Abstract
Spx of Bacillus subtilis is a redox-sensitive protein, which, under disulfide stress, interacts with RNA polymerase to activate genes required for maintaining thiol homeostasis. Spx orthologs are highly conserved among low %GC Gram-positive bacteria, and often exist in multiple paralogous forms. In this study, we used B. anthracis Sterne, which harbors two paralogous spx genes, spxA1 and spxA2, to examine the phenotypes of spx null mutations and to identify the genes regulated by each Spx paralog. Cells devoid of spxA1 were sensitive to diamide and hydrogen peroxide, while the spxA1 spoxA2 double mutant was hypersensitive to the thiol-specific oxidant, diamide. Bacillus anthracis Sterne strains expressing spxA1DD or spxA2DD alleles encoding protease-resistant products were used in microarray and quantitative real-time polymerase chain reaction (RT-qPCR) analyses in order to uncover genes under SpxA1, SpxA2, or SpxA1/SpxA2 control. Comparison of transcriptomes identified many genes that were upregulated when either SpxA1DD or SpxA2DD was produced, but several genes were uncovered whose transcript levels increased in only one of the two SpxADD-expression strains, suggesting that each Spx paralog governs a unique regulon. Among genes that were upregulated were those encoding orthologs of proteins that are specifically involved in maintaining intracellular thiol homeostasis or alleviating oxidative stress. Some of these genes have important roles in B. anthracis pathogenesis, and a large number of upregulated hypothetical genes have no homology outside of the B. cereus/thuringiensis group. Microarray and RT-qPCR analyses also unveiled a regulatory link that exists between the two spx paralogous genes. The data indicate that spxA1 and spxA2 are transcriptional regulators involved in relieving disulfide stress but also control a set of genes whose products function in other cellular processes.
Bacillus anthracis harbors two paralogs of the global transcriptional regulator of stress response, SpxA. SpxA1 and SpxA2 contribute to disulfide stress tolerance, but only SpxA1 functions in resistance to peroxide. Transcriptome analysis uncovered potential SpxA1 and SpxA2 regulon members, which include genes activated by both paralogs. However, paralog-specific gene activation was also observed. Genes encoding glutamate racemase, CoA disulfide reductase, and products functioning in bacillithiol biosynthesis, are among the genes activated by the SpxA paralogs.
Keywords: Bacillus anthracis, oxidative stress, SpxA1, SpxA2, transcriptomic
Introduction
Bacillus anthracis is a spore-forming, nonmotile Gram-positive bacterium that is the causative agent of the zoonotic infectious disease, anthrax (Beyer and Turnbull 2009). It is an effective pathogen because the infectious agent of anthrax is the metabolically dormant and highly resistant spore. Upon ingestion by a professional phagocytic cell (e.g., activated macrophage), the spore undergoes germination and outgrowth to generate a vegetative cell that is capable of reproduction within the infected host, as it produces plasmid-encoded toxins and protective capsule material for evading immune capture and destruction (Fouet et al. 1999; Koehler 2009; Moayeri and Leppla 2009; Tournier et al. 2009). Germination and outgrowth in the macrophage takes place within a hostile environment, made so by the phagocyte's oxidative burst, which generates a toxic combination of reactive oxygen species (ROS), nitric oxide (NO), and hypochlorous acid (HOCl), as well as phospholipase, and antimicrobial peptides (Piris-Gimenez et al. 2005; Passalacqua and Bergman 2006; Passalacqua et al. 2006; Dawson and Liu 2008; Welkos et al. 2011). Successful establishment of infection involves mechanisms of oxidant resistance (Shatalin et al. 2008; Welkos et al. 2011). Such systems in bacteria are activated by encounters with a variety of toxic agents, not only components of the oxidative burst, but antibiotics and other chemical and physical insults (Gusarov et al. 2009; Mols and Abee 2011).
