Abstract
Bacteriocins of the LlpA family have previously been characterized in the γ-proteobacteria Pseudomonas and Xanthomonas. These proteins are composed of two MMBL (monocot mannose-binding lectin) domains, a module predominantly and abundantly found in lectins from monocot plants. Genes encoding four different types of LlpA-like proteins were identified in genomes from strains belonging to the Burkholderia cepacia complex (Bcc) and the Burkholderia pseudomallei group. A selected recombinant LlpA-like protein from the human isolate Burkholderia cenocepacia AU1054 displayed narrow-spectrum genus-specific antibacterial activity, thus representing the first functionally characterized bacteriocin within this β-proteobacterial genus. Strain-specific killing was confined to other members of the Bcc, with mostly Burkholderia ambifaria strains being susceptible. In addition to killing planktonic cells, this bacteriocin also acted as an antibiofilm agent.
Bacteriocins mediate highly selective antagonism among closely related bacteria but such antimicrobial proteins have not yet been reported in Burkholderia. We identified a lectin-like protein of the LlpA family in a Burkholderia cenocepacia human isolate that strain-specifically and selectively kills planktonic and biofilm cells of other Burkholderia cepacia complex members.
Keywords: Antagonism, Burkholderia cepacia complex, lectin-like bacteriocin, LlpA, MMBL family, planktonic, sessile cells
Introduction
While some members of the β-proteobacterial genus Burkholderia exhibit attractive properties for biodegradation of environmental pollutants or growth promotion of plants (Suárez-Moreno et al. 2012), several species represent a threat to animal and human health. The Burkholderia pseudomallei group includes the causative agents of human melioidosis, B. pseudomallei, and of animal glanders, Burkholderia mallei (Galyov et al. 2010). The Burkholderia cepacia complex (Bcc), encompassing 17 species, is home to opportunistic pathogens, such as Burkholderia multivorans and Burkholderia cenocepacia, that cause respiratory infections in cystic fibrosis patients and immunocompromised individuals (Sousa et al. 2011; Vial et al. 2011; Suárez-Moreno et al. 2012). Bcc bacteria are difficult to combat due to high intrinsic antibiotic and biocide resistance, biofilm-forming behavior, and prevalence of multidrug-resistant strains (Horsley and Jones 2012).
A possible strategy to devise alternative anti-Burkholderia strategies is to exploit the antibacterial activity of molecules involved in competition among Burkholderia strains and the potentially novel molecular targets involved (Chandler et al. 2012). Production of the polyketide enacyloxins by Burkholderia ambifaria AMMD enables inhibition of some other Bcc species (Burkholderia dolosa, B. multivorans) (Mahenthiralingam et al. 2011). Certain Bcc species (Burkholderia ubonensis, Burkholderia vietnamiensis) are susceptible to capistruin, a lasso peptide ribosomally synthesized by Burkholderia thailandensis E264 (a member of the B. pseudomallei group) that most strongly inhibits the plant-associated Burkholderia caledonica (Knappe et al. 2008). Recently, contact-dependent inhibition systems mediating competition among Burkholderia strains were characterized in B. pseudomallei and B. thailandensis (Anderson et al. 2012; Nikolakakis et al. 2012). The role of bacteriocin-mediated antagonism among cystic fibrosis isolates has been investigated by Bakkal et al. (2010). A study of B. pseudomallei antagonism displayed by B. ubonensis provided indications of the production of a pepsin-sensitive bacteriocin-like compound (Marshall et al. 2010). However, antagonistic molecules involved in these interactions have not been identified yet.
Bacteriocins have the potential of selectively killing target cells and some of these molecules deserve further scrutiny as candidate alternative antibacterials (Brown et al. 2012; Lukacik et al. 2012; Riley et al. 2012; Cotter et al. 2013). Here, we report on the bacteriocin activity of a lectin-like protein encoded in the genome of a B. cenocepacia human isolate.
MATERIALS AND METHODS
Strains and culture conditions
Bacterial strains and plasmids used in this study are listed in Table S1. Bordetella, Escherichia coli, and Ralstonia were routinely grown in shaken LB broth (MP Biomedicals, Brussels, Belgium) at 37°C. Burkholderia strains were grown in LB broth or Tryptic Soy Broth (TSB, BD Biosciences, Erembodegem, Belgium), at 37°C with shaking. Achromobacter, Pseudomonas, and Xanthomonas were grown in TSB, Alcaligenes in 869 medium, Azoarcus in medium 1 LMG, Chromobacterium and Variovorax in LB, and Herbaspirillum in Nutrient Broth, at 30°C with shaking. Alternative media to initiate LlpA production in B. cenocepacia AU1054 are listed in Table S2. Media were solidified with 1.5% agar (Invitrogen, Ghent, Belgium) and supplemented with filter-sterilized kanamycin (Sigma-Aldrich, Diegem, Belgium) at 50 μg/mL when required.
Plasmids used for sequencing were propagated in E. coli TOP10F' (Invitrogen). E. coli BL21 (DE3) (Novagen, Darmstadt, Germany) was taken as a host for recombinant protein expression. Genomic DNA from Burkholderia strains was isolated using the Puregene Yeast/Bact. Kit B (Qiagen, Venlo, Netherlands). Plasmid DNA was extracted using the QIAprep Spin Miniprep Kit (Qiagen). Bacterial stocks were stored at −80°C in the appropriate medium in 25% (v/v) glycerol.
Recombinant DNA methods
Standard methods were used for the preparation of competent E. coli cells and heat shock transformation of E. coli (Green and Sambrook 2012). DNA ligation was performed using T4 DNA ligase (Invitrogen). Restriction enzymes were used according to the supplier's specifications (Roche Diagnostics, Vilvoorde, Belgium). Plasmid sequencing was performed by GATC Biotech (Constance, Germany).
