Abstract
Human subjects consumed biscuits containing either galacto-oligosaccharides or fructo-oligosaccharides in a double-blinded, crossover study. The impact of supplementing the diet with three biscuits per day on the fecal microbiota was evaluated by selective culture of particular bacterial groups, measurement of β-galactosidase activity, and nucleic acid-based analytical methods (PCR-denaturing gradient gel electrophoresis [PCR-DGGE] and fluorescent in situ hybridization). The composition of the bifidobacterial populations was monitored at the level of species (PCR-DGGE) and strains (pulsed-field gel electrophoresis of DNA digests), and representative cultures were tested quantitatively for their ability to use galacto-oligosaccharides. Technical improvements to DGGE analysis of the microbiota were made by the use of an internal standard that allowed valid comparisons of fragment staining intensities to be made between profiles, the use of S1 nuclease digestion to remove single-stranded DNA to facilitate cloning of DNA sequences cut from gels, and the extraction of RNA to be used as the template in reverse transcription-PCR-DGGE. RNA-DGGE profiles were markedly different (Dice's similarity coefficient, 58.5%) from those generated by DNA-DGGE. Neither the sizes of the bacterial populations nor the DNA-DGGE profiles of the microbiota were altered by the consumption of the biscuits, but the RNA-DGGE profiles were altered by the detection or increased staining intensity of 16S rRNA gene sequences originating from Bifidobacterium adolescentis and/or Colinsella aerofaciens in the feces of 11 of 15 subjects. β-Galactosidase activity was elevated in the feces of some subjects as a result of biscuit consumption. Subjects differed in the ability of the bifidobacterial strains harbored in their feces to use galacto-oligosaccharides. Our observations suggest that a phylogenetic approach to analysis of the gut ecosystem may not always be optimal and that a more physiological (biochemical) method might be more informative.
The distal intestinal tract of humans harbors a complex bacterial community, referred to as the microbiota, the composition of which is represented in the feces (24). The intestinal microbiota impacts markedly on the immunology, biochemistry, physiology, and nonspecific disease resistance of the host (14). Additionally, in genetically susceptible hosts, the microbiota is an essential factor in the pathogenesis of inflammatory diseases of the bowel (29). These observations have promoted the view that modification of the composition of the intestinal microbiota by means of dietary supplements might promote health (12, 13, 27).
Probiotics (products containing living bacteria) transiently alter the composition of Lactobacillus or Bifidobacterium populations during the period of administration (8, 30, 32, 33), whereas the consumption of prebiotics (potential substrates for bacterial inhabitants of the intestine) has been reported to selectively increase the number of bifidobacteria (4, 6, 11, 16, 20, 25, 34). Inulin and fructo-oligosaccharides (FOS) have been studied in terms of their prebiotic activity in previous studies, but galacto-oligosaccharides (GOS), derived from lactose in milk, have received little attention (2, 6). Most studies of prebiotics have involved the consumption of inulin- or oligosaccharide-containing powders (4, 6, 11, 16, 20, 25), which may not be relevant to everyday life because the substrates, for commercial use, would be incorporated into a food product. Therefore, we were interested in determining the impact of GOS consumed in biscuits on the composition and activities of the fecal microbiota.
The impact of the ingestion of these biscuits on the fecal microbiota of humans was measured by culture-based enumeration of selected bacterial populations, monitoring the bifidobacterial species and strain composition by nucleic acid-based methods, fecal β-galactosidase activity, comparison of fecal microbiota profiles generated by denaturing gradient gel electrophoresis (DGGE) following amplification of 16S ribosomal DNA (rDNA) sequences from fecal DNA by PCR or RNA by reverse transcription-PCR (RT-PCR), and by enumeration of Bifidobacterium adolescentis and Colinsella aerofaciens by fluorescent in situ hybridization.
MATERIALS AND METHODS
Study design.
A double-blinded, crossover study that involved 15 healthy human subjects (eight female and seven male) who maintained their usual lifestyles was carried out. The study was approved by the Otago Ethics Committee, reference 01/02/001. The biscuit code was broken when all of the analytical procedures had been completed. The subjects consumed three chocolate chip biscuits at one sitting per day, giving a daily dose of 2.5 g of GOS, FOS, or no oligosaccharides (NONE). All of the biscuits contained lactose to balance for the nonhydrolyzed lactose present in the GOS preparation. The subjects consumed each type of biscuit for 3 weeks, interspersed with washout periods of 2 weeks as well as a final washout period of 2 weeks. The subjects were divided into three groups: group A consumed biscuits in the order GOS, FOS, NONE; group B consumed biscuits in the order FOS, NONE, GOS; and group C consumed biscuits in the order NONE, GOS, FOS. A fecal sample was obtained at the beginning of the study before dietary supplementation commenced (control) and at the end of each week of the study. Thus, a total of 16 fecal samples were obtained per subject.
Preparation of biscuits.
Chocolate chip biscuits were prepared at Fonterra Research, Palmerston North, New Zealand, and sent in coded containers to the University of Otago. Three batches of biscuits that contained the same quantities of butter, sugar, brown sugar, vanilla, eggs, flour, chocolate chips, and baking powder were prepared. Each biscuit weighed 20 g. The batches of biscuits differed in that GOS biscuits contained Oligovite (Fonterra), FOS biscuits contained FOS (Cosucra, Brussels, Belgium), and NONE biscuits did not contain oligosaccharides.
Culture-based enumeration of selected bacterial populations.
Enterobacteria, lactobacilli, enterococci, and bifidobacteria were enumerated in selective media and incubation conditions that have been described previously (33). All of the fecal samples collected from each subject were examined within 1 h of collection.
Extraction of DNA and RNA from feces.
