Abstract
The aim of this study was to initiate autecological studies on uncultivated natural populations of diazotrophic bacteria by examining the distribution of specific diazotrophs in the Chesapeake Bay. By use of quantitative PCR, the abundance of two nifH sequences (907h22 and 912h4) was quantified in water samples collected along a transect from the head to the mouth of the Chesapeake Bay during cruises in April and October 2001 and 2002. Standard curves for the quantitative PCR assays demonstrated that the relationship between gene copies and cycle threshold was linear and highly reproducible from 1 to 107 gene copies. The maximum number of 907h22 gene copies detected was approximately 140 ml−1 and the maximum number of 912h4 gene copies detected was approximately 340 ml−1. Sequence 912h4 was most abundant at the mouth of the Chesapeake Bay, and in general, its abundance increased with increasing salinity, with the highest abundances observed in April 2002. Overall, the 907h22 phylotype was most abundant at the mid-bay station. Additionally, 907h22 was most abundant in the April samples from the mid-bay and mouth of the Chesapeake Bay. Despite the fact that the Chesapeake Bay is rarely nitrogen limited, our results show that individual nitrogen-fixing bacteria have distinct nonrandom spatial and seasonal distributions in the Chesapeake Bay and are either distributed by specific physical processes or adapted to different environmental niches.
The abundance and biogeochemical importance of marine bacteria has been realized for decades (19). Facilitated by advances in molecular biology techniques, in recent years the taxonomic and phylogenetic identities of marine bacteria have been investigated. Using rRNA genes as phylogenetic markers (18), Giovannoni and coworkers identified previously unknown bacterial phylogenetic lineages in the Sargasso Sea (12), indicating that natural communities of bacteria were not well represented in culture collections. Since that time, studies of the diversity of marine microbial communities have revealed that the majority of bacteria in the ocean are uncultivated and many belong to unexpected taxa (8, 11, 14, 20).
With similar molecular methods but aimed at genes encoding enzymes involved in key biogeochemical transformations (sometimes termed functional genes), studies have investigated the diversity and roles of microorganisms involved in specific processes, such as sulfate reduction (9) and nitrogen cycling (31). Biological nitrogen fixation, the reduction of atmospheric dinitrogen to ammonium, is an important process in maintaining the availability of fixed inorganic nitrogen in the biosphere, balancing the opposing process of denitrification. A recent modeling study illustrated the importance of nitrogen fixers to oceanic primary production and argued that nitrogen fixation is the main reason nitrogen availability limits primary production only on short time scales, while phosphorus limits oceanic production over longer time scales (26).
Nitrogenase is the multisubunit enzyme that catalyzes nitrogen fixation and is encoded by the nifHDK genes. In the late 1980s, studies directed at the Fe protein subunit (nifH) of the nitrogenase protein complex were initiated by designing PCR primers for the cyanobacterial genus Trichodesmium (29). Since that time, information gathered on the diversity of nitrogenase genes has revealed that nifH is encoded by a wide range of autotrophic and heterotrophic prokaryotes, including Archaea, Firmicutes, spirochetes, and α-, β-, γ-, and δ-Proteobacteria (28). Diverse nitrogen-fixing microorganisms have been detected in aquatic habitats ranging from microbial mats (22, 30) to lakes (15), salt marshes (2), estuaries (1, 6), and hypersaline lakes (23). Although patterns of distribution of nitrogen-fixing microorganisms across habitats and ecosystems are appearing (28), the factors that drive the distribution and diversity of nitrogen-fixing microorganisms in aquatic environments are not understood.
Recently, Burns and coworkers (6) discovered that the phylogenetic composition of diazotrophs in Chesapeake Bay and Neuse River sediments varied and concluded that environmental conditions selected for different diazotroph communities. Heidelberg and coworkers examined the seasonality of Chesapeake Bay bacterioplankton (13) and concluded that the distribution of specific taxa along the salinity gradient was patchy but relatively homogeneous. On the other hand, changes in the bacterial response to nutrient amendments (21) and changes in the composition of free-living and particle-associated bacterial communities (5, 7) have been associated with environmental changes along the estuarine gradient.
To investigate how the environment affects the distribution of nitrogen-fixing microorganisms and initiate autecological studies on uncultivated natural populations of diazotrophs, we developed a quantitative PCR approach to study the abundance and distribution of specific nifH phylotypes. Ultimately, the goal of this study was to determine if specific bacteria encoding nifH are randomly distributed throughout the Chesapeake Bay.
MATERIALS AND METHODS
Sample collection.
Water samples were collected with Go-Flo bottles (General Oceanics) mounted on a rosette during a series of cruises aboard the R/V Cape Henelopen. Samples were collected from the surface (1 to 2 m), mid-depth (4 to 10 m), and near-bottom (10 to 20 m) water at three stations in the Chesapeake Bay (Fig. 1) during the spring (April) and fall (October) of 2001 and 2002. Depth, temperature, and salinity data were recorded at each station (Table 1). Water samples were filtered onto 0.22-μm Sterivex-GV filters (Millipore) with a peristaltic pump until the filtrate flow decreased to a trickle. For each sample, the volume filtered was measured with a graduated cylinder. Immediately after filtration, the Sterivex capsules were purged with air, frozen in liquid nitrogen, and stored at −80°C until extraction.