Much of what is known about the oxidative stress response in Bacilli has come from studies of Bacillus subtilis, a nonpathogen, which is a model genetic system used in studying Gram-positive physiology and the bacterial response to harsh conditions. Recent studies of oxidant sensitivity indicated that B. subtilis is more sensitive to the lethal effects of peroxide and superoxide-generating agents than is B. anthracis (Pohl et al. 2011). The findings suggest that more robust processes of oxidant detoxification and tolerance have evolved in the pathogen, which is in keeping with its developmental cycle involving reproduction within phagocytic hosts. Several regulatory proteins govern the oxidative stress response in B. subtilis, including the peroxide sensor PerR (Lee and Helmann 2006), organic hydroperoxide-sensing MarR family protein, OhrR (Fuangthong et al. 2001), HypR, which senses thiol-reactive HOCl stress (Palm et al. 2012), and SpxA (Zuber 2004). All of these proteins have orthologs in B. anthracis (PerR [BA0537], HypR [BA3379], OhrR [BA4699]). Both species possess the general stress response sigma subunit, σB, which controls a large regulon that becomes activated by starvation and reduced energy-generating capability, as well as by stress brought about through encounters with toxic chemical and physical agents (Hecker et al. 2007; van Schaik et al. 2007). Peroxide induces the σB regulon in B. subtilis, but σB is poorly activated by peroxide stress in B. anthracis, which is likely due to the different regulatory architectures that operate in the two organisms (Pohl et al. 2011; Tu et al. 2012b). The response of B. anthracis to superoxide stress, which is likely encountered within the infected macrophage, is the elevated expression of genes within the Fur (Ferric uptake regulator) regulon specifying iron uptake mechanisms, which is also observed in B. subtilis (Mostertz et al. 2004; Passalacqua et al. 2007; Tu et al. 2012a). The response seems maladaptive as it would expose macromolecules to potentially damaging, hydroxyl radicals generated by the Fenton reaction (Liochev and Fridovich 1999; Imlay 2008). Superoxide is known to cause decomposition of enzyme iron centers, which could trigger an iron starvation response through inactivation of the Fur transcriptional regulator and stimulation of the Fur regulon (Mostertz et al. 2004; Passalacqua et al. 2007). However, it has been proposed that superoxide is a germination signal for B. anthracis spores (Fisher and Hanna 2005), and accelerated iron uptake during subsequent outgrowth may assist in coping with the iron-poor environment that characterizes the infected host. In contrast to B. subtilis, the response of B. anthracis to superoxide is limited involving around 40 genes, which might reflect the signaling role of superoxide in B. anthracis infection rather than an agent of general stress generation (Tu et al. 2012b).
Peroxide stress induces over 200 genes in B. anthracis that specify a variety of activities related to detoxification, macromolecular damage repair, and disposal of damaged protein (Pohl et al. 2011). Thus, genes encoding DNA repair enzymes, and other members of the LexA regulon, are induced. Genes functioning in redox homeostasis, including those encoding components of the bacillithiol biosynthesis pathway, are also activated. A significant change in the B. anthracis transcriptome is evident from the observation that several genes encoding regulatory proteins are activated after peroxide treatment. These include genes specifying the PerR and SpxA transcriptional regulators (Bergman et al. 2007; Passalacqua et al. 2007; Pohl et al. 2011).
SpxA is a global regulator of the stress response that is activated upon thiol stress. Over 250 genes, or 144 operons, are controlled by SpxA in B. subtilis (Nakano et al. 2003; Rochat et al. 2012). The protein is a direct transcriptional activator through an interaction with the RNA polymerase alpha subunit in B. subtilis (Nakano et al. 2005; Newberry et al. 2005). Among the genes activated by SpxA are those required to establish the reduced state of thiols in the cytoplasm. Such genes include those that encode thioredoxin (trxA), thioredoxin reductase (trxB), methionine sulfoxide reductase (You et al. 2008), enzymes required for synthesis of the low-molecular-weight thiol bacillithiol (Gaballa et al. 2010; Chi et al. 2011; Gaballa and Helmann 2011), and genes that function in cysteine biosynthesis (Nakano et al. 2003; Choi et al. 2006; Zuber et al. 2011; Rochat et al. 2012). A null mutation in spx causes sensitivity to thiol reactive compounds, partial cysteine auxotrophy, and causes disruption of iron uptake control. The spx gene is under complex transcriptional control that is responsive to stress caused by variety of physical and chemical agents (Helmann et al. 2001; Petersohn et al. 2001; Jervis et al. 2007; Leelakriangsak et al. 2007; Eiamphungporn and Helmann 2008). In B. subtilis and in other Gram-positive species, spxA is transcriptionally induced by mechanisms responsive to cell envelope stress. SpxA can undergo stress-induced thiol oxidation of a CxxC disulfide center, which is necessary for its productive interaction with RNA polymerase (Nakano et al. 2005). The SpxA protein is under proteolytic control that requires the ATP-dependent protease, ClpXP, and a substrate recognition factor, YjbH (Larsson et al. 2007; Garg et al. 2009). SpxA, as might be expected given its role in regulating the oxidative stress response, has been found to be required for virulence in Streptococci and Enterococcus (Kajfasz et al. 2010, 2012; Chen et al. 2012).