Nucleic acids were extracted from fecal samples by preparing a 1/10 (wt/vol) homogenate in sterile phosphate-buffered saline (pH 7.0). Samples were mixed by vortexing and stored frozen at −80°C until further use. DNA extraction followed a previously described method (33). For RNA extraction, all labware and solutions were made RNase-free with diethyl pyrocarbonate (DEPC)-treated water; 500 μl of fecal homogenates was centrifuged at low speed (200 × g, 4°C) for 5 min, and the upper phase was collected. This was centrifuged at 14,000 × g for 5 min at 4°C to harvest the bacterial cells. These were washed once with 1 ml of phosphate-buffered saline and resuspended in 200 μl of phosphate-buffered saline, and the samples were then transferred to screw-cap tubes containing 0.1 g of glass beads (0.1-mm diameter). After the addition of 200 μl of lysis buffer containing lysozyme (10 mg/ml; 10 mM Tris-HCl, 10 mM EDTA, pH 7.9, prepared just before use), samples were homogenized by vortexing and incubated for 30 min at room temperature. Then 50 μl of 20% sodium dodecyl sulfate-300 μl of 50 mM sodium acetate-10 mM EDTA (pH 5.1) and 300 μl of phenol saturated with sodium acetate-EDTA buffer (pH 5.1) were added to the tubes. The samples were shaken at 5,000 rpm for 2 min in a Mini-Bead Beater. After centrifugation at 14,000 × g for 10 min at 4°C, the supernatants were transferred to microcentrifuge tubes, and 600 μl of phenol saturated with sodium acetate-EDTA buffer (pH 5.1) was added to the tubes. Samples were mixed by vortexing for 1 min and centrifuged under the conditions described above; 600 μl of phenol-chloroform-isoamyl alcohol (25:24:1) was added to the supernatants, and the mixtures were vortexed for 1 min and centrifuged. This step was repeated once. Then 600 μl of chloroform-isoamyl alcohol (24:1) was added to the supernatants, which were vortexed for 1 min and centrifuged for 5 min at 14,000 × g. RNA was precipitated in 1 ml of isopropanol overnight at −20°C.
Residual DNA was removed by DNase I treatment as follows. Samples that had been stored overnight at −20°C were centrifuged at 14,000 × g for 10 min at 4°C, and the pellets were washed with 80% ethanol. Air-dried pellets were dissolved in 50 μl of DEPC-treated water and incubated for 1 h at 37°C in a solution containing 20 μl of 5× DNase I buffer (30 mM MgCl2, 10 mM NaCl, 200 mM Tris-HCl, pH 7.9), 1.5 μl of RNase-free DNase I (Roche; 10 U/μl), and 28.5 μl of DEPC-treated water. Samples were then processed with Qiagen RNA/DNA columns according to the manufacturer's instructions. Eluted RNA was isopropanol precipitated on ice for 10 min and centrifuged at 15,000 × g for 30 min at 4°C. Pellets were washed twice with 80% ethanol and air dried for 10 min at room temperature. Finally, RNA pellets were dissolved in 60 μl of DEPC-treated water and stored at −80°C. RNA integrity was assessed by electrophoresis of each sample in a 1.2% agarose gel in which 23S and 16S RNA were observed. RNA concentration and purity were determined spectrophotometrically.
PCR and RT-PCR.
Amplification of total bacterial community DNA was carried out by targeting 16S rRNA gene sequences with universal bacterial primers HDA1-GC and HDA-2 and a previously described program (33). The PCR products were checked by electrophoresis in a 2% agarose gel stained with ethidium bromide (5 μg/ml) and viewed by UV transillumination.
RT-PCR used the Qiagen OneStep RT-PCR kit. Each mixture (50 μl) was prepared according to the manufacturer's instructions with HDA primers at a concentration of 0.6 μM and 10 to 40 ng of template RNA. RT-PCRs were carried out with the following amplification program: one cycle consisting of 30 min at 50°C (reverse transcription), one cycle of 15 min at 95°C, and then 35 cycles consisting of 30 s at 94°C, 30 s at 56°C, and 45 s at 72°C. The amplification products were checked by electrophoresis in 2% agarose and ethidium bromide staining prior to DGGE analysis.
Analysis of fecal microbiota by DGGE.
Amplicons from either PCR or RT-PCR were analyzed by DGGE with a DCode apparatus (Bio-Rad, Hercules, Calif.) on a 6% polyacrylamide gel with a 30 to 55% gradient of 7.0 M urea and 40% (vol/vol) formamide that increased in the direction of the electrophoresis. Electrophoresis was carried out in 1× TAE (40 mM Tris, 20 mM acetic acid, 1 mM EDTA) buffer at 130 V and 60°C for 4 h. Gels were stained with ethidium bromide solution (5 μg/ml) for 20 min, washed with deionized water for 20 min, and viewed by UV transillumination. DGGE profiles were compared with Quantity One version 4.2 software, which is part of the Discovery Series (Bio-Rad Laboratories), and similarities were expressed with Dice's similarity coefficient.
Cloning of DNA fragments eluted from DGGE gels.
Fragments of interest were cut from the polyacrylamide gel with a sterile scalpel blade after the stained gel had been photographed. DNA was eluted by the addition of 100 μl of diffusion buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA [pH 8.0], and 0.1% sodium dodecyl sulfate) and incubation for 30 min at 50°C. After vortexing and brief centrifugation, the supernatants were transferred to a QIAquick gel extraction kit spin column (Qiagen). Purification of nucleic acid was done according to the manufacturer's instructions. Purified fragments were digested with S1 nuclease (Roche) to remove single-stranded DNA; 33 μl of eluted DNA was digested with 3 μl of S1 nuclease (400 U/μl) and 4.0 μl of 10× buffer (330 mM sodium acetate, 500 mM NaCl, 0.3 mM ZnSO4, pH 4.5) for 1 h at 37°C. The reaction was stopped by heating the mixture at 95°C for 5 min, and samples were stored frozen at −80°C until further use. S1-treated DNAs were used as templates in PCRs with the HDA primers prior to ligation.