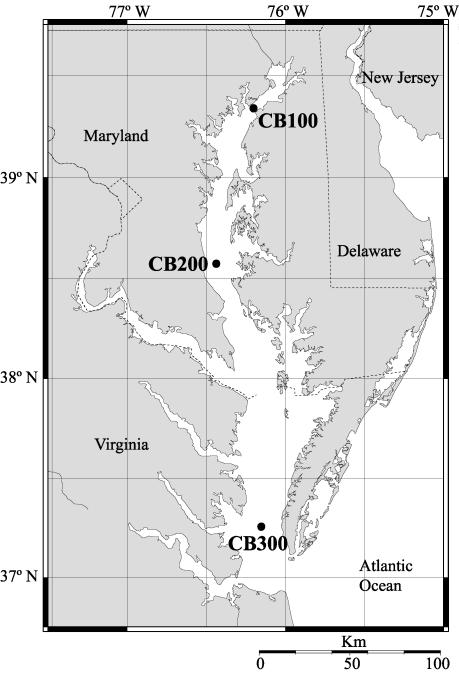
FIG. 1.
Map of sampling locations in the Chesapeake Bay. The locations of the upper, mid-, and lower bay sampling sites are labeled CB100, CB200, and CB300, respectively.
TABLE 1.
Physical data for Chesapeake Bay samplesa
| Site | Date | Surface
|
Mid-depth
|
Deep
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Depth (m) | Temp (°C) | Salinity (PSU) | Depth (m) | Temp (°C) | Salinity (PSU) | Depth (m) | Temp (°C) | Salinity (PSU) | ||
| CB100 | 4 Apr 01 | 1.8 | 7.3 | 0.4 | 4.8 | 7.2 | 0.5 | 9.7 | 6.7 | 4.3 |
| 2 Oct 01 | 1.3 | 17.5 | 5.2 | 5.7 | 18.6 | 5.0 | 10.6 | 19.2 | 4.8 | |
| 4 Apr 02 | 2.0 | 9.9 | 0.1 | 4.0 | 9.8 | 0.1 | 8.0 | 9.8 | 0.1 | |
| 2 Oct 02 | 1.0 | 22.2 | 7.9 | 5.5 | NA | 10.0 | 10.0 | NA | 11.0 | |
| CB200 | 5 Apr 01 | 1.8 | 8.3 | 10.1 | 11.2 | 7.4 | 16.5 | 17.6 | 7.2 | 18.6 |
| 3 Oct 01 | 1.6 | 19.2 | 15.4 | 7.7 | 19.7 | 16.8 | 18.0 | 22.0 | 21.0 | |
| 5 Apr 02 | 2.1 | 9.9 | 15.0 | 13.8 | 9.8 | 17.4 | 18.9 | 8.9 | 21.2 | |
| 3 Oct 02 | 1.0 | 23.5 | 18.7 | 10.0 | 23.5 | 18.8 | 13.0 | 23.5 | 19.5 | |
| CB300 | 6 Apr 01 | 1.9 | 9.3 | 20.5 | 8.3 | 8.8 | 23.1 | 11.3 | 8.7 | 24.1 |
| 4 Oct 01 | 1.7 | 19.6 | 21.8 | 5.5 | 19.4 | 24.6 | 11.9 | 18.9 | 28.6 | |
| 6 Apr 02 | 1.9 | 11.4 | 24.5 | 7.6 | 11.5 | 25.2 | 9.1 | 11.7 | 25.9 | |
| 4 Oct 02 | 1.0 | 24.7 | 23.7 | 5.8 | 24.0 | 24.7 | 10.0 | 21.0 | 26.3 | |
NA, not available; PSU, practical salinity units
DNA extraction.
DNA was extracted from Sterivex filters by a modified xanthogenate protocol (25). Briefly, 1 ml of XS buffer (1% potassium ethyl xanthogenate [Fluka], 100 mM Tris-HCl [pH 7.4], 1 mM EDTA [pH 8.0], 1% sodium dodecyl sulfate, 800 mM ammonium acetate) was added to the filters with a 3-ml syringe, and the filters with syringes attached were then incubated at 70°C for 80 min. The filters were rotated every 20 min to ensure that DNA was extracted from cells on the entire filter surface. After extraction, the buffer was withdrawn with the syringe, transferred to a 2-ml centrifuge tube, vortexed for 10 s, and placed on ice for 30 min. Cell debris was removed by centrifugation at room temperature at 10,000 × g for 12 min in a Marathon 21K/BR bench-top centrifuge (Fisher Scientific), and supernatants were decanted into clean 2-ml centrifuge tubes containing 1 ml of room temperature isopropanol. Samples were incubated at room temperature for 10 min, and the precipitated DNA was pelleted by centrifugation at 10,000 × g for16 min at room temperature. Isopropanol was decanted and DNA pellets were washed with 70% ethanol, air dried, and resuspended in 100 μl of 10 mM Tris-HCl (pH 8.5). Samples were stored at −80°C until further use. Sample DNA yields were quantified with a PicoGreen double-stranded DNA quantification kit (Molecular Probes) following the manufacturer's protocol and a Cary Eclipse fluorescence spectrophotometer (Varian). Lambda phage genomic DNA (Invitrogen) was used for the standard curves for DNA quantification.