Burkholderia llpA genes were amplified by polymerase chain reaction (PCR) with Platinum Pfx DNA polymerase (Invitrogen), using a C1000 Thermal Cycler (Bio-Rad, Nazareth Eke, Belgium). Genomic DNA from B. cenocepacia AU1054 and B. ambifaria MEX-5 was taken as a template; PCR primers are listed in Table S3. Amplicons were purified using the QIAquick PCR Purification Kit (Qiagen), digested with NdeI and XhoI, ligated in pET28a(+), and transformed to E. coli TOP10F'. Transformants were verified for the presence of insert by PCR using Taq Polymerase (BIOKÉ, Leiden, Netherlands) with primers PGPRB-5104 and PGPRB-5105. Insert confirmed plasmids (pCMPG6192 (Bcen_1091) and pCMPG6196 (Bcen_1092) from B. cenocepacia AU1054; pCMPG6200 (Bamb_0926) from B. ambifaria MEX-5) were purified and checked by sequencing. The three llpA genes were cloned without their predicted N-terminal secretory motifs (SignalP; http://www.cbs.dtu.dk/services/SignalP), with the His6-tag at the N-terminus (Parret et al. 2004; Ghequire et al. 2012a).
Overexpression and purification of recombinant B. cenocepacia LlpA
Induction of expression, lysis of harvested cells by sonication, and protein extraction of N-terminal His-tagged LlpAs from E. coli BL21 (DE3), carrying expression constructs pCMPG6192, pCMPG6196, and pCMPG6200, have been described previously (Parret et al. 2004). Recombinant protein was purified by nickel affinity chromatography, using an Äkta Purifier (GE Healthcare Life Sciences, Amersham Bioscience, Diegem, Belgium) with a 5-mL HisTrap column (Ghequire et al. 2012a).
The presence of His-tagged protein was checked via immunodetection by Western blot, using monoclonal anti-His6 (IgG1 from mouse; Roche Diagnostics) as primary antibody. Fractions free of other proteins, as verified by SDS-PAGE and subsequent Coomassie Blue staining, were dialyzed against bis-TRIS propane buffer (20 mmol/L, 200 mmol/L NaCl, pH 7.0). Concentrations of purified recombinant proteins were determined by absorbance measurement at 280 nm with molar extinction coefficients of 56,505 mol/L−1 cm−1 for Bcen_1091 (with calculated molecular weight 30,566 Da), 58,120 mol/L−1 cm−1 for Bcen_1092 (29,144 Da), and 74,620 mol/L−1 cm−1 for Bamb_0926 (30,046 Da) (Pace et al. 1995). N-terminal amino acid sequences of proteins on a blot were determined by Edman degradation, using a Procise 491 cLC protein sequencer (Applied Biosystems, Foster City, CA).
Bacteriocin assay
Antibacterial activity of purified recombinant His-tagged protein was detected by spot assay (Ghequire et al. 2012a). Briefly, plates were overlaid with 5 mL of soft agar (0.5%) of the appropriate medium, seeded with 50 μL of an indicator culture (16 h, ~108–109 CFU/mL) to generate a cell lawn. After drying, a 10-μL spot of purified recombinant protein (conc. 1 mg/mL) was applied on the plate. Bis-TRIS propane buffer was used as a negative control. Plates were incubated overnight at 30°C or 37°C (depending on the strains tested) and evaluated for the presence of zones of growth inhibition (halos) next day.
Determination of the MIC of LlpA
The minimum inhibitory concentration (MIC) of LlpA was determined in triplicate following the broth microdilution procedure using flat-bottom 96-well microtiter plates (TPP, Trasadingen, Switzerland) (European committee for antimicrobial susceptibility testing (EUCAST) 2003; Brackman et al. 2009). Briefly, an overnight inoculum (TSB medium, 24 h) of B. ambifaria LMG 19182 was diluted to ~5.105 CFU/mL, supplemented with a twofold dilution series of recombinant His6-tagged LlpA (Bcen_1091), and incubated overnight at 37°C (20 h). Bis-TRIS propane buffer was used as a control. The MIC was determined as the minimum concentration of LlpA at which no growth of the indicator strain was observed (OD590 <0.1) after 24 h of incubation.
Determination of the inhibition and eradication of Burkholderia biofilms
The minimal biofilm inhibitory concentration (MBIC) of LlpA was determined by assaying the growth-inhibitory effect on freshly adhered sessile cells (Peeters et al. 2009). In brief, an overnight culture was diluted to ~108 CFU/mL and the suspension was added to the wells of a round-bottomed 96-well microtiter plate (TPP). After 4 h of adhesion, the supernatant was removed and the plates were rinsed with physiological saline (PS, 0.9% NaCl). One hundred microliter volumes of LlpA at selected concentrations (in bis-TRIS propane buffer) were added to the wells and the plates were further incubated at 37°C. After 20 h of treatment, wells were rinsed a final time with PS. Cells were removed from the wells of the plates by vortexing and sonication, and suspended in 100-μL volumes of PS. The number of CFU/biofilm was determined by plating the resulting suspension on Tryptic Soy Agar (TSA). Results were averaged over 8 technical repeats. The MBIC was defined as the minimum concentration of LlpA necessary to prevent biofilm growth and maturation.
To determine the biofilm eradication effect of LlpA, biofilms of B. ambifaria were initially grown in the wells of round-bottomed 96-well plates (Brackman et al. 2009). After formation of the biofilm (24 h), planktonic cells were washed away by rinsing the plates with PS. Subsequently, 100-μL volumes of LlpA at selected concentrations (in bis-TRIS propane buffer) were added to interact with the biofilm, submerged in 100-μL volumes of PS. Plates were incubated for another 24 h and rinsed again with PS. Sessile cells were removed from the wells of the plates by vortexing and sonication, suspended in 100-μL volumes of PS. The number of CFU/biofilm was determined by plating the resulting suspension on TSA. Results were averaged over 16 repeats (2 biological repeats, 8 technical repeats) (Brackman et al. 2011).