The resulting PCR products were cleaned with the QIAquick PCR purification kit (Qiagen) and cloned in Escherichia coli DH5α with the pGEM-T Easy vector system (Promega, Madison, Wis.). Recombinant plasmids were purified from colonies as described previously (19), and the migration of the amplified inserts was checked by DGGE by comparison to the original sample from which the fragments had been obtained. Only sequences that migrated as a single fragment to the correct position were amplified from the cloning vector with T7 and SP6 primers (19), purified (QIAquick PCR purification kit), and sent to the sequencing facility (Center of Gene Research, University of Otago). T7 was used as the sequencing primer. Sequences were compared to those in the GenBank database with the BLASTn algorithm (3).
Comparison of DGGE profiles with an internal standard.
The intensity of staining of selected DNA fragments in RNA-DGGE gel profiles was compared to that of an internal standard of known concentration; 1 μl of a 100-pg/ml solution of DNA from Bifidobacterium infantis DSM 20088 was subjected to PCR amplification under the conditions described above together with 1 μl of a 1:100 dilution of the RT-PCR product from the appropriate fecal sample. PCR products were used in DGGE with a gradient of 30 to 55% and the conditions described above. Gels were stained with ethidium bromide, and fragment intensities were analyzed from gel images with Quantity One software (version 4.2; Bio-Rad Laboratories). The ratio of the fragment intensity of B. adolescentis or C. aerofaciens to the fragment intensity of B. infantis DSM 20088 was calculated.
Direct detection of bifidobacterial species by PCR-DGGE.
Bifidobacterial species present in the feces were detected by PCR (amplification of a region of the transaldolase gene that varies in nucleotide base composition between species) and DGGE, as described previously (26). Species were identified by reference to an identification ladder or by sequencing and alignment of sequences with reference strains (26).
Differentiation of bifidobacterial strains.
For each sample, 10 bifidobacterial colonies were picked randomly from plates of selective culture medium, as described previously (20). DNA was extracted from the cells of pure cultures of each isolate in agarose plugs. The DNA was digested with XbaI, and the restriction fragment length profile was generated by pulsed-field gel electrophoresis (PFGE) (33). Representative strains were identified to species level by PCR amplification of a region of the transaldolase gene and DGGE (26).
Degradation of GOS by bifidobacterial strains.
Representative strains of bifidobacteria isolated from the feces of six selected subjects were tested for their ability to use the GOS present in Oligovite. This subset of subjects was chosen according to whether their RNA-DGGE profiles showed alteration with respect to the detection and intensity of staining of bifidobacterial or colinsellal DNA fragments. The strains were cultured anaerobically in peptone-yeast extract medium (18), to which was added Oligovite syrup to give a GOS concentration of 0.5%. After 72 h of anaerobic incubation at 37°C, the cultures were centrifuged, and the supernatants were analyzed by high-pressure liquid chromatography to determine the utilization of the components of the Oligovite syrup (21).
β-Galactosidase activity.
β-Galactosidase activity was assayed spectrometrically at 420 nm as described by Smart et al. (31). Briefly, a fecal sample suspension (1/10, wt/vol) prepared in sodium phosphate buffer (pH 7.5) was sonicated as described previously (33), and 16 μl of lysate was used in the assay. Spectrophotometric readings were recorded 5, 10, and 20 min after the addition of substrate (2 mM o-nitrophenyl-β-d-galactopyranoside) to warmed preparations (37°C). The reaction was stopped at the appropriate times by the addition of sodium bicarbonate. The amount of nitrophenol released was measured by reference to a standard curve and standardized per milligram of protein in the fecal lysate, which was measured by the method of Lowry et al. (22).
Fluorescent in situ hybridization.
The cells of B. adolescentis and C. aerofaciens were enumerated in fecal samples with specific, fluorescently labeled oligonucleotide probes (15). The specificity of the newly designed B. adolescentis probe (Badol1255; 5′-CCCCTCACGAGGTCGCAT-3′) was validated with nine bifidobacterial species and eight other bacterial reference strains as described previously (15). C. aerofaciens populations were measured with probe COR653 (17). Total bacterial cell numbers were determined in fecal preparations that had been stained with 4′,6′-diamidino-2-phenylindole, and the B. adolescentis and C. aerofaciens populations were recorded as a percentage of the total cell count.
RESULTS
Enumeration of bacterial populations by selective culture.
Statistical analysis of bacteriological culture results (Friedman test and nonparametric repeated-measures analysis of variance) showed that the populations of lactobacilli, lactose-fermenting (LF) enterobacteria, total enterobacteria, and enterococci did not differ throughout the study (P, 0.4413, 0.5047, 0.0643, and 0.1455, respectively). Bifidobacteria were not detected in the feces of two subjects. There was no significant difference between the means of bifidobacterial populations for the remaining 13 subjects (P = 0.1162). We concluded that consumption of the biscuits did not alter the size of the selected bacterial populations, as determined by quantitative culture.
Comparison of fecal microbiota profiles generated by DNA-DGGE and RNA-DGGE.