5′-Nuclease assays.
For all 5′-nuclease assays, the 25-μl reactions contained 1× TaqMan buffer (Applied Biosystems), 2.0 mM MgCl2, 200 μM each dATP, dGTP, and dCTP, 400 μM dUTP, 400 nM each forward and reverse primers, 200 nM fluorogenic probe, 0.25 U of AmpErase uracyl N-glycosylase (Applied Biosystems), 0.625 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), and 2 μl of template DNA; although the concentration of DNA depended on the type of assay, the volume added to each reaction was consistent. For the no-template controls, 2 μl of-nuclease-free water (Ambion) was added to each reaction. For every sample, 5′-nuclease reactions were replicated at least three times, and for every reaction, the no-template controls were run in triplicate. Thermal cycling was conducted in a GeneAmp 5700 sequence detection system (PE Applied Biosystems) with the following parameters: 50°C for 2 min, 95°C for 10 min, and 45 cycles of 95°C for 15 s followed by 60°C for 1 min.
Primers and probe design.
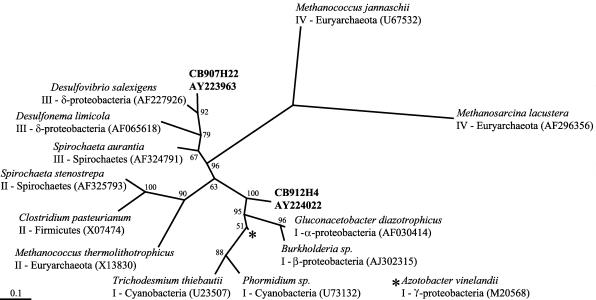
As there is little information on the molecular ecology of diazotrophs in the Chesapeake Bay, we compared the distribution of two nifH sequences. Sequences 907h22 and 912h4 were amplified and cloned from Chesapeake Bay samples collected in April 2001. Because of high nifH diversity, none of the clone libraries from the Chesapeake Bay was exhaustively sequenced. Nonetheless, sequence 907h22 was recovered seven times (100% nucleotide identity) from a library of 72 clones from the surface at CB200 and was not closely related to any other sequences in our Chesapeake Bay nifH library of 207 clones. Sequence 912h4 was recovered once from a library of eight clones from the deep water at CB300 and was more than 99% identical to four other sequences in our Chesapeake Bay nifH library. We chose these particular sequences because they were not closely related and were detected at different stations. Compared to nifH sequences in GenBank, the sequences clustered within different nifH groups (Fig. 2). PCR primers and TaqMan probes were designed with Primer Express software (Applied Biosystems) from the nifH sequences 907h22 and 912h4 (Table 2). The probe was 5′ labeled with the fluorescent reporter FAM (6-carboxyfluorescein) and 3′ labeled with TAMRA (6-carboxytetramethylrhodamine) as a quenching dye.
FIG. 2.
Maximum-likelihood tree of inferred amino acids of selected nifH sequences. The sequences and GenBank accession numbers in bold were used to design primers and probes (CB907h22 and CB912h4). Other sequences used for comparison are labeled with genus and species names, nifH group, phylogenetic affiliation, and GenBank accession numbers.
TABLE 2.
Oligonucleotide primers and probes and the number of target copies with the resulting Ct values from specificity experiments
| Primers and probes | nifH clone | Oligonucleotide | Sequence (5′-3′)a | No. of target copies | Ct ± SDb |
|---|---|---|---|---|---|
| 907h22 | 907h22 | Forward | ACGGCGGAACTTGGTGTGT | 1.7 × 105 | 21.98 ± 0.07 |
| Reverse | AATACCGCGACCTGCACAAC | ||||
| Probe | CGGTGGTCCTGAGCCGGGAGTT | ||||
| 911h7 | Forward | ATGGCGGAACCTGGTGTGT | 1.5 × 105 | 43.84 ± 2.01 | |
| Reverse | GATGCCACGACCGGCACAGC | ||||
| Probe | CGGAGGTCCGGAACCGGGGGTT | ||||
| 912h4 | 912h4 | Forward | GGTTATGGACAAGGTCCGTGAA | 1.6 × 105 | 22.04 ± 0.05 |
| Reverse | AGCCGCGTTTACAGACATCTTC | ||||
| Probe | AGTTCCAGATCCTCAACGGTGCCGA | ||||
| 914h6 | Forward | GGTCATGGACAAAGTCCGTGAC | 1.4 × 105 | 45.00 ± 0.00 | |
| Reverse | AGCCCCACTTCAACACATCTTC | ||||
| Probe | AGGTCCAGATCCTCGACGGTTCCGA | ||||
| 910h3 | Forward | GGTTATGGACAAGGTCCGTGAA | 1.6 × 105 | 45.00 ± NA | |
| Reverse | AACCCTTGCGACAAACATCTTC | ||||
| Probe | AACTCGAGGTCCTCAACGGTACCGA | ||||
| 911h10 | Forward | GGTTATGGACAAGGTCCGCGAA | 1.8 × 105 | 25.66 ± NA | |
| Reverse | AGCCGCGTTTACAGACATCTTC | ||||
| Probe | AGTTCCAGGTCCTCTACGGTGCCGA |
Mismatched bases are shown underlined in the negative control sequences.