Several of the low %GC Gram-positive bacteria possess multiple paralogs of Spx (Veiga et al. 2007; Turlan et al. 2009). The genome of B. anthracis bears two spxA genes, spxA1 and spxA2 (Fig. 1A). The spxA1-linked genes show syntenic similarity to those in B. subtilis, but B. anthracis also contains an additional paralogous spx gene in a part of the genome that shows no synteny with B. subtilis (Fig. 1B). Previous transcriptomic studies have shown that spxA1 is expressed in early log phase of a B. anthracis culture, while spxA2 transcript is detected during stationary phase (Bergman et al. 2006). The spxA2 gene is one of the most highly induced genes in B. anthracis cells following germination in the host macrophage (Bergman et al. 2007). The differences in expression patterns exhibited by the two paralogous genes suggest differences in their roles within the stress response network of B. anthracis.
Figure 1.
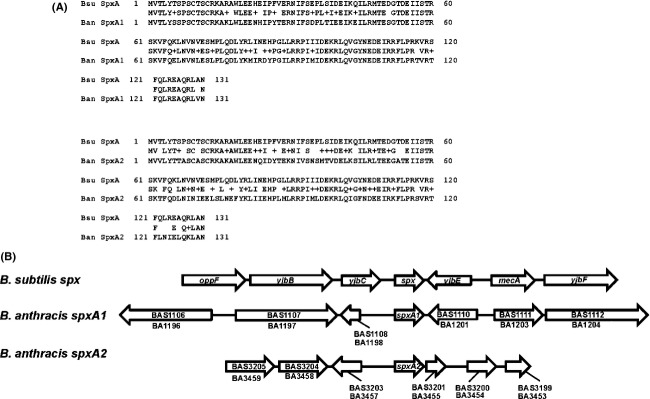
Bacillus anthracis paralogs, SpxA1 and SpxA2, are orthologs of B. subtilis Spx. (A) Comparison of the primary structures of SpxA1 and SpxA2 in B. anthracis with Spx in B. subtilis. ClpXP protease-resistant forms of Spx, SpxA1DD and SpxA2DD, were constructed by replacing the two C-terminal residues with DD. (B) Synteny between the paralogous spx genes in B. anthracis and spx in B. subtilis. Bacillus anthracis Sterne nomenclature is used. BAS1110 and BAS1111 encode, respectively, YjbE and MecA orthologs. Also shown are the gene designations according to the B. anthracis Ames nomenclature.
To uncover the roles of the two paralogous genes of spxA in B. anthracis, a study was conducted to examine the phenotype conferred by null mutations in the genes encoding the two paralogs and to identify the genes that are regulated by SpxA1 and SpxA2. The work reported herein shows that the two paralogous SpxA proteins oversee two large overlapping regulons. While both function in the oxidative stress response, SpxA1 plays an essential role in the bacterium's resistance to peroxide and disulfide stress.
Experimental Procedures
Bacterial strains and growth conditions
Bacterial strains and plasmids are listed in Table 1. The B. anthracis Sterne strains used in this study are derivatives of 7702 (pXO1+ pXO2−) and, in most cases, were grown in Lysogeny Broth (LB) medium at 37°C or sporulation medium (SM) at 30°C (Barua et al. 2009). The B. subtilis strains used in this study are derivatives of JH642 (trpC2 pheA1) and were grown at 37°C in Difco sporulation medium (DSM) unless otherwise indicated. Escherichia coli DH5α was used for plasmid construction and was grown at 37°C in LB liquid or on LB solid medium containing 1.2% agar (Difco, BD Biosciences, San Jose, CA). Appropriate antibiotics were added: B. subtilis, 75 μg/mL spectinomycin, 1 μg/mL erythromycin/25 μg/mL lincomycin, 5 μg/mL kanamycin; B. anthracis, 100 μg/mL streptomycin, 100 μg/mL spectinomycin, 20 μg/mL kanamycin; E. coli, 100 μg/mL ampicillin, 100 μg/mL spectinomycin, 20 μg/mL kanamycin.