Phylogenetic analysis
Putative LlpA-like proteins in Burkholderia spp. were identified using the National Center for Biotechnology (NCBI) non-redundant database, via Blast searches. Sequences of previously identified LlpAs from Pseudomonas and Xanthomonas were used as a query. Multiple sequence alignments and phylogenetic analysis were performed with the Geneious 5.4 software (Drummond et al. 2011). Predicted N-terminal signal peptide sequences (SignalP), if present, were removed before alignment.
Results
Genes encoding LlpA-like proteins in Burkholderia genomes
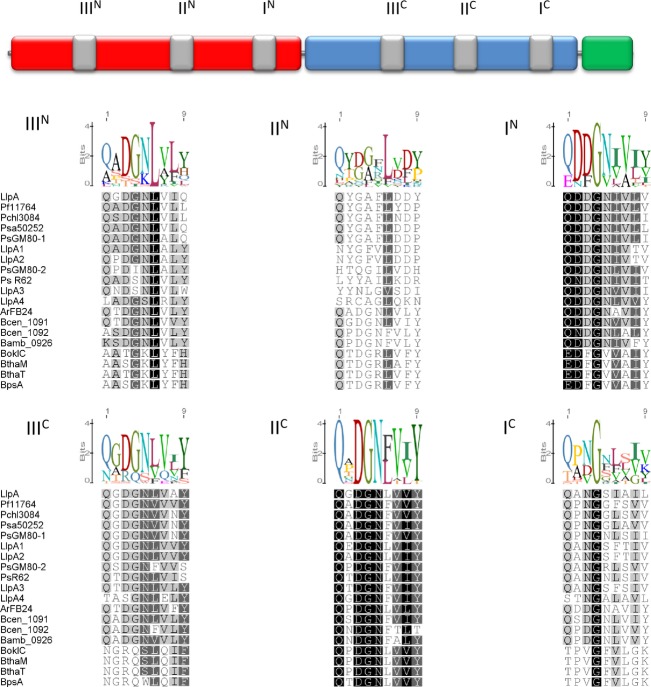
Lectin-like bacteriocins, called LlpAs, represent a novel class of antibacterial proteins identified in the γ-proteobacterial genera Pseudomonas and Xanthomonas (Ghequire et al. 2012b). Being composed of a MMBL (monocot mannose-binding lectin) domain tandem, they show remarkable structural similarities with plant lectins (Ghequire et al. 2013). The prototypical MMBL domain is characterized by the presence of three potential mannose-binding pockets, identifiable by an amino acid stretch corresponding to the motif QxDxNxVxY (with x representing any amino acid). Carbohydrate-binding capacity of the C-terminal domain of LlpA is crucial for killing, whereas the N-terminal domain represents the main specificity determinant (Ghequire et al. 2013).
Through homology searches we identified several genes encoding potential new members of the LlpA family in genomic sequences of the genus Burkholderia. Amino acid sequence alignment of the derived gene products with functionally characterized LlpA bacteriocins from pseudomonads and xanthomonads, displayed the characteristic tandem MMBL organization, although the level of amino acid sequence identity with these LlpAs is low (<30%; Fig. S1). The second MMBL domain is followed by an equivalent of the β-hairpin in Pseudomonas putida LlpA (Ghequire et al. 2013), a poorly conserved carboxyterminal extension varying considerably in length (from 24 to 52 amino acids) among different Burkholderia strains. The presence of a typical aminoterminal signal sequence suggests that the Burkholderia proteins are proteolytically processed upon Sec-dependent secretion, similarly to LlpA4 translocation in Xanthomonas but differently from the unknown export route of pseudomonad LlpA proteins, which all lack a cleavable aminoterminal signal peptide (Ghequire et al. 2012b, 2013).
Phylogenetic comparison of the LlpA-like Burkholderia proteins revealed two distinct clusters, with separate grouping of sequences from four Bcc strains (B. ambifaria MEX-5, and B. cenocepacia AU1054, HI2424, and MC0-3) and those from a large number of B. pseudomallei group strains: Burkholderia oklahomensis C6786 (with an orthologue in strain EO147), B. pseudomallei (with orthologues in >45 strains), B. thailandensis MSMB4, and B. thailandensis TXDOH (with orthologues in strains Bt4 and E264) (Fig. 1). As the B. pseudomallei group sequences share high amino acid sequence identity (>87%), essentially four different types of LlpA-like proteins can be distinguished. The three Bcc types share 43–60% mutual identity but show poor homology with the conserved B. pseudomallei group sequences, apart from the presence of MMBL signature motifs (Fig. S1; Fig. 2). In contrast to the B. pseudomallei group coding sequences (~64% G+C), the GC content of the Bcc genes (50–55% G+C) is considerably lower than the respective genomic values (66–67% G+C). An additional indication for acquisition by lateral gene transfer is the nearby presence of a tRNA gene and integrase in the B. cenocepacia genomes.
Figure 1.