Triplicate reactions; NA, replicate reactions were not conducted.
To determine the specificity of the TaqMan primers and probes, reactions with plasmids containing the appropriate positive control fragment were compared to reactions with plasmids containing closely related fragments. In our current database of over 2,000 nifH sequences, the closest relatives to the positive control fragments were also recovered from Chesapeake Bay clone libraries. For the 907h22 primers and probe, fragment 911h7 was the closest relative to 907h22 and was 81% identical in DNA sequence. Several fragments closely related to 912h4 were available and were used to test the specificity of the 912h4 primers and probe; fragments 911h10, 910h3, and 914h6 were 89, 84, and 81% identical, respectively, to 912h4 in DNA sequence.
Standard curves.
For each primer and probe set, standard curves were produced with triplicate 10-fold dilution series ranging from 0.6 ng to 0.6 ag of linearized plasmid containing the 907h22 or 912h4 nifH insert. To test the precision of the 5′-nuclease assays, two of the triplicate dilution series were analyzed during the same experiment, while the third replicate dilution series was analyzed separately several days later. Regression analysis of the standard curves was performed with Sigma Plot version 6.0 (SPSS).
Assay optimization.
Experiments were conducted to determine if background DNA would affect 5′-nuclease assays. Reactions with a 10-fold dilution series (range, 0.6 ng to 0.6 fg) of linearized plasmid with the 912h4 insert and reactions containing the same targets plus 100 ng of Escherichia coli genomic DNA (Sigma-Aldrich) were run in parallel. The resulting cycle threshold values (Ct) from the two treatments were compared with a paired t test.
Before 5′-nuclease reactions were conducted on the environmental DNA extracts, the amount of template added to the reaction mixtures was optimized. Reactions with the 912H4 primers and TaqMan probe and 0.6 pg of the positive control plasmid were compared to reactions containing plasmid plus 10, 5, 2, 1, or 0.5 ng of DNA from different environmental extracts; the environmental DNA samples were diluted so that the volume added to each reaction was constant (2 μl). Among all samples, 2 ng of template DNA consistently gave the highest signal with the least evidence of inhibition. Therefore, all environmental DNA samples were diluted to a concentration of 1 ng μl−1, and 2 μl was added to each reaction.
The PCR efficiency was estimated for each sample by comparing the Cts from amplification of 0.6 pg of the appropriate linearized plasmid plus 2 ng of environmental template to Cts from reactions with 0.6 pg of plasmid alone. Amplification efficiency was calculated with the formula Xn = X0 × (1 + Ex)n, where Xn is the number of target molecules at cycle n, X0 is the initial number of target molecules, Ex is the efficiency of target amplification, and n is the number of cycles (Ct). Assuming an efficiency of 1, the Ct value from the amplification of plasmid alone was used to calculate Xn. To estimate sample amplification efficiency, the calculated Xn and the Ct value from the amplification of plasmid plus sample were then used to solve the above formula for Ex. The Ex value was converted to a percentage, and samples that amplified with less than 95% efficiency were noted.
RESULTS
The average water temperature and standard deviation among all of the stations and depths sampled in October was 9.1 ± 1.5°C, while in April it was 20.9 ± 2.4°C. The average salinity and standard deviation at stations CB100, CB200, and CB300 was 4.1 ± 4.0, 17.4 ± 3.0, and, 24.4 ± 2.1 practical salinity units, respectively. As expected, the water in the Chesapeake Bay was warmer in October than in April, and the salinity among all depths sampled increased from the head to the mouth of the estuary.
The closely related fragment used to test the 907h22 primers and probe specificity was not detected in the 5′-nuclease assay. Reactions with the plasmid containing the 907h22 fragment produced a strong signal (low Ct), while the plasmid containing the 911h7 fragment produced no signal (Table 2); Ct values of >40 indicate no signal. However, reactions with fragments used to test the 912h4 primers and probe specificity revealed that some closely related sequences were detected in the 5′-nuclease assay. Reactions with the plasmid containing the 912h4 fragment produced a strong signal, and the plasmid with the 911h10 fragment also produced a strong signal, yet plasmids with the 914h6 and 910h3 fragments produced no signal (Table 2). It should be noted that the efficiency of the reaction with the clone 911h10 was reduced relative to that of clone 912h4; the slope of the regression of Ct versus gene copies was higher for 911h10 (4.0) compared to 912h4 (3.4), indicating reduced amplification efficiency (data not shown).