Bacterial strains and plasmids used in this study
| Strain | Genotype | Derivation | Antibiotic resistance | Source | ||||
|---|---|---|---|---|---|---|---|---|
| Bacillus anthracis Sterne 7702 derivative strains | ||||||||
| B. anthracis Sterne 7702 | Parent | None | Pasteur Institute Cataldi et al. (1990) | |||||
| B. anthracis Sterne 7702 SR1 | Streptomycin resistant | Streptomycin-resistant isolate of B. anthracis Sterne 7702 | Strep | This study | ||||
| ORB7863 | ICEBs1::Pspank(hy)-spxA1DD | ICEBs1::pCSZ35 (Pspank(hy)-spxA1DD) × B. anthracis Sterne 7702 SR1 | Spec, Strep | This study | ||||
| ORB7864 | ICEBs1::Pspank(hy)-spxA2DD | ICEBs1::pCSZ36 (Pspank(hy)-spxA2DD) × B. anthracis Sterne 7702 SR1 | Spec, Strep | This study | ||||
| ORB8092 | ICEBs1 | pJMA402 (ICEBs1) × B. anthracis Sterne 7702 SR1 | Spec, Strep | This study | ||||
| ORB8115 | ΔspxA2 | pSB3 (ΔspxA2) × B. anthracis Sterne 7702 SR1 | Strep | This study | ||||
| ORB8170 | ΔspxA1 | pSB2 (ΔspxA1) × B. anthracis Sterne 7702 | None | This study | ||||
| ORB8285 | ΔspxA1 ΔspxA2 | pSB3 (ΔspxA2) × ORB8170 | None | This study | ||||
| ORB8390 | ΔspxA2 ICEBs1::Pspank(hy)-spxA1DD | ICEBs1::pCSZ35 (Pspank(hy)-spxA1DD) × ORB8115 | Spec, Strep | This study | ||||
| ORB8391 | ΔspxA2 ICEBs1::Pspank(hy)-spxA2DD | ICEBs1::pCSZ36 (Pspank(hy)-spxA2DD) × ORB8115 | Spec, Strep | This study | ||||
| ORB8398 | ΔspxA1 | Streptomycin-resistant isolate of ORB8170 | Strep | This study | ||||
| ORB8404 | ΔspxA1 ICEBs1::Pspank(hy)-spxA1DD | ICEBs1::pCSZ35 (Pspank(hy)-spxA1DD) × ORB8170 | Spec, Strep | This study | ||||
| ORB8405 | ΔspxA1 ICEBs1::Pspank(hy)-spxA2DD | ICEBs1::pCSZ36 (Pspank(hy)-spxA2DD) × ORB8170 | Spec, Strep | This study | ||||
| ORB8438 | ΔspxA2 | pSB3 (ΔspxA2) × B. anthracis Sterne 7702 | None | This study | ||||
| ORB8481 | ΔspxA1 ΔspxA2 | Streptomycin-resistant isolate of ORB8285 | Strep | This study | ||||
| ORB8485 | ΔspxA1 ΔspxA2 ICEBs1::Pspank(hy)-spxA1DD | ICEBs1::pCSZ35 (Pspank(hy)-spxA1DD) × ORB8481 | Spec, Strep | This study | ||||
| ORB8486 | ΔspxA1 ΔspxA2 ICEBs1::Pspank(hy)-spxA2DD | ICEBs1::pCSZ36 (Pspank(hy)-spxA2DD) × ORB8481 | Spec, Strep | This study | ||||
| Bacillus subtilis JH642 (trpC2 pheA1) derivative strains | ||||||||
| JH642 | Parent | None | J. Hoch | |||||
| JMA475 | cgcD::Pspank(hy)-rapI kan458 | Kan | A. Grossman Auchtung et al. (2005) | |||||
| ORB3834 | Δspx::neo | Δspx::neo × JH642 | Neo | Nakano et al. (2001) | ||||
| ORB7262 | Δspx::neo thrC::PBA5387-lacZ | thrC::pDYR9 (PBA5387-lacZ) × ORB8384 | Neo, Erm | This study | ||||
| ORB7854 | ICEBs1::Pspank(hy)-spxA1DD | ICEBs1::pCSZ35 (Pspank(hy)-spxA1DD) × JH642 | Spec | This study | ||||
| ORB7860 | ICEBs1::Pspank(hy)-spxA2DD | ICEBs1::pCSZ36 (Pspank(hy)-spxA2DD) × JH642 | Spec | This study | ||||