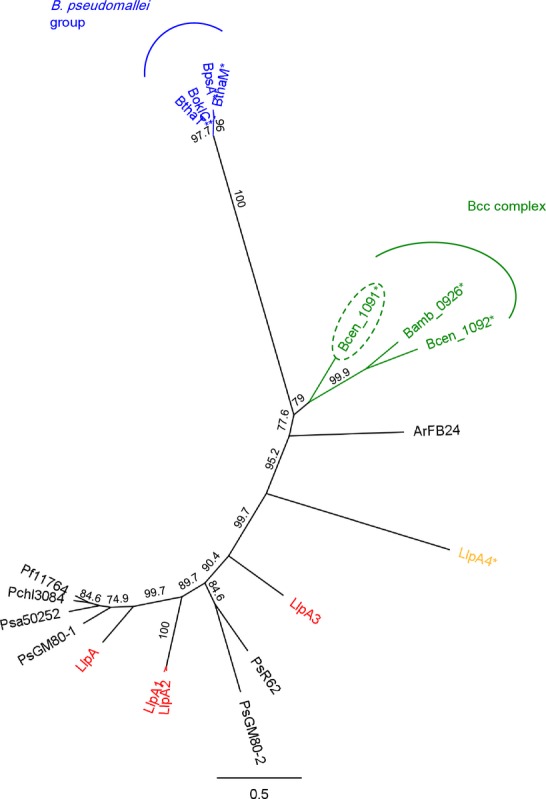
Phylogenetic analysis of LlpA-like proteins. Unrooted maximum-likelihood tree (PhyML; JTT matrix, [Guindon and Gascuel 2003]) inferred from a multiple amino acid alignment (Fig. S1) of bacteriocins from Pseudomonas putida BW11M1 (LlpA, [Parret et al. 2003]), P. fluorescens Pf-5 (LlpA1 = PFL_1229, LlpA2 = PFL_2127 [P. fluorescens Wayne1, CADW01000205], [Parret et al. 2005]), P. syringae pv. syringae 642 (LlpA3, [Ghequire et al. 2012a]), and Xanthomonas axonopodis pv. citri str. 306/Xanthomonas citri pv. malvacearum LMG 761 (LlpA4 [X. axonopodis pv. malvacearum GSPB1386, AHIB01000114], [Ghequire et al. 2012a]), with hypothetical proteins from Arthrobacter sp. FB24 (ArFB24, YP_829274), Burkholderia ambifaria MEX-5 (Bamb_0926, ZP_02905572), B. cenocepacia AU1054 (Bcen_1091, ABF75998; Bcen_1092, ABF75999 [strains MC0-3, HI2424]), B. oklahomensis C6786 (BoklC, ZP_02366769 [strain EO147], B. pseudomallei 1710a (BpsA, ZP_04953366 [46 strains]), B. thailandensis TXDOH (BthaT, ZP_02369690 [strains Bt4, E264]), and B. thailandensis MSMB43 (BthaM, ZP_02467384), Pseudomonas chlororaphis subsp. aureofaciens 30-84 (Pchl3084, ZP_18874430), P. fluorescens NCIMB 11764 (Pf11764, ALWP01000740), P. syringae pv. aptata DSM 50252 (Psa50252, EGH77666), Pseudomonas sp. GM80 (PsGM80-1, ZP_10606046; PsGM80-2, ZP_10606131), and Pseudomonas sp. R62 (PsR62, AHZM01000533). Orthologues (99–100% amino acid identity) of the listed proteins are mentioned in square brackets (sequences not included in Fig. S1 and tree). Before alignment, predicted N-terminal signal peptide sequences (SignalP), if present, were removed (names marked with asterisk). Underlined accession numbers represent unannotated nucleotide sequences from unfinished microbial genomes encoding LlpA-like proteins identified by Blast searches. The scale bar represents 0.5 substitutions per site. Bootstrap values (percentage of 1000 replicates) are shown at the branches. Values lower than 50 are not displayed. LlpA from B. cenocepacia AU1054, subject of this study, is indicated by a dotted ellipse. Previously characterized LlpAs from γ-proteobacteria are indicated in red (Pseudomonas) and orange (Xanthomonas). Nodes of LlpA-like proteins from Burkholderia belonging to the Bcc complex are colored in green and those from the B. pseudomallei group in blue.
Figure 2.
General domain structure of LlpA proteins with potential carbohydrate-binding motifs (gray). The N-domain is colored red, the C-domain blue, and the C-terminal extension green. The respective potential mannose-binding motifs corresponding to the consensus motif QxDxNxVxY in LlpA-like proteins (derived from sequence alignment in Fig. S1) are aligned. The protein codes and accession numbers are specified in Figure 1. Sequence conservation is visualized by differential shading and the sequence logo graph visualizes the degree of consensus for each residue.
The phylogenetic tree further shows that the Bcc LlpA-like protein sequences are more similar to the bacteriocins from γ-proteobacteria than to the proteins found in the B. pseudomallei group. This is also apparent from the relative sequence conservation of the QxDxNxVxY motif corresponding to the six potential carbohydrate-binding pockets, three in each MMBL domain (Fig. 2). Best conserved across all known sequences, including those of the B. pseudomallei group, is pocket IIC for which an additive role in bacteriocin toxicity of P. putida LlpA was demonstrated (Ghequire et al. 2013). However, unlike the Bcc proteins, the B. pseudomallei group sequences lack motif conservation equivalent to the crucial carbohydrate-binding site IIIC in P. putida LlpA. A similar dichotomy among the Burkholderia sequences is evident for three LlpA sites lacking carbohydrate-binding capacity (IIIN, IN, IC). Whereas the canonical mannose-binding motif IIN is essentially absent in pseudomonad LlpAs, this stretch is well conserved in the burkholderiad sequences. These observations suggest that carbohydrate-binding properties of the Burkholderia LlpA-like proteins may differ between both groups, but also diverge from the P. putida prototype LlpA.
Bacteriocin activity of B. cenocepacia LlpA
To enable testing of the potential antibacterial activity of the Bcc LlpA-like proteins, heterologous expression in E. coli was attempted. The B. cenocepacia AU1054 genes encoding Bcen_1091 and Bcen_1092, and Bamb_0926 from B. ambifaria MEX-5 were cloned into pET28a(+) with an N-terminal His6-tag immediately fused to the mature protein, lacking the respective predicted signal peptides for Sec-dependent secretion (Fig. S1). Detectable expression could only be achieved for Bcen_1091 (Fig. 3A). The protein was purified by Ni-NTA affinity chromatography (Fig. 3A) and its identity was confirmed by N-terminal sequencing and Western Blotting with anti-His6 antibodies. The Burkholderia LlpA was found to be cross-reactive with antibodies raised against LlpABW11M1 from P. putida (Fig. 3B; Parret et al. 2005). The purified LlpA showed no detectable binding to glycan array slides (Fig. S2, Blixt et al. 2004). This was also observed for Pseudomonas LlpAs (Ghequire et al. 2012a, 2013; M. Ghequire and R. De Mot, unpubl. data) and may be due to the absence of appropriate prokaryotic target carbohydrates.