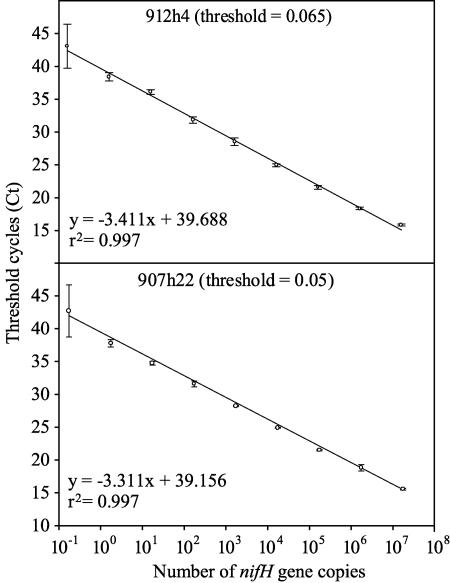
The relationship between Ct and gene copies was linear from 1 to 107 target molecules detected with the 5′-nuclease assay (Fig. 3). The r2 value for the regression of Ct versus target copies was 0.997 for the standard curve for both 912h4 and 907h22. The error of triplicate samples was small, indicating that the standard curves were very reproducible. The average standard deviation of Ct for all concentrations except 10−1 was 0.27 and 0.36 for the 907h22 and 912h4 standards, respectively. The standard deviations of triplicate Cts from the 10−1 dilutions were 3.97 and 3.35 for the 907h22 and 912h4 standards, respectively.
FIG. 3.
Standard curves for quantitative PCR. The regression of threshold cycles (Ct) versus the logarithm of the number of nifH gene copies was calculated for the positive-control plasmids for each primer and probe set. The error bars represent the standard deviations of triplicate reactions.
Experiments comparing amplification of a dilution series of plasmid 912h4 plus 100 ng of genomic nontarget DNA to amplification of plasmid alone demonstrated that nontarget DNA did not affect the amplification of target in the quantitative PCRs. A paired t test of the two treatments revealed that the difference in Ct between plasmid alone versus plasmid plus background genomic DNA was not significant (P = 0.3661, n = 7) for targets ranging from ca. 101 to 107 copies (data not shown).
Comparison of amplification with plasmid controls alone versus plasmids plus sample revealed that some samples reduced the amplification efficiency of the reaction (Fig. 4). For the 907h22 primers and probe, four samples were between 90 and 95% efficient and three samples (CB100D, April 2002; CB200S, October 2002; and CB200D, October 2002) were between 80 and 90% efficient. Similarly, for the 912h4 primers and probe, five samples were between 90 and 95% efficient and three samples (CB100M, October 2001; CB100D, April 2002; and CB200S, October 2002) were between 80 and 90% efficient.
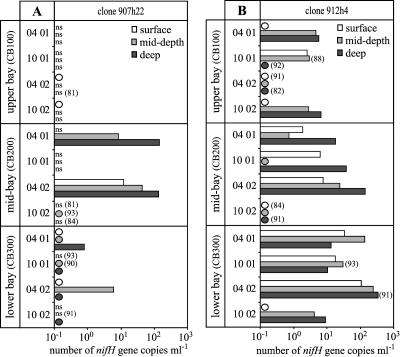
FIG. 4.
Number of 907h22 nifH gene copies per ml (A) and number of 912h4 nifH gene copies per ml (B) in the Chesapeake Bay. Circles indicate samples with a detectable signal that were not quantifiable; i.e., at least one but not all replicate reactions resulted in detectable amplification. ns, no amplification signal was detected in any of the replicate reactions. For reactions that were less than 95% efficient, the calculated percent efficiency is indicated in parentheses. Note that the scale along the x axis is logarithmic.
The number of 907h22 and 912h4 nifH genes detected in the Chesapeake Bay was spatially and temporally variable. The abundance of the 912h4 nifH phylotype ranged from detected but not quantifiable to 340 copies per ml, while the 907h22 phylotype ranged from undetectable to 143 copies per ml (Fig. 4). Although 907h22 and 912h4 nifH gene types were detected in many samples, the distribution of the two genes was very different.
At the head of the Chesapeake Bay, the 907h22 gene type was detected only in the surface samples collected in 2002. Overall, the highest abundance of 907h22 was observed in the mid-depth and deep samples collected from the mid-bay station (CB200) in the spring. In contrast, the 912h4 genes were most abundant in the samples collected from the mouth of the Chesapeake Bay (station CB300) during the spring. At the mid-bay station, these genes were most abundant in the deep samples, while at the mouth of the Chesapeake Bay, differences between the samples taken at different depths were less apparent. Overall, the abundance of the 912h4 clone increased along the salinity gradient from the head to the mouth of the Chesapeake Bay (Fig. 4).