| ORB7861 | ICEBs1::Pspank(hy)-spxA1DD cgcD::Pspankhy-rapI kan458 | JMA475 genomic DNA × ORB7854 | Spec, Kan | This study | ||||
| ORB7862 | ICEBs1::Pspank(hy)-spxA2DD cgcD::Pspank(hy)-rapI kan458 | JMA475 genomic DNA × ORB7860 | Spec, Kan | This study | ||||
| ORB7871 | Δspx thrC::PBA5387-lacZ amyE::Pspank(hy)-spxA1DD | amyE::pMMN818 (Pspank(hy)-spxA1DD) × ORB7262 | Neo, Erm, Spec | This study | ||||
| ORB7872 | Δspx thrC::PBA5387-lacZ amyE::Pspank(hy)-spxA2DD | amyE::pMMN819 (Pspank(hy)-spxA2DD) × ORB7262 | Neo, Erm, Spec | This study | ||||
| ORB8343 | Δspx::neo thrC::PBA0847-lacZ | thrC::PBA0847-lacZ × ORB3834 | Neo, Erm | This study | ||||
| ORB8344 | Δspx::neo thrC::PBA1119-lacZ | thrC::PBA1119-lacZ × ORB3834 | Neo, Erm | This study | ||||
| ORB8354 | Δspx::neo thrC::PBA1951-lacZ | thrC::PBA1951-lacZ × ORB3834 | Neo, Erm | This study | ||||
| ORB8356 | Δspx::neo thrC::PBA5387-lacZ | thrC::PBA5387-lacZ × ORB3834 | Neo, Erm | This study | ||||
| ORB8359 | Δspx::neo thrC::PBA0847-lacZ amyE::Pspank(hy)-spxA1DD | amyE:: Pspank(hy)-spxA1DD × ORB8343 | Neo, Erm, Spec | This study | ||||
| ORB8360 | Δspx::neo thrC::PBA0847-lacZ amyE:: Pspank(hy)-spxA2DD | amyE:: Pspank(hy)-spxA2DD × ORB8343 | Neo, Erm, Spec | This study | ||||
| ORB8361 | Δspx::neo thrC::PBA1119-lacZ amyE:: Pspank(hy)-spxA1DD | amyE:: Pspank(hy)-spxA1DD × ORB8344 | Neo, Erm, Spec | This study | ||||
| ORB8362 | Δspx::neo thrC::PBA1119-lacZ amyE:: Pspank(hy)-spxA2DD | amyE:: Pspank(hy)-spxA2DD × ORB8344 | Neo, Erm, Spec | This study | ||||
| ORB8363 | Δspx::neo thrC::PBA1951-lacZ amyE:: Pspank(hy)-spxA1DD | amyE:: Pspank(hy)-spxA1DD × ORB8354 | Neo, Erm, Spec | This study | ||||
| ORB8364 | Δspx::neo thrC::PBA1951-lacZ amyE:: Pspank(hy)-spxA2DD | amyE:: Pspank(hy)-spxA2DD × ORB8354 | Neo, Erm, Spec | This study | ||||
| ORB8367 | Δspx::neo thrC::PBA5387-lacZ amyE:: Pspank(hy)-spxA1DD | amyE:: Pspank(hy)-spxA1DD × ORB8356 | Neo, Erm, Spec | This study | ||||
| ORB8368 | Δspx::neo thrC::PBA5387-lacZ amyE:: Pspank(hy)-spxA2DD | amyE:: Pspank(hy)-spxA2DD × ORB8356 | Neo, Erm, Spec | This study | ||||
| ORB8373 | Δspx::neo thrC::PBA1200-lacZ | thrC::PBA1200-lacZ × ORB3834 | Neo, Erm | This study | ||||
| ORB8380 | Δspx::neo thrC::PspxA1-lacZ amyE:: Pspank(hy)-spxA1DD | amyE:: Pspank(hy)-spxA1DD × ORB8373 | Neo, Erm, Spec | This study | ||||
| ORB8381 | Δspx::neo thrC::PBA1200-lacZ amyE:: Pspank(hy)-spxA2DD | amyE:: Pspank(hy)-spxA2DD × ORB8373 | Neo, Erm, Spec | This study | ||||
| ORB8389 | thrC::PBA3868-lacZ | thrC::PBA3868-lacZ × ORB3834 | Neo, Erm | This study | ||||
| ORB8396 | thrC::PBA3868-lacZ amyE:: Pspank(hy)-spxA2DD | amyE:: Pspank(hy)-spxA2DD × ORB8389 | Neo, Erm, Spec | This study | ||||
| ORB8397 | thrC::PBA3868-lacZ amyE:: Pspank(hy)-spxA1DD | amyE:: Pspank(hy)-spxA1DD × ORB8389 | Neo, Erm, Spec | This study | ||||