Figure 3.
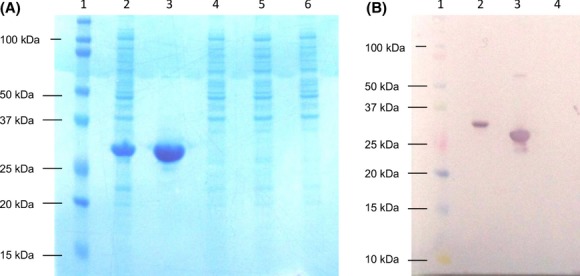
Purification and activity of recombinant Burkholderia cenocepacia AU1054 LlpA. (A) SDS-PAGE analysis of His-tagged Bcen_1091 expression and purification. Proteins present in the lysate soluble fraction of Escherichia coli BL21 (DE3) cells carrying the specified plasmids. Lane 1: Kaleidoscope protein ladder; lane 2, lysate of pCMPG6192 (Bcen_1091 from B. cenocepacia AU1054); lane 3, purified LlpA (Bcen_1091 from B. cenocepacia AU1054); lane 4, lysate of pCMPG6196 (Bcen_1092 from B. cenocepacia AU1054); lane 5, lysate of pCMPG6200 (Bamb_0926 from B. ambifaria MEX-5); and lane 6, lysate of pET28a(+) (no insert). (B) Western Blot analysis with anti-LlpA antibodies. Lane 1: Kaleidoscope protein ladder; lane 2: 2.5 ng of purified Pseudomonas LlpA (Parret et al. 2004); lane 3: 2.5 ng of purified Burkholderia LlpA; lane 4: 2.5 ng of bovine serum albumin (negative control).
Bacteriocin activity was assayed by applying spots of pure protein on indicator-seeded agar plates (Ghequire et al. 2012a) to visualize growth inhibition as a halo formed in a confluent lawn of bacterial target cells. The test panel included representative members of the Bcc complex and clinical isolates belonging to the B. pseudomallei group (Table 1). In addition, a set of other β-proteobacteria and several γ-proteobacteria (Pseudomonas and Xanthomonas spp.) were tested (Table S1). Antibacterial activity was only observed against strains belonging to the Bcc complex (16.7% of strains in the test panel). Whereas none of the B. pseudomallei group strains and only one of the B. cenocepacia strains proved susceptible, several B. ambifaria strains were found to be sensitive to the LlpA-like protein. Such patchy strain-dependent pattern of sensitivity is in line with the activity spectra of the corresponding Pseudomonas and Xanthomonas bacteriocins (Parret et al. 2003, 2005; Ghequire et al. 2012a). Other β-proteobacteria were not susceptible, nor were the tested γ-proteobacteria affected, confirming that this Bcc LlpA is a narrow-spectrum genus-specific bacteriocin that acts across species borders.
Antibacterial activity of purified recombinant LlpA (Bcen_1091 from Burkholderia cenocepacia AU1054) against a panel of Burkholderia strains
| Indicator strain | Growth inhibition by LlpA | Indicator strain | Growth inhibition by LlpA | ||
|---|---|---|---|---|---|
| Bcc complex | B. seminalis | LMG 24272 | − | ||
| B. ambifaria | LMG 17828 | + | B. stabilis | LMG 14294 | − |
| LMG 17829 | + | LMG 14086 | − | ||
| LMG 19182 | + | B. ubonensis | LMG 20358 | − | |
| LMG 19466 | + | LMG 24263 | − | ||
| LMG 19467 | − | B. vietnamensis | LMG 18835 | − | |
| LMG 26702 | − | LMG 10927 | − | ||
| B. anthina | LMG 20980 | + | LMG 10929 | − | |
| LMG 20983 | − | ||||
| B. arboris | LMG 24066 | − | B. Pseudomallei group | ||
| R-132 | − | B. mallei | NCTC 120 | − | |
| B. cenocepacia | LMG 6986 | − | B. mallei | NCTC 3708 | − |
| LMG 16656 | − | B. mallei | NCTC 3709 | − | |
| LMG 16659 | − | B. mallei | NCTC 10229 | − | |
| LMG 18826 | − | B. mallei | NCTC 10230 | − | |
| LMG 18827 | − | B. mallei | NCTC 10245 | − | |
| LMG 18828 | − | B. mallei | NCTC 10247 | − | |
| LMG 18829 | − | B. mallei | NCTC 10248 | − | |
| LMG 18830 | − | B. mallei | NCTC 10260 | − | |
| LMG 18863 | − | B. pseudomallei | ATCC11668 | − | |
| LMG 19230 | + | B. pseudomallei | Bengla 01 | − | |
| LMG 21461 | − | B. pseudomallei | ID 1476 | − | |
| LMG 21462 | − | B. pseudomallei | NCTC 1688 | − | |
| B. cepacia | LMG 1222 | − | B. pseudomallei | NCTC 4845 | − |
| LMG 18821 | − | B. pseudomallei | NCTC 4846 | − | |
| B. contaminans | LMG 16227 | − | B. pseudomallei | NCTC 6700 | − |
| R-12710 | + | B. pseudomallei | NCTC 7383 | − | |
| B. diffusa | LMG 24065 | − | B. pseudomallei | NCTC 7431 | − |
| LMG 24266 | − | B. pseudomallei | NCTC 8016 | − | |
| B. dolosa | LMG 18941 | − | B. pseudomallei | NCTC 8707 | − |
| LMG 18943 | − | B. pseudomallei | NCTC 8708 | − | |
| B. lata | LMG 6992 | − | B. pseudomallei | NCTC 10274 | − |
| R-9940 | − | B. pseudomallei | NCTC 10276 | − | |
| B. latens | LMG 24064 | − | B. pseudomallei | NCTC 11642 | − |
| R-11768 | − | B. pseudomallei | UCl 467 | − | |
| B. metallica | LMG 24068 | + | B. thailandensis | CIP 106301 | − |
| R-2712 | − | B. thailandensis | CIP 106302 | − | |
| B. multivorans | LMG 18825 | − | |||
| LMG 13010 | − | Other strains | |||
| B. pyrrocinia | LMG 14191 | − | B. glumae | LMG 2196 | − |
| LMG 21824 | − | B. gladioli | LMG 2216 | − | |
| B. seminalis | LMG 24067 | − | B. plantarii | LMG 9035 | − |
Antibiofilm activity of B. cenocepacia LlpA
The MIC for planktonic cells was determined using a microdilution assay. For B. ambifaria LMG 19182, a representative strain from the indicator panel, a MIC value of 7.0 μg/mL (0.23 μmol/L) was obtained. Previously identified compounds-mediating antagonism between Burkholderia species, enacyloxin IIa and capistruin, displayed MIC values of 9.3 and 12 μmol/L, respectively (Knappe et al. 2008; Mahenthiralingam et al. 2011), 40- to 50-fold higher than LlpA.