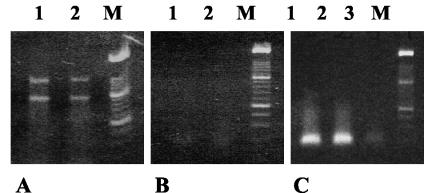
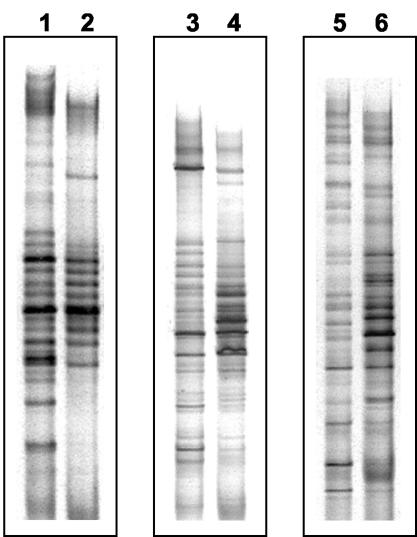
RNA of high quality was extracted from all samples (Fig. 1A). Fragments corresponding to the 23S and 16S rRNA molecules were visible in all extracts. The absence of a PCR product when the RNA preparations were tested with HDA primers demonstrated the absence of contaminating DNA (Fig. 1B). RT-PCRs performed with the same preparations generated amplicons of the correct size (≈200 bp) and of suitable intensity for DGGE analysis (Fig. 1C). Strikingly, whereas the DNA-DGGE profiles were composed of fragments distributed throughout the length of the gel, the RNA-DGGE profiles showed intensely staining fragments concentrated in the central portion of the gel (Fig. 2 and 3). Weighted comparisons of DNA- and RNA-DGGE profiles of the subjects showed that they differed markedly within subjects (Dice similarity coefficient mean, 58.5%, standard error of the mean, 3.90%; t test with Welch correction, P = 0.0029). Alterations to DNA-DGGE profiles as a result of supplementing the diet with biscuits were not observed.
FIG. 1.
Assessment of RNA extractions. (A) RNA extracts from two subjects electrophoresed in a 1.2% agarose gel. Lanes 1 and 2, fecal extracts; lane M, molecular size markers. (B) Absence of amplified DNA from RNA preparations in panel A when tested with PCR primers. (C) Presence of amplified DNA after RT-PCR.
FIG. 2.
Comparison of DGGE profiles generated with fecal RNA as the template (RT-PCR) or fecal DNA (PCR) from three human subjects. Lanes 1 and 2, DNA- and corresponding RNA-DGGE profiles for subject 1; lanes 3 and 4, DNA- and corresponding RNA-DGGE profiles for subject 2; lanes 5 and 6, DNA- and corresponding RNA-DGGE profiles for subject 3. Note intensely staining fragments in the central region of RT-PCR profiles relative to PCR profiles.
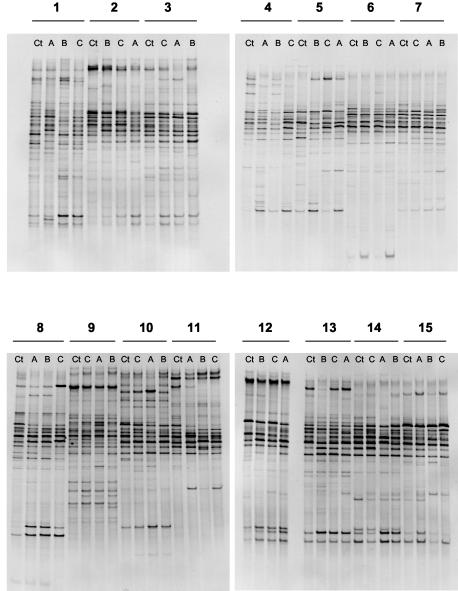
FIG. 3.
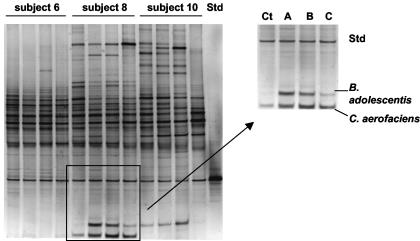
RNA-DGGE gels of the 15 subjects. Ct, control fecal sample (no biscuits); A, GOS biscuits; B, FOS biscuits; C, NONE biscuits. Note altered profiles associated with biscuit consumption in the lower region of the gels (all subjects except 6, 7, 9, and 11).
In contrast, the RNA-DGGE profiles of 11 of the 15 subjects showed altered profiles during one or more of the periods of dietary supplementation relative to control samples either through the detection of additional fragments or more intense staining of fragments. Alterations in profiles were most frequent in the lower part of the gels (Fig. 3). Such altered profiles were detected in relation to GOS biscuit consumption (10 subjects), FOS biscuit consumption (9 subjects), and NONE biscuit consumption (8 subjects). We focused on the bacterial species represented by fragments in the lower part of the gels because alterations to profiles were commonly detected there. Furthermore, bifidobacterial DNA fragments would be likely to migrate to this region of the gel because they are high-G+C bacteria. Bifidobacterial populations have been reported to be selectively stimulated by prebiotic consumption.
Efficacy of S1 nuclease treatment and identification of bacterial origin of DNA fragments.
The DNA fragments excised from DGGE gels are commonly contaminated with single-stranded DNA originating from many of the bacteria represented in the sample, and consequently several erroneous fragments or even complete profiles can sometimes be generated from DNA extracted from a single region of the DGGE gel (unpublished observation). Failing to check the migration of cloned fragments in comparison to the fecal profile could lead to incorrect identifications. To overcome this problem, we treated eluted DNA with S1 nuclease to remove contaminating single-stranded DNA prior to amplification and cloning. Through this strategy, cloning of the correct fragment was increased from a frequency of 50% to a frequency of 75 to 100%. Alterations in profiles observed in the lower part of the gels were due to changes in the presence or absence or increased or decreased staining intensity of DNA fragments originating from B. adolescentis and C. aerofaciens (BLASTn identities: B. adolescentis, 100% identity over 183 bp, AF275882; C. aerofaciens, 99% identity over 175 bp; AJ245920).
Confirmation of increased staining intensity of bifidobacterial and colinsellal fragments in RNA-DGGE gels by use of an internal standard.