| Plasmid | Genotype | Selection | ||||||
| pCSZ35 | pJMA402::Pspank(hy)-spxA1DD | Spec | ||||||
| pCSZ36 | pJMA402::Pspank(hy)-spxA2DD | Spec | ||||||
| pSB2 | pRP1028::ΔspxA1 | Spec | ||||||
| pSB3 | pRP1028::ΔspxA2 | Spec | ||||||
| pSB10 | pPROEX-1::spxA2 | Amp | ||||||
| pMMN818 | pDR111::Pspank(hy)-spxA1DD | Spec | ||||||
| pMMN819 | pDR111::Pspank(hy)-spxA2DD | Spec | ||||||
| pDG793 | thrC integration vector with promoter-less lacZ reporter | Erm Guerout-Fleury et al. (1996) | ||||||
| pSS1827 | Conjugation helper strain | Amp | ||||||
| pSS4332 | Harbors I-SceI gene and cyan fluorescent protein reporter construct | Kan | ||||||
| pRP1028 | Cloning vector for conjugation | Spec | ||||||
| pJMA402 | Cloning vector for ICEBs1 conjugation | Spec | ||||||
| pDR111 | amyE integration vector with Pspank(hy) promoter | Spec | ||||||
| pPROEX-1 | E. coli expression vector | Amp | ||||||
| pDRY9 | thrC integration plasmid carrying B. subtilis trxB (−115 to +47)-lacZ | Erm | ||||||
Spec, spectinomycin; Strep, streptomycin; Kan, kanamycin; Amp, ampicillin; Erm, erythromycin; Neo, neomycin.
Construction of B. anthracis spx mutants
Two kilobytes DNA amplicons containing 1 kb up- and 1 kb downstream flanking DNA regions of either spxA1 or spxA2 were generated by overlapping fusion polymerase chain reaction (PCR) using chromosomal DNA from B. anthracis 7702 (see Table S6 for primer sequences). The DNA amplicons were digested with HindIII and KpnI, purified, and ligated with similarly digested and purified pRP1028 using T4 DNA ligase to construct pSB2 (pRP1028::ΔspxA1) and pSB3 (pRP1028::ΔspxA2). The plasmids containing in-frame deletion mutations of spxA1 and spxA2 were introduced into B. anthracis 7702 (or a derivative 7702 SR1) using a markerless allelic replacement technique developed by Janes and Stibitz (Janes and Stibitz 2006). Escherichia coli DH5α strains harboring pRP1028::ΔspxA1 (pSB2) or pRP1028::ΔspxA2 (pSB3) were conjugated at room temperature for 24 h with B. anthracis Sterne 7702 in the presence of an E. coli helper strain containing pSS1827 on brain-heart infusion (BHI) plates lacking antibiotic. The transconjugate diploid intermediates (Δspx spx+) were conjugated for a second time at 37°C with E. coli strains containing pSS4332 (encoding nuclease I-SceI) and pSS1827. Conjugants were screened for the loss of pRP1028 and were subsequently subjected to several rounds of plating on BHI agar with incubation at 37°C to eliminate plasmid pSS4332. Spectinomycin and kanamycin sensitive isolates were obtained and screened by PCR (using primers that anneal outside of the sequence used in the construction of the mutant) and nucleotide sequence analysis of these PCR products was performed to ensure that the final ΔspxA1 (ORB8170) and ΔspxA2 (ORB8115 and ORB8438) constructs were correct.