Possible antibiofilm activity of LlpA against strain LMG 19182 was studied by monitoring the biofilm inhibitory and eradicatory effects at super- and sub-MIC concentrations. Treatment at 5.8 μmol/L LlpA (25 × MIC) at the onset of biofilm formation (biofilm inhibition) resulted in a 52% reduction of the biofilm, whereas at 58 nmol/L LlpA (0.25 × MIC) a 36% reduction could be observed. Addition of LlpA to a mature biofilm (biofilm eradication) reduced CFU count by 62% (with a standard deviation of 24%) when LlpA was applied at a concentration of 5.8 μmol/L. No CFU reduction was observed at 58 nmol/L LlpA.
Discussion
Whereas numerous bacteriocins from other Proteobacteria, such as E. coli and Pseudomonas, have been characterized, this article constitutes the first description of such a protein-mediating antagonism among members of the Burkholderia genus. This lectin-like protein from B. cenocepacia AU1054 displays an intrinsic killing activity significantly higher than the previously identified Burkholderia molecules with intragenus toxicity, the polyketide enacyloxin IIa, and the lasso peptide capistruin. Notably, the B. cenocepacia protein was also active against sessile cells growing in a biofilm. Gram-negative bacteriocins exerting killing activity on biofilm cells have also been described for Pseudomonas aeruginosa (Smith et al. 2012) and Citrobacter freundii (Shanks et al. 2012). As observed for several other bacteriocins from Gram-negative bacteria, the susceptibility to this B. cenocepacia protein is highly strain dependent, making it not readily suitable as an alternative for antibiotics. As strain selectivity and killing activity appear to be linked with different parts of the LlpA protein (Ghequire et al. 2013), insight in its mode of action may nevertheless reveal alternative targets for killing bacteria. As the Burkholderia LlpA-like proteins differ from the pseudomonad LlpAs in relative sequence conservation of the potential carbohydrate-binding pockets present in both MMBL domains, determinants of toxicity and selectivity may also deviate from those identified in the prototype P. putida LlpA (Ghequire et al. 2013).
This type of antibacterial protein likely plays a role in the social life of Burkholderia strains in their natural environment by mediating antagonism among strains colonizing similar niches. Expression of the tandemly organized llpA-like genes from human isolate B. cenocepacia AU1054 and soil isolate B. cenocepacia HI2424, both belonging to the PHDC lineage (LiPuma et al. 2002), was observed by RNA-seq, both in conditions mimicking a soil environment and in synthetic cystic fibrosis sputum medium (Yoder-Himes et al. 2009). However, our attempts to trigger detectable bacteriocin production on different media, including rhizosphere soil extract media (Table S2), were not successful (data not shown).
Among bacteriocins, LlpA takes a unique position by virtue of its lectin-like properties. A number of “genuine” lectins with different carbohydrate-binding specificities have been identified in B. cenocepacia (Lameignere et al. 2010; Sulák et al. 2011) and B. ambifaria (Audfray et al. 2012). A possible role in biofilm formation has been proposed for them (Inhülsen et al. 2012), but they have no known antimicrobial activities. Conceivably, the more distantly related MMBL-like proteins from the B. pseudomallei group representatives may have evolved toward a similar biofilm-supporting function by promoting physical interbacterial interaction rather than antagonism. Compared to the Bcc cluster proteins which harbor bacteriocin activity, those of the B. pseudomallei group show little sequence divergence, have conserved a different set of potential carbohydrate pockets, and are encoded by genes lacking characteristics of (recent) acquisition, suggesting a more general, possibly virulence-related physiological role, although the latter remains speculative at this point.
Acknowledgments
The authors wish to thank J. Tiedje (Microbiology and Molecular Genetics, Michigan State University) for kindly supplying strain B. ambifaria MEX-5, and P. Proost (Rega Institute for Medical Research, KU Leuven) for N-terminal amino acid sequence determination. The Protein-Glycan Interaction Resource of the Consortium for Functional Glycomics (grant GM098791) is acknowledged for glycan array analysis of LlpA.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Table S1. List of strains and plasmids used in this study.
Table S2. List of bacterial growth media tested to trigger LlpA production in B. cenocepacia AU1054. Media were solidified with 1.5% agar. Overlay assays with LlpA-sensitive bacteria after chloroform killing of B. cenocepacia AU1054 were done in LB soft agar (0.5% agar). Media were also tested in coculture, in the latter case cell lawns of indicator bacteria were made in soft agar, after which AU1054 was spotted on top. In parallel, the different growth media were also tested when B. cenocepacia AU1054 was supplemented with mitomycin (1 μg/mL final concentration) prior to spotting. Plates were scored after overnight incubation at 37°C. Generated halos were not pronase sensitive, and not thought to be bacteriocin derived.