DISCUSSION
Quantitative PCR is a sensitive, robust technique that can be used to estimate the abundance of nifH sequences against a variable background of environmental genomic DNA. Experiments with fragments closely related to the target demonstrated that a sequence 89% identical to the target sequence was detected with one of the primer and probe sets, indicating that caution must be exercised when interpreting our results. Currently, it is not possible to determine if the different sequences detected with a single probe and primer set represent different strains or species of bacteria. It is likely that this degree of divergence represents very closely related bacteria because nifH sequences are more divergent than 16S ribosomal DNA sequences (26). Nonetheless, quantitative PCR revealed nonrandom trends in the distribution of two distantly related DNA phylotypes in the Chesapeake Bay.
Because it is impossible to transfer less than one gene copy into a reaction tube, the practical detection limit of the quantitative PCR is one gene copy per reaction. However, the lowest-concentration replicates of the standard curves (Fig. 3) revealed that it was possible to detect less than one target molecule per reaction. Previous work has shown that the Poisson distribution describes the probability that one or more target molecules will be transferred into a reaction tube when the average target concentration is less than one molecule per volume pipetted, and thus, with enough replication, it is possible to quantify samples containing less than a single target per volume used in a PCR (27). Because quantification of samples with an average concentration of less than one copy per volume requires substantial replication, samples with detectable amplification in at least one but not all replicate reactions were interpreted as indicating presence only (Fig. 4).
The number of gene copies per ml of sample was calculated as the product of the number of gene copies per nanogram of DNA and the amount of DNA recovered per volume filtered. Because the volumes filtered and the amounts of DNA recovered for each sample varied, the theoretical detection limit of quantitative PCR was also variable for each sample. For example, the amount of DNA recovered per volume filtered was 1 ng ml−1 and 16 ng ml−1 for the April 2001 CB300 deep and April 2002 CB200 mid-depth samples, respectively. A detection limit of one copy per reaction (as stated above, 2 ng of template was added to each reaction) corresponds to 0.5 and 8.0 copies ml−1 for the above samples, respectively. Thus, assuming one gene per cell and a bacterial abundance of 106 ml−1, the detection limit of the described quantitative techniques ranged from 0.00005 to 0.0008% of the total bacterioplankton. This detection limit estimate is conservative, as the assumed bacterial abundance is probably low; previous studies have reported that bacterial abundances in the Chesapeake Bay and Choptank River range from 1.5 × 106 to 25 × 106 cells ml−1 (4, 13, 21). Moreover, assuming one gene per cell is conservative, as plasmids exhibiting homology to a nifH probe have been observed. Beeson and others (3) reported that 26% of 521 diazotrophs isolated from salt marsh grass rhizoplanes contained plasmids, and 4% of the plasmids exhibited homology to nifH. Although documented examples of plasmids encoding nifH are rare, their occurrence could affect the interpretation of our results, and therefore, we have avoided reporting the abundance of the 907h22 and 912h4 phylotypes as cells per milliliter and instead have used gene copies per ml. Regardless, the estimated detection limits illustrate the sensitivity of quantitative PCR.
The maximum number of 907h22 nifH gene copies detected in the Chesapeake Bay was ca. 140 ml−1 for the April 2001 CB200 deep sample, and the maximum number of 912h4 nifH gene copies detected was ca. 340 ml−1 for the April 2002 CB300 deep sample. Although these phylotypes are not numerically abundant compared to the entire bacterial community, they may represent important components of Chesapeake Bay diazotroph communities. In addition, as there was evidence for reduced PCR efficiency in some samples (Fig. 4), the estimated number of gene copies in these samples was an underestimate. Because amplification efficiency was calculated for each sample, it is possible to apply a correction for PCR efficiency to the gene copy number estimate. For example, the calculated amplification efficiency for the deep sample collected in April 2002 from CB300 was 91%, and the average Ct was 33.09. Adjusting efficiency to 100%, the calculated Ct becomes 30.89, and the estimated number of 912h4 nifH gene copies increases from ca. 340 to 1,500 copies ml−1. However, some samples that were less than 95% efficient produced no detectable amplification, and therefore, it was not possible to apply the correction. Hence, corrections were not applied to all samples, and samples that reduced PCR efficiency relative to plasmid controls were noted. These results indicate that for some samples, PCR inhibition was problematic. In the future it may be worthwhile to individually optimize the amount of DNA added to each reaction to minimize inhibitory effects. Nonetheless, the majority of samples analyzed in this study did not show any evidence of PCR inhibition, and the most important trends observed in this study were not affected by these PCR artifacts.
It is also possible that DNA extraction efficiencies may vary from sample to sample and influence our results. To circumvent variable extraction efficiencies, Suzuki et al. (24) normalized the abundance of genes of interest to the abundance of total small-subunit ribosomal DNA and reported the abundance of particular genes as proportions of total bacterial genomes. In this study we reported nifH abundances normalized to the DNA extraction yield per volume of water filtered (Fig. 4). When the number of nifH gene copies was not normalized to yield and volume (the amount of DNA added to each reaction was constant), the distributions of the two nifH phylotypes examined did not change, suggesting that our conclusions were robust (data not shown). In addition, the fact that the distribution of the two gene types was different (and often inverse) suggests that factors such as sample extraction efficiency cannot explain our data.
