. Ecophysiology of the aerobic halophilic archaebacteria. Syst. Appl. Microbiol. 7:202-212. [Google Scholar]
- 61.Urbach, E., K. L. Vergin, L. Young, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 62.van der Maarel, M. J. E. C., R. R. E. Artz, R. Haanstra, and L. J. Forney. 1998. Association of marine archaea with the digestive tracts of two marine fish species. Appl. Environ. Microbiol. 64:2894-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vreeland, R. H., and J. H. Huval. 1991. Phenotypic characterization of halophilic bacteria from ground water sources in the United States, p. 53-62. In F. Rodriguez Valera (ed.), General and applied aspects of halophilic microorganisms. Plenum Press, New York, N.Y.
- 65.Ward, D. M. 1978. Thermophilic methanogenesis in a hot-spring algal-bacterial mat (71 to 30°C). Appl. Environ. Microbiol. 35:1019-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watanabe, K., Y. Kodama, N. Hamamura, and N. Kaku. 2002. Diversity, abundance, and activity of archaeal populations in oil-contaminated groundwater accumulated at the bottom of an underground crude oil storage cavity. Appl. Environ. Microbiol. 68:3899-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitman, W. B., T. L. Bowen, and D. R. Boone. 1999. The methanogenic bacteria. In M. Dworkin, N. Falkow, H. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The Prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.0. [Online.] Springer-Verlag, New York, N.Y. www.prokaryotes.com.
- 68.Younger, P. 1986. Barite travertine from Southwestern Oklahoma and Western-Central Colorado. M.S. thesis. Oklahoma State University, Stillwater.
- 69.Zellner, G., E. Stackebrandt, P. Messner, B. J. Tindall, E. Conway de Macario, H. Kneifel, U. B. Sleytr, and J. Winter. 1989. Methanocorpusculaceae fam. nov., represented by Methanocorpusculum parvum, Methanocorpusculum sinense spec. nov. and Methanocorpusculum bavaricum spec. nov. Arch. Microbiol. 151:381-390. [DOI] [PubMed]