Table S3. List of primers used in this study.
Figure S1. Multiple amino acid sequence alignment used to construct the phylogenetic tree of LlpA homologues (Fig. ). The protein codes and accession numbers are specified in Figure 1. Sequence conservation is visualized by differential shading. The position of the sequences corresponding to QxDxNxVxY-like motifs of MMBL proteins, representing potential carbohydrate-binding pockets, and the separate domains (N- and C-domain with MMBL fold; β-hairpin extension) present in P. putida BW11M1 LlpA (PDB 3M7H) are indicated.
Figure S2. Glycan array profile of LlpA (Bcen_1091 from B. cenocepacia AU1054) as measured by fluorescence intensity. A complete list of tested carbohydrates (array version PA v5) is available from the Consortium of Functional Glycomics (CFG, www.functionalglycomics.org).
References
- Anderson MS, Garcia EC, Cotter PA. The Burkholderia bcpAIOB genes define unique classes of two-partner secretion and contact dependent growth inhibition systems. PLoS Genet. 2012;8:e1002877. doi: 10.1371/journal.pgen.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audfray A, Claudinon J, Abounit S, Ruvoën-Clouet N, Larson G, Smith DF, et al. Fucose-binding lectin from opportunistic pathogen Burkholderia ambifaria binds to both plant and human oligosaccharidic epitopes. J. Biol. Chem. 2012;287:4335–4347. doi: 10.1074/jbc.M111.314831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkal S, Robinson SM, Ordonez CL, Waltz DA, Riley MA. Role of bacteriocins in mediating interactions of bacterial isolates taken from cystic fibrosis patients. Microbiology. 2010;156(Pt 7):2058–2067. doi: 10.1099/mic.0.036848-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackman G, Hillaert U, Nelis S, van Calenbergh HJ, Coenye T. Use of quorum sensing inhibitors to interfere with biofilm formation and development in Burkholderia multivorans and Burkholderia cenocepacia. Res. Microbiol. 2009;160:144–151. doi: 10.1016/j.resmic.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 2011;55:2655–2661. doi: 10.1128/AAC.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CL, Smith K, McCaughey L, Walker D. Colicin-like bacteriocins as novel therapeutic agents for the treatment of chronic biofilm-mediated infection. Biochem. Soc. Trans. 2012;40:1549–1552. doi: 10.1042/BST20120241. [DOI] [PubMed] [Google Scholar]
- Chandler JR, Heilmann S, Mittler JE, Greenberg EP. Acyl-homoserine lactone-dependent eavesdropping promotes competition in a laboratory co-culture model. ISME J. 2012;6:2219–2228. doi: 10.1038/ismej.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Ross RP, Hill C. Bacteriocins – a viable alternative to antibiotics? Nat. Rev. Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, et al. 2011. Geneious v5.4. Available at http://www.geneious.com (accessed 21 March 2013)
- European committee for antimicrobial susceptibility testing (EUCAST) Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. 2003;9:ix–xv. [Google Scholar]
- Galyov EE, Brett PJ, DeShazer D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu. Rev. Microbiol. 2010;64:495–517. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- Ghequire MGK, Li W, Proost P, Loris R, de Mot R. Plant lectin-like antibacterial proteins from phytopathogens Pseudomonas syringae and Xanthomonas citri. Environ. Microbiol. Rep. 2012a;4:373–380. doi: 10.1111/j.1758-2229.2012.00331.x. [DOI] [PubMed] [Google Scholar]
- Ghequire MGK, Loris R, de Mot R. MMBL proteins: from lectin to bacteriocin. Biochem. Soc. Trans. 2012b;40:1553–1559. doi: 10.1042/BST20120170. [DOI] [PubMed] [Google Scholar]
- Ghequire MGK, Garcia-Pino A, Lebbe EKM, Spaepen S, Loris R, de Mot R. Structural determinants for activity and specificity of the bacterial toxin LlpA. PLoS Pathog. 2013;9:e1003199. doi: 10.1371/journal.ppat.1003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Sambrook JR. Molecular cloning: a laboratory manual. 4th ed. New York: Cold Spring Harbor Laboratory Press; 2012. p. 2028. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Horsley A, Jones AM. Antibiotic treatment for Burkholderia cepacia complex in people with cystic fibrosis experiencing a pulmonary exacerbation. Cochrane Database Syst. Rev. 2012;10:CD009529. doi: 10.1002/14651858.CD009529.pub2. doi: 10.1002/14651858.CD009529.pub2. [DOI] [PubMed] [Google Scholar]
- Inhülsen S, Aguilar C, Schmid N, Suppiger A, Riedel K, Eberl L. Identification of functions linking quorum sensing with biofilm formation in Burkholderia cenocepacia H111. Microbiologyopen. 2012;1:225–242. doi: 10.1002/mbo3.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe TA, Linne U, Zirah S, Rebuffat S, Xie X, Marahiel MA. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J. Am. Chem. Soc. 2008;130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- Lameignere E, Shiao TC, Roy R, Wimmerova M, Dubreuil F, Varrot A, et al. Structural basis of the affinity for oligomannosides and analogs displayed by BC2L-A, a Burkholderia cenocepacia soluble lectin. Glycobiology. 2010;20:87–98. doi: 10.1093/glycob/cwp151. [DOI] [PubMed] [Google Scholar]
- LiPuma JJ, Spilker T, Coenye T, Gonzalez CF. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet. 2002;359:2002–2003. doi: 10.1016/S0140-6736(02)08836-0. [DOI] [PubMed] [Google Scholar]