Our results show that individual nitrogen-fixing bacteria have distinct nonrandom spatial and seasonal distributions in the Chesapeake Bay. The sequence 912H4 was most abundant at CB300, and in general, its abundance increased with increasing salinity; it is notable that both NH4 and NO3 concentrations decreased with increasing salinity (data not shown). This correlation between phylotype distribution and salinity is consistent with previous observations on the distribution of specific bacterial groups based on low-molecular-weight RNA analysis (4) or fluorescence in situ hybridization (5) in the Chesapeake Bay. However, the relationship between salinity and the distribution of specific phylotypes must be interpreted cautiously, as other physical and chemical parameters covary with salinity. On the other hand, the distribution of sequence 907h22 was not correlated with salinity and was most abundant at CB200. Interestingly, the source of this sequence was a clone library of PCR products amplified from CB200. Although the correlation of phylotype distribution to specific environmental factors is speculative, it is apparent that the phylotypes examined were not homogenously distributed throughout the Chesapeake Bay.
The abundance of the nifH phylotypes appeared to be seasonal. The 907h22 phylotype was most abundant in the spring at the stations CB200 and CB300. Similarly, at CB200 and CB300, the highest abundance of 912h4 was observed in the spring of 2002. In fact, the highest overall abundance of the 912h4 phylotype was observed at CB300 in the spring of 2002. However, at station CB100, the highest abundance was observed in the fall of 2002; this pattern of abundance may be correlated to salinity, as the highest salinity at CB100 was observed at the same time. Neither nifH phylotype was evenly abundant throughout the year at any location. Whatever environmental factor(s) determines the distribution of 907h22 and 912h4 in the Chesapeake Bay, a seasonal signal was apparent.
This study demonstrates that diazotrophs are not randomly distributed in the Chesapeake Bay and are either distributed by specific physical processes or adapted to different environmental niches. This conclusion is supported by previous work demonstrating that differences in environmental characteristics select for different types of sediment diazotrophs (6) or that microenvironmental heterogeneity promoted diazotroph diversity by selecting for physiologically specialized populations in Spartina alterniflora rhizospheres (2). Clearly, the distribution of diazotrophs in the Chesapeake Bay must be determined either by environmental selection and/or physical processes related to hydrodynamics and sediment resuspension. Although the Choptank River and Chesapeake Bay may be N limited at certain times of year (10), especially at the mouth of the Bay (16), negligible rates of nitrogen fixation have been detected in the water column, and only low rates have been detected in sediments (17). Thus, it seems unlikely that N-limiting conditions selected for the nifH phylotypes in the water column of the estuary. Nonetheless, it is possible that some microorganisms use nitrogen fixation as a means of obtaining nitrogen even when fixed nitrogen is available, but their activity is not detected since they are a small fraction of the community.
It may be that nitrogen-fixing microorganisms are being selected by other components of their genomes in the estuary environment, or that they are simply in transit from watershed soils or benthic sediments. With the ability to detect and quantify these organisms, it is now possible to investigate possible sources of these microorganisms as well as determine the conditions under which nitrogenase is expressed. This study represents the first autecological study of uncultivated natural populations of diazotrophs, and the methods presented can be easily adapted to studies of gene expression via reverse transcription and quantitative PCR. If no evidence is found for nitrogenase expression in environments rife with fixed nitrogen, the enigma of the wide distribution of nitrogen fixation genes in non-N-limited environments will need to be addressed. On the other hand, evidence for nitrogen fixation in waters replete with fixed nitrogen will pose a new set of questions about the function of nitrogenase genes and the ecology of nitrogen fixation. Future research with the techniques outlined in this study will help resolve the mechanisms driving the diversity and distribution of nitrogen-fixing microbes in the Chesapeake Bay, with implications for the factors controlling the distribution of genes and microorganisms in the environment in general.
Acknowledgments
We thank the crew of the R/V Cape Henlopen for their help collecting samples. We also thank the staff of UMCES Horn Point Lab, J. C. Collier, P. A. Gilbert, G. A. Jackson, J. D. Kirshtein, G. Taroncher-Oldenburg, M. A. Voytek, and B. B. Ward, for help with sample collection, processing, and management. We are grateful for the assistance with statistical analysis provided by M. J. Church.
This research was supported by an NSF Biocomplexity grant (OCE-9981437) and an NSERC postdoctoral fellowship to S. M. Short.