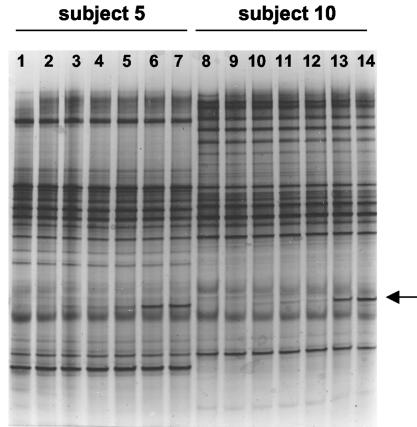
The PCR amplicon generated from Bifidobacterium infantis DSM 20088 DNA with HDA primers migrated to a position that did not overlap any other fragments in the profiles. In order to optimize the concentration of standard DNA to be added to the samples, increasing concentrations of standard DNA were added to decreasing concentrations of fecal RT-PCR cDNA obtained from two different subjects, and PCRs were performed. Either 10 or 100 pg/ml of standard DNA (1 μl) was added to 1:10, 1:50, and 1:100 dilutions of RT-PCR products (1 μl) and used in PCR. Visual inspection of the resulting gel showed that 100 pg of DNA B. infantis per ml and a 1:100 dilution of RT-PCR product from the samples gave the best combination (Fig. 4).
FIG. 4.
Optimization of internal standard concentration in DGGE gels. The arrow indicates the internal standard fragment generated from B. infantis DSM 20088 DNA. Lane 1, no internal standard; lanes 2 to 6 show the result of addition of the standard (10 and 100 pg/ml in alternate lanes) to 10-fold-, 50-fold-, and 100-fold-diluted RT-PCR amplicons from the feces of subject 5. Lanes 8 to 14 show the results obtained from the same experiment with feces from subject 10. Note the optimal detection of the standard when the RT-PCR amplicon was diluted 100-fold and that the fecal profiles were otherwise unaltered.
Figure 5 shows the results obtained for three of the subjects. The intensity of the internal standard fragment was similar within the samples to be compared. Results from the analysis of B. adolescentis and C. aerofaciens for seven subjects are shown in Table 1. The preparations from the remaining four subjects in which alterations to RNA-DGGE profiles were detected were not satisfactory for fragment intensity measurements because the internal standard did not amplify evenly. In the case of the seven subjects for whom fragment intensities were measured, consumption of biscuits containing FOS resulted in an increase in the staining intensity of the bands corresponding to B. adolescentis (P < 0.05, Friedman test, nonparametric repeated-measures analysis of variance) and C. aerofaciens (P < 0.05, Friedman test, nonparametric repeated-measures analysis of variance) relative to that of control samples. Overall values obtained from profiles of samples collected after the consumption of GOS or NONE biscuits were not statistically different from those for the controls. Nevertheless, seven subjects within these groups showed a twofold increase or more, similar to that seen with FOS biscuits, with respect to the control (Table 1).
FIG. 5.
Example of different staining intensities of DNA fragments relative to biscuit consumption and internal standard. Ct, control fecal sample; A, GOS biscuit consumption period; B, FOS biscuit consumption period; C, NONE biscuit consumption period; Std, internal standard.
TABLE 1.
Ratio of staining intensities of bifidobacterial and colinsellal DNA fragments relative to the internal standard in RNA-DGGE gels
| Subject no. | Relative staining intensitya
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
Bifidbacterium adolescentis
|
Colinsella aerofaciens
|
|||||||
| Control | GOS3 | FOS3 | NONE3 | Control | GOS3 | FOS3 | NONE3 | |
| 1 | 0.78 | 0.40 | 7.96 | 2.30 | 0.26 | 0.38 | 4.23 | 0.90 |
| 5 | ND | ND | 0.25 | ND | 0.51 | 1.22 | 1.52 | 0.29 |
| 8 | ND | 2.17 | 1.21 | 0.24 | 0.98 | 1.99 | 3.15 | 1.12 |
| 10 | 1.24 | 2.86 | ND | 2.26 | ND | ND | ND | ND |
| 12 | ND | 1.00 | 1.85 | 0.49 | ND | 0.91 | 1.05 | 0.74 |
| 13 | 0.20 | 0.82 | 6.00 | 2.74 | 0.45 | 0.56 | 1.29 | 1.33 |
| 14 | 0.40 | 1.19 | 1.45 | 0.28 | 0.65 | 0.80 | 0.95 | 0.43 |
| Median | 0.20 | 1.00 | 1.45b | 0.49 | 0.45 | 0.80 | 1.29b | 0.74 |
Control, fecal sample collected prior to first period of supplementation of diet; other fecal samples were collected at the end of the third week of consumption of biscuits. ND, none detected.
Significantly different from control value, P < 0.05.
β-Galactosidase activity in feces.
For comparative purposes, the mean control and washout values were compared to those obtained from samples collected at the end of each period of dietary supplementation. Four subjects had higher (twofold or greater) β-galactosidase activities in their feces when GOS biscuits were consumed, compared to two subjects when FOS or NONE biscuits were eaten. One of these subjects showed altered enzyme activity during all three periods of supplementation (subject 4), and one subject showed higher activity when GOS or NONE biscuits were consumed (subject 12).
Detection of bifidobacterial strains and species.
Five of the 13 subjects who had detectable bifidobacteria in the feces harbored a population in which one strain predominated throughout the study, whereas the remainder of the subjects had populations comprised of multiple strains. On average, subjects harbored five unique strains, as determined by pulsed-field gel electrophoresis of DNA digests. The strain composition of the bifidobacterial population was not altered in any of the subjects by dietary supplementation. The species composition of the bifidobacterial populations was not altered by dietary supplementation, and considerable stability of the DGGE profiles generated from fecal DNA with primers that targeted the transaldolase gene was observed. Comparison of bifidobacterial species by direct detection in feces by PCR-DGGE or by identification of bifidobacterial isolates (representative strains) cultured from feces showed that B. adolescentis was the most commonly detected species (Table 2). Direct detection did not reveal as much species diversity as identification of representative strains cultured from the feces (Table 2).
TABLE 2.