- Lukacik P, Barnard TJ, Buchanan SK. Using a bacteriocin structure to engineer a phage lysin that targets Yersinia pestis. Biochem. Soc. Trans. 2012;40:1503–1506. doi: 10.1042/BST20120209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E, Song L, Sass A, White J, Wilmot C, Marchbank A, et al. Enacyloxins are products of an unusual hybrid modular polyketide synthase encoded by a cryptic Burkholderia ambifaria genomic island. Chem. Biol. 2011;18:665–677. doi: 10.1016/j.chembiol.2011.01.020. [DOI] [PubMed] [Google Scholar]
- Marshall K, Shakya S, Greenhill AR, Padilla G, Baker A, Warner JM. Antibiosis of Burkholderia ubonensis against Burkholderia pseudomallei, the causative agent for melioidosis. Southeast Asian J. Trop. Med. Public Health. 2010;41:904–912. [PubMed] [Google Scholar]
- Nikolakakis K, Amber S, Wilbur JS, Diner EJ, Aoki SK, Poole SJ, et al. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol. Microbiol. 2012;84:516–529. doi: 10.1111/j.1365-2958.2012.08039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parret AHA, Schoofs G, Proost P, de Mot R. Plant lectin-like bacteriocin from a rhizosphere-colonizing Pseudomonas isolate. J. Bacteriol. 2003;185:897–908. doi: 10.1128/JB.185.3.897-908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parret AHA, Wyns L, De Mot R, Loris R. Overexpression, purification and crystallization of bacteriocin LlpA from Pseudomonas sp. BW11M1. Acta Crystallogr. D Biol. Crystallogr. 2004;60(Pt 10):1922–1924. doi: 10.1107/S0907444904020153. [DOI] [PubMed] [Google Scholar]
- Parret AHA, Temmerman K, de Mot R. Novel lectin-like bacteriocins of biocontrol strain Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 2005;71:5197–5207. doi: 10.1128/AEM.71.9.5197-5207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters E, Nelis HJ, Coenye T. In vitro activity of ceftazidime, ciprofloxacin, meropenem, minocycline, tobramycin and trimethoprim/sulfamethoxazole against planktonic and sessile Burkholderia cepacia complex bacteria. J. Antimicrob. Chemother. 2009;64:801–809. doi: 10.1093/jac/dkp253. [DOI] [PubMed] [Google Scholar]
- Riley MA, Robinson SM, Roy CM, Dennis M, Liu V, Dorit RL. Resistance is futile: the bacteriocin model for addressing the antibiotic resistance challenge. Biochem. Soc. Trans. 2012;40:1438–1442. doi: 10.1042/BST20120179. [DOI] [PubMed] [Google Scholar]
- Shanks RM, Dashiff A, Alster JS, Kadouri DE. Isolation and identification of a bacteriocin with antibacterial and antibiofilm activity from Citrobacter freundii. Arch. Microbiol. 2012;194:575–587. doi: 10.1007/s00203-012-0793-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Martin L, Rinaldi A, Rajendran R, Ramage G, Walker D. Activity of pyocin S2 against Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2012;56:1599–1601. doi: 10.1128/AAC.05714-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa SA, Ramos CG, Leitão JH. Burkholderia cepacia complex: emerging multihost pathogens equipped with a wide range of virulence factors and determinants. Int. J. Microbiol. 2011;2011:607575. doi: 10.1155/2011/607575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonça-Previato L, James EK, Venturi V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 2012;63:249–266. doi: 10.1007/s00248-011-9929-1. [DOI] [PubMed] [Google Scholar]
- Sulák O, Cioci G, Lameignère E, Balloy V, Round A, Gutsche I, et al. Burkholderia cenocepacia BC2L-C is a super lectin with dual specificity and proinflammatory activity. PLoS Pathog. 2011;7:e1002238. doi: 10.1371/journal.ppat.1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial L, Chapalain A, Groleau MC, Déziel E. The various lifestyles of the Burkholderia cepacia complex species: a tribute to adaptation. Environ. Microbiol. 2011;13:1–12. doi: 10.1111/j.1462-2920.2010.02343.x. [DOI] [PubMed] [Google Scholar]
- Yoder-Himes DR, Chain PS, Zhu Y, Wurtzel O, Rubin EM, Tiedje JM, et al. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc. Natl. Acad. Sci. USA. 2009;106:3976–3981. doi: 10.1073/pnas.0813403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of strains and plasmids used in this study.
Table S2. List of bacterial growth media tested to trigger LlpA production in B. cenocepacia AU1054. Media were solidified with 1.5% agar. Overlay assays with LlpA-sensitive bacteria after chloroform killing of B. cenocepacia AU1054 were done in LB soft agar (0.5% agar). Media were also tested in coculture, in the latter case cell lawns of indicator bacteria were made in soft agar, after which AU1054 was spotted on top. In parallel, the different growth media were also tested when B. cenocepacia AU1054 was supplemented with mitomycin (1 μg/mL final concentration) prior to spotting. Plates were scored after overnight incubation at 37°C. Generated halos were not pronase sensitive, and not thought to be bacteriocin derived.
Table S3. List of primers used in this study.
Figure S1. Multiple amino acid sequence alignment used to construct the phylogenetic tree of LlpA homologues (Fig. ). The protein codes and accession numbers are specified in Figure 1. Sequence conservation is visualized by differential shading. The position of the sequences corresponding to QxDxNxVxY-like motifs of MMBL proteins, representing potential carbohydrate-binding pockets, and the separate domains (N- and C-domain with MMBL fold; β-hairpin extension) present in P. putida BW11M1 LlpA (PDB 3M7H) are indicated.
Figure S2. Glycan array profile of LlpA (Bcen_1091 from B. cenocepacia AU1054) as measured by fluorescence intensity. A complete list of tested carbohydrates (array version PA v5) is available from the Consortium of Functional Glycomics (CFG, www.functionalglycomics.org).