REFERENCES
- 1.Affourtit, J., J. P. Zehr, and H. W. Paerl. 2001. Distribution of nitrogen fixing microorganisms along the Neuse River Estuary, North Carolina. Microb. Ecol. 41:114-123. [DOI] [PubMed] [Google Scholar]
- 2.Bagwell, C. E., and C. R. Lovell. 2000. Microdiversity of culturable diazotrophs from the rhizoplanes of the salt marsh grasses Spartina alterniflora and Juncus roemerianus. Microb. Ecol. 39:128-136. [DOI] [PubMed] [Google Scholar]
- 3.Beeson, K. E., D. L. Erdner, C. E. Bagwell, C. R. Lovell, and P. A. Sobecky. 2002. Differentiation of plasmids in marine diazotroph assemblages determined by randomly amplified polymorphic DNA analysis. Microbiology 148:179-189. [DOI] [PubMed] [Google Scholar]
- 4.Bidle, K. D., and M. Fletcher. 1995. Comparison of free-living and particle-associated bacterial communities in the Chesapeake Bay by stable low-molecular-weight RNA analysis. Appl. Environ. Microbiol. 61:944-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 6.Burns, J. A., J. P. Zehr, and D. G. Capone. 2002. Nitrogen-fixing phylotypes of Chesapeake Bay and Neuse River estuary sediments. Microb. Ecol. 44:336-343. [DOI] [PubMed] [Google Scholar]
- 7.del Giorgio, P. A., and T. C. Bouvier. 2002. Linking the physiologic and phylogenetic successions in free-living bacterial communities along an estuarine salinity gradient. Limnol. Oceanogr. 47:471-486. [Google Scholar]
- 8.DeLong, E. F. 2001. Microbial seascapes revisited. Curr. Opin. Microbiol. 4:290-295. [DOI] [PubMed] [Google Scholar]
- 9.Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher, T. R., A. B. Gustafson, K. Sellner, R. Lacouture, L. W. Haas, R. L. Wetzel, R. Magnien, D. Everitt, B. Michaels, and R. Karrh. 1999. Spatial and temporal variation of resource limitation in Chesapeake Bay. Mar. Biol. 133:763-778. [Google Scholar]
- 11.Fuhrman, J. A., K. Mccallum, and A. A. Davis. 1993. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl. Environ. Microbiol. 59:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 13.Heidelberg, J. F., K. B. Heidelberg, and R. R. Colwell. 2002. Seasonality of Chesapeake Bay bacterioplankton species. Appl. Environ. Microbiol. 68:5488-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karner, M. B., E. F. Delong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 15.Macgregor, B. J., B. Van Mooy, B. J. Baker, M. Mellon, P. H. Moisander, H. W. Paerl, J. Zehr, D. Hollander, and D. A. Stahl. 2001. Microbiological, molecular biological and stable isotopic evidence for nitrogen fixation in the open waters of Lake Michigan. Environ. Microbiol. 3:205-219. [DOI] [PubMed] [Google Scholar]
- 16.Marshall, H. G., M. F. Lane, and K. K. Nesius. 2003. Long-term phytoplankton trends and related water quality trends in the lower Chesapeake Bay, Virginia, USA. Environ. Monit. Assess. 81:349-360. [PubMed] [Google Scholar]
- 17.Marsho, T. V., R. P. Burchard, and R. Fleming. 1975. Nitrogen fixation in Rhode River Estuary of Chesapeake Bay. Can. J. Microbiol. 21:1348-1356. [DOI] [PubMed] [Google Scholar]
- 18.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 19.Pomeroy, L. R. 1974. The ocean's food web, a changing paradigm. BioScience 24:499-504. [Google Scholar]
- 20.Rappé, M. S., K. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 21.Smith, E. M., and W. M. Kemp. 2003. Planktonic and bacterial respiration along an estuarine gradient: responses to carbon and nutrient enrichment. Aquat. Microb. Ecol. 30:251-261. [Google Scholar]
- 22.Steppe, T. F., and H. W. Paerl. 2002. Potential N2 fixation by sulfate-reducing bacteria in a marine intertidal microbial mat. Aquat. Microb. Ecol. 28:1-12. [Google Scholar]
- 23.Steward, G. F., J. P. Zehr, R. Jellison, J. P. Montoya, and J. T. Hollibaugh. 2003. Vertical distribution of nitrogen-fixing phylotypes in a meromictic, hypersaline lake. Microb. Ecol. 10.1007/s00248-003-1017-8. [DOI] [PubMed]
- 24.Suzuki, M. T., L. T. Taylor, and E. F. Delong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tillett, D., and B. A. Neilan. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36:251-258. [Google Scholar]
- 26.Tyrrell, T. 1999. The relative influences of nitrogen and phosphorus on oceanic primary production. Nature 400:525-531. [Google Scholar]
- 27.Wang, Z., and J. Spadoro. 1998. Determination of target copy number of quantitative standards used in PCR-based diagnostic assays, p. 31-43. In F. Ferre (ed.), Gene quantification. Birkhauser, Boston, Mass.
- 28.Zehr, J. P., B. D. Jenkins, S. M. Short, and G. F. Steward. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539-554. [DOI] [PubMed] [Google Scholar]
- 29.Zehr, J. P., and L. A. McReynolds. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 55:2522-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zehr, J. P., M. Mellon, S. Braun, W. Litaker, T. Steppe, and H. W. Paerl. 1995. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl. Environ. Microbiol. 61:2527-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zehr, J. P., and B. B. Ward. 2002. Nitrogen cycling in the ocean: New perspectives on processes and paradigms. Appl. Environ. Microbiol. 68:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]