Bifidobacterial species detected directly in feces (PCR-DGGE) or by identification of representative strains cultured from feces
| Subject no. | Species detected
|
|
|---|---|---|
| PCR-DGGE | Culture | |
| 1 | B. adolescentis | B. adolescentis |
| 2 | B. adolescentis | B. adolescentis |
| 3 | B. longum | B. longum and B. adolescentis |
| 4 | B. catenulatum, B. adolescentis | B. catenulatum, B. adolescentis |
| 5 | B. adolescentis | B. adolescentis, B. bifidum |
| 6 | B. adolescentis | B. adolescentis, B. bifidum |
| 7 | B. adolescentis | B. adolescentis, B. longum |
| 8 | B. adolescentis | B. adolescentis |
| 9 | None | None |
| 10 | B. adolescentis | B. adolescentis, B. bifidum, B. longum |
| 11 | None | None |
| 12 | B. adolescentis | B. adolescentis, B. longum |
| 13 | B. adolescentis | B. adolescentis, B. bifidum |
| 14 | B. pseudocatenulatum | B. pseudocatenulatum, B. adolescentis, B. longum |
| 15 | B. adolescentis | B. adolescentis, B. bifidum |
GOS utilization by bifidobacterial strains.
The utilization of the components of the Oligovite syrup used in the preparation of GOS biscuits by representative strains isolated from the feces of selected subjects is shown in Table 3. At least one strain isolated from four subjects whose RNA-DGGE profile was altered following GOS biscuits consumption used more than 20% of the disaccharide, trisaccharide, or longer-chain oligosaccharide components of the Oligovite syrup used in the preparation of the biscuits. In contrast, the strains isolated from two subjects whose profiles were unaltered used less than 20% of these components (Table 3).
TABLE 3.
Utilization of Oligovite components by representative bifidobacterial strains (single assay for each strain) cultured from human feces
| Subject no.a | Bifidobacterial strain | Utilization (% of total) of Oligovite components
|
||
|---|---|---|---|---|
| Disaccharides | Trisaccharides | Tetrasaccharides and larger oligosaccharides | ||
| 2 | 1 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | |
| 3 | 30.0 | 29.0 | 29.0 | |
| 3 | 1 | 29.0 | 29.0 | 30.0 |
| 2 | 70.0 | 72.5 | 73.2 | |
| 3 | 39.6 | 40.0 | 41.1 | |
| 4 | 85.9 | 87.1 | 91.9 | |
| 4 | 1 | 0 | 0 | 0 |
| 2 | 42.0 | 43.3 | 33.2 | |
| 3 | 8.2 | 0 | 5.8 | |
| 4 | 5.1 | 4.5 | 6.0 | |
| 5 | 21.3 | 10.4 | 31.2 | |
| 6 | 1 | 8.0 | 8.0 | 6.0 |
| 2 | 3.0 | 0 | 0 | |
| 3 | 15.0 | 11.0 | 14.0 | |
| 7 | 1 | 15.0 | 18.0 | 14.0 |
| 2 | 13.0 | 14.0 | 14.0 | |
| 3 | 15.0 | 16.0 | 14.0 | |
| 4 | 9.0 | 11.0 | 14.0 | |
| 5 | 6.0 | 4.0 | 3.0 | |
| 8 | 1 | 12.9 | 0 | 12.0 |
| 2 | 35.7 | 37.7 | 38.0 | |
| 3 | 21.3 | 20.8 | 6.0 | |
Subjects 2, 3, 4, and 8 had altered RNA-DGGE profiles after consuming GOS biscuits. The profiles of subjects 6 and 7 were unaltered.
Enumeration of B. adolescentis and C. aerofaciens cells by FISH.
C. aerofaciens was detected in all of the samples that were examined (Table 3). The Colinsella populations did not differ as a percentage of the total microbiota after consumption of the biscuits (P = 0.2078, Friedman test, nonparametric repeated-measures analysis of variance). B. adolescentis was present in detectable numbers (>106/g) in the feces of seven subjects. The bifidobacterial populations did not differ as a percentage of the total microbiota through the consumption of the biscuits (P = 0.1222, Friedman test, nonparametric repeated-measures analysis of variance).
DISCUSSION
PCR of 16S rDNA sequences combined with analysis by DGGE has provided an efficient means of screening bacterial communities for changes in composition in response to allochthonous factors (19). Nevertheless, the method has some disadvantages, notably that it is semiquantitative, it is difficult to standardize and hence to make valid comparisons of staining intensity of DNA fragments, and only the predominant populations (comprising 90 to 99% of the total community) are monitored when universal bacterial primers are used (36). The last difficulty has been overcome through the use of group- or species-specific PCR primers that allow even minority populations in the community to be detected (26, 35). We have devised two important technical advances to improve analysis of fecal microbiota with PCR-DGGE. First, we used an internal standard by which DGGE profiles can be standardized to enable accurate comparison of profiles. The use of the internal standard permitted quantitative measurements of fragment staining intensities to be made that could then be evaluated statistically. Second, we used S1 nuclease treatment of DNA eluted from gels in order to remove single-stranded DNA. This resulted in a much higher success rate in cloning the DNA fragment of choice. Both of these technical improvements enhance PCR-DGGE as an analytical method.
A striking feature of our PCR-DGGE observations was the marked difference in bacterial community profiles generated with RNA compared to DNA as the PCR template. Profiles generated from bacterial RNA showed intensely stained fragments clustered in the middle of the denaturing gradient and were markedly different from those generated from DNA. RNA extracted from bacterial cells is mostly rRNA and can be used as an indicator of metabolic activity because the ribosome-per-cell ratio is roughly proportional to the growth rate of the bacteria (9). While DNA-based analytical procedures provide a phylogenetic picture of the community, they do not reflect metabolic activity because the DNA could originate from living active cells, living dormant cells, lysed cells, or dead cells. On the basis of our observations, it can be proposed that the human gut provides a habitat for a diversity of bacterial species, as seen in the DNA-DGGE profiles, the composite of which varies from one human to another. The “core” or “true” gut microflora may be comprised of relatively few populations that provide the major metabolic activities. This is clearly a topic that requires further investigation.
The diets of our subjects were supplemented with biscuits that contained oligosaccharides at concentrations that produced edible products of commercial quality. The consumption of three biscuits provided a dose of oligosaccharides that did not cause flatus or abdominal pain in the subjects. The daily dosage was lower than that used in most prebiotic studies reported in the literature (4, 6, 11, 16, 20, 25, 34), yet changes to fecal microbiota profiles were observed. Alterations to the bacterial community profiles in response to dietary supplementation were only detected in profiles that were generated from bacterial RNA. Therefore, we detected changes in bacterial metabolic activity in response to the consumption of the biscuits, but not changes in the population structure of the community. This seems a reasonable outcome considering the highly regulated nature of bacterial communities in which the proportions of the constituent populations are strictly maintained (1, 10).
Although oligosaccharides added to the gut ecosystem are generally expected to increase the number of bifidobacterial cells in particular, we did not observe any changes in population sizes by selective culture or nucleic acid-based analysis (FISH). Increased metabolic activity without an increase in the number of bacterial cells is not contradictory, since it has been shown in studies with rumen bacteria that “energy spillage” occurs in bacterial communities (28). The correlation between ATP and biomass formation is often poor, and while some of this variation in growth efficiency can be explained by maintenance energy expenditure, bacteria have other mechanisms of nongrowth energy dissipation (futile cycles) (28). Such energy spilling may occur when growth is limited by nutrients other than energy sources, as has been described for a rumen species, Streptococcus bovis (5).
DNA fragments representing B. adolescentis and C. aerofaciens were present in RNA-DGGE gel profiles, especially when biscuits had been consumed. It can be argued that these bacterial cells were in a metabolically quiescent state prior to dietary supplementation with the biscuits. When oligosaccharides and/or lactose supplemented the host diet, a substrate for metabolism became available for the bacteria in the large bowel, but because other nutrients were growth limiting in the highly regulated bacterial community, an increase in bifidobacterial and colinsellal cell numbers did not occur. Therefore, bifidobacterial and colinsellal populations remained constant in size and the composition of the bifidobacterial population at the level of species and strains did not differ, but RNA-DGGE provided evidence of increased metabolic activity. The significance of this increased metabolism to the bacterial community and to the human host, even if it occurs to a greater magnitude in the proximal large bowel (7), is not obvious but could provide important topics for future research.
β-Galactosidase activity was markedly increased in the feces of some subjects when the diet was supplemented with biscuits. At least twofold increases in activity were recorded only in samples collected at the end of periods during which biscuits had been consumed. The increases did not match alterations in RNA-DGGE profiles (increased staining of bifidobacterial and colinsellal fragments) and must therefore be considered to be due to alterations in the collective metabolism, including enzyme induction, of the ecosystem as a result of GOS and/or lactose entering the ecosystem.
We were interested in comparing the effect of consuming GOS in biscuits relative to FOS because most emphasis in the prebiotic literature has been placed on the latter substrate. Eleven of the 15 subjects showed alterations to RNA-DGGE profiles in association with biscuit consumption. Most of these changes were in profiles related to GOS biscuits (10 of 15), but the increased intensity of staining of B. adolescentis and C. aerofaciens fragments was more marked in samples collected at the end of the FOS biscuit consumption period. Some subjects showed altered profiles when NONE biscuits had been eaten, demonstrating that lactose itself had a “prebiotic” effect. The variation among subjects with respect to DGGE profiles and NONE biscuits could relate to the degree of lactose utilization in the small bowel and proximal large bowel of each individual.
As noted in a previous study (23), each subject harbored a unique collection of bifidobacterial strains in the feces. High-pressure liquid chromatography analysis of culture residues of representative strains showed that they varied greatly in their ability to use the Oligovite (GOS) components. This method could provide a basis of selection for bifidobacterial strains suitable for use in synbiotics, which are products containing a combination of living bacterial cells and a substrate that they can metabolize (27). Interestingly, strains that used a major portion of the oligosaccharide components of Oligovite were only isolated from subjects whose RNA-DGGE profiles were altered following GOS biscuit consumption.
Our study has provided important knowledge about the microbial ecology of the gut. First, we developed technical innovations for DGGE analysis of the bacterial community in feces (S1 nuclease activity and an internal DNA standard). Second, we obtained a new perspective on the composition of the fecal community by the use of RNA as the template in PCR-DGGE, which, after further investigation, may reveal the identity of the core, metabolically active members of the distal gut ecosystem. The use of highly purified RNA is of course critical to this analysis; otherwise, contaminating DNA will give the same profile obtained with a DNA extract. The RNA extraction method that we have described satisfies this criterion. Third, we demonstrated that evidence of increased metabolic activity in response to the consumption of GOS or FOS does not necessarily equate to increased numbers of bifidobacteria in a complex community, an observation that may explain the very small changes to bifidobacterial population sizes reported in other oligosaccharide studies, even when relatively large doses of prebiotics were administered (2, 4, 6, 11, 16, 20, 25, 34).
Our observations suggest that a phylogenetic approach to analysis of the gut ecosystem may not always be optimal and that a more physiological (biochemical) methodology might be more informative. Overall, our study has provided new perspectives to the analysis of the fecal microbiota in relation to the consumption of oligosaccharides in a concentration that is relevant to the food industry.
Acknowledgments
The dedication of the human subjects who participated in this study is gratefully acknowledged. We thank Greg Cook for review of the manuscript during its preparation.
We thank Fonterra for support of this study. R. Bibiloni was the recipient of a postdoctoral fellowship from the Consejo Nacional de Investigaciones Cientificas y Tecnicas, Argentina.
REFERENCES
- 1.Alexander, M. 1971. Microbial ecology. John Wiley and Sons, New York, N.Y.
- 2.Alles, M. S., R. Hartemink, S. Meyboom, J. L. Harryman, K. M. J. Van Laere, F. M. Nagengast, and J. G. A. J. Hautvast. 1999. Effect of transgalactooligosaccharides on the composition of the human intestinal microflora and on the putative risk markers for colon cancer. Am. J. Clin. Nutr. 69:980-991. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and L. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bouhnik, Y., B. Flourie, M. Riottot, N. Bisetti, M.-F. Gailing, A. Guibert, F. Bornet and J.-C. Rambaud. 1996. Effects of fructo-oligosaccharides ingestion on fecal bifidobacteria and selected metabolic indexes of colon carcinogenesis in healthy humans. Nutr. Cancer 26:21-29. [DOI] [PubMed] [Google Scholar]
- 5.Cook, G. M., and J. B. Russell. 1994. Energy spilling reactions of Streptococcus bovis and resistance of its membrane to proton conductance. Appl. Environ. Microbiol. 60:1942-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crittenden, R. G. 1999. Prebiotics, p. 141-156. In G. W. Tannock (ed.), Probiotics: a critical review. Horizon Scientific Press, Wymondham, England.
- 7.Cummings, J. H., and G. T. Macfarlane. 1991. The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70:443-459. [DOI] [PubMed] [Google Scholar]
- 8.Dunne, C., L. Murphy, S. Flynn, L. Omahory, S. O. O'Halloran, et al. 1999. Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie van Leeuwenhoeek 76:279-292. [PubMed] [Google Scholar]
- 9.Felske, A., H. Rheims, A. Wolerink, E. Stackebrandt, and A. D. L. Akkermans. 1997. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 143:2983-2989. [DOI] [PubMed] [Google Scholar]
- 10.Freter, R. 1988. Mechanisms of bacterial colonization of the mucosal surfaces of the gut, p. 45-60. In J. A. Roth (ed.), Virulence mechanisms of bacterial pathogens. American Society for Microbiology, Washington, D.C.
- 11.Gibson, G. R., E. R. Beatty, X. Wang, and J. H. Cummings. 1995. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 108:975-982. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 13.Goldin, B. R. and S. L. Gorbach. 1992. Probiotics for humans, p. 355-376. In R. Fuller (ed.), Probiotics: the scientific basis. Chapman and Hall, London, England.
- 14.Gordon, H. A., and L. Pesti. 1971. The gnotobiotic animal as a tool in the study of host-microbial relationships. Bacteriol. Rev. 35:390-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmsen, H. J. M., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmsen, H. J. M., G. C. Raangs, A. H. Franks, A. C. M. Wildeboer-Veloo, and G. W. Welling. 2002. The effect of the prebiotic inulin and the probiotic Bifidobacterium longum on the fecal microflora of healthy volunteers measured by FISH and DGGE. Microb. Ecol. Health Dis. 14:211-219. [Google Scholar]
- 17.Harmsen, H. J. M., A. C. M. Wildeboer-Veloo, J. Grijpstra, J. Knol, J. E. Degener, and G. W. Welling. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 66:4523-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holdeman, L. V., E. P. Cato and W. E. C. Moore. 1973. Anaerobe laboratory manual. VPI Anaerobe Laboratory, Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 19.Knarreborg, A., M. A. Simon, R. C. Engberg, B. B. Jensen, and G. W. Tannock. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens of various ages. Appl. Environ. Microbiol. 68:5918-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse, H.-P., B. Kleesen, and M. Blaut. 1999. Effects of inulin on faecal bifidobacteria in human subjects. Br. J. Nutr. 82:375-382. [DOI] [PubMed] [Google Scholar]
- 21.Lee, Y. C. 1990. High performance anion-exchange chromatography for carbohydrate analysis. Anal. Biochem. 189:151-162. [DOI] [PubMed] [Google Scholar]
- 22.Lowry, O. M., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 23.McCartney, A. L., W. Wang, and G. W. Tannock. 1996. Molecular analysis of the composition of the bifidobacterial and lactobacillus microflora of humans. Appl. Environ. Microbiol. 62:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, W. E. C., E. P. Cato, and L. V. Holdeman. 1978. Some current concepts in intestinal bacteriology. Am. J. Clin. Nutr. 31:S33-S42. [DOI] [PubMed] [Google Scholar]
- 25.Rao, A. V. 1999. Dose-response effects of inulin and oligofructose on intestinal bifidogenesis effects. J. Nutr. 129:1442S-1445S. [DOI] [PubMed] [Google Scholar]
- 26.Requena, T., J. Burton, T. Matsuki, K. Munro, M. A. Simon, R. Tanaka, K. Watanabe, and G. W. Tannock. 2002. Identification, detection, and enumeration of human Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 68:2420-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberfroid, M. B. 1998. Prebiotics and synbiotics: concepts and nutritional properties. Br. J. Nutr. 80:S197-S202. [PubMed] [Google Scholar]
- 28.Russell, J. B., and G. M. Cook. 1995. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 59:48-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sartor, R. B. 1997. The influence of the normal microbial flora on the development of chronic mucosal inflammation. Res. Immunol. 148:567-576. [DOI] [PubMed] [Google Scholar]
- 30.Satokari, R. M., E. E. Vaughan, A. D. L. Akkermans, M. Sareela, and W. M. De Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smart, J. B., V. L. Crow, and T. D. Thomas. 1985. Lactose hydrolysis in milk and whey using β-galactosidase from Streptococcus thermophilus. N. Z. J. Dairy Sci. Technol. 20:43-56. [Google Scholar]
- 32.Spanhaak, S., R. Havenaar, and G. Schaafsma. 1998. The effect of consumption of milk fermented by Lactobacillus casei strain Shirota on the intestinal microflora and immune parameters in humans. Eur. J. Clin. Nutr. 52:899-907. [DOI] [PubMed] [Google Scholar]
- 33.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuohy, K. M., S. Kolida, A. M. Lustenberger, and G. R. Gibson. 2001. The prebiotic effects of biscuits containing partially hydrolysed guar gum and fructo-oligosaccharides — a human volunteer study. Br. J. Nutr. 86:341-348. [DOI] [PubMed] [Google Scholar]
- 35.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoetendal, E. G., A. D. L. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoretic analysis of 16S rRNA from fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]