Construction of B. anthracis strains that produce ClpXP-insensitive SpxA1 and SpxA2
The spxA1DD and spxA2DD genes were amplified by PCR from B. anthracis 7702 chromosomal DNA using oMN10-535 and oMN10-536 (for spxA1DD) or oMN10-537 and oMN10-528 (for spxA2DD). The PCR products were digested with HindIII and SphI and ligated into pDR111 (Britton et al. 2002) digested with the same enzymes. The resultant plasmids pMMN818 and pMMN819 contain spxA1DD and spxA2DD under the control of an IPTG (isopropyl β–D-1-thiogalactopyranoside)-inducible Pspank(hy) promoter. pMMN818 and pMMN819 were digested with BamHI and were filled-in using T4 DNA polymerase and dNTPs, followed by EcoRI digestion. The released 2.3-kb fragment containing lacI and spxADD was subcloned into pJMA402 (kindly provided by C. Lee and A. D. Grossman), digested with EcoRI and HincII to generate pCSZ35 and pCSZ36. pCSZ35 and pCSZ36 were used to transform B. subtilis JH642 with selection for spectinomycin resistance, resulting in strains ORB7854 and ORB7860, respectively. Donor strains used for ICEBs1-mediated conjugation, namely ORB7861 and ORB7862, were generated by transformation of ORB7854 and ORB7860 with chromosomal DNA isolated from JMA475 with selection for kanamycin resistance. ICEBs1-mediated conjugation was carried out as previously described (Auchtung et al. 2005) using ORB7861 or ORB7862 as a donor and 7702 SR1 as a recipient with selection for both streptomycin and spectinomycin resistance. spxA1DD and spxA2DD in B. anthracis conjugants ORB7863 and ORB7864 were verified by sequence analysis of the PCR product amplified from the chromosomal DNA using oligonucleotides spac-up and spac-down (Table S6).
Culture preparation and RNA extraction
Bacillus anthracis Sterne 7702 SR1 and overexpression derivatives bearing Pspank(hy)-spxA1DD (ORB7863), Pspank(hy)-spxA2DD (ORB7864), and empty vector control strain bearing Pspank(hy)-empty (ORB8092), were grown in 60 mL LB 37°C, 200 rpm until mid-exponential phase (OD600 0.2–0.4) and then split into two equal volumes. One millimoles per liter IPTG was added to one set of cultures and the cultures were incubated for an additional 15 or 45 min, after which the pellets from 10 mL culture samples were harvested by centrifugation (5180g, 4°C, 10 min.), and frozen at −80C until use. For diamide-treated cultures, B. anthracis Sterne 7702 and deletion derivatives, ΔspxA1 (ORB8170) and ΔspxA1 ΔspxA2 (ORB8285), were grown in 40 mL LB at 37°C, 200 rpm until mid-exponential phase (OD600 0.2–0.4) and split into two equal volumes. One mmol/L diamide (freshly prepared, Sigma-Aldrich, St. Louis, MO) was added to one set of cultures and, following incubation for an additional 20 min, were harvested by centrifugation, as above. RNA was purified as previously described (Igo and Losick 1986).The resulting RNA concentrations were measured by ultraviolet spectrophotometry. RNA quality was assessed by measuring the ratio of absorbance at 260 nm to absorbance at 280 nm, as well as by visualization in agarose gels.
Microarray design, data collection, and data analysis
cDNA for microarray experiments were generated by adding 2 μg of total RNA in a mixture containing 6 μg of random hexamers (Life Technologies, Carlsbad, CA), 0.01 mol/L dithiothreitol, an aminoallyl-deoxynucleoside triphosphate mixture containing 25 mmol/L each dATP, dCTP, and dGTP, 15 mmol/L dTTP, and 10 mmol/L amino-allyl-dUTP (aa-dUTP) (Sigma), reaction buffer, and 400 units of SuperScript III reverse transcriptase (RT) (Life Technologies) at 42°C overnight. The RNA template then was hydrolyzed by adding NaOH and ethylenediaminetetraacetic acid to a final concentration of 0.2 and 0.1 mol/L, respectively, and incubating at 70°C for 15 min. Unincorporated aa-dUTP was removed with a Minelute column (Qiagen, Gaithersburg, MD). The probe was eluted with a phosphate elution buffer (4 mmol/L potassium phosphate buffer, pH 8.5, in ultrapure water), dried, and resuspended in 0.1 mol/L sodium carbonate buffer (pH 9.0). To couple the amino-allyl cDNA with fluorescent labels, normal human serum-Cy3 or normal human serum-Cy5 (GE Healthcare Biosciences, Pittsburgh, PA) was added at room temperature for 1 h. Uncoupled label was removed using a Minelute column (Qiagen). Aminosilane-coated slides printed with a set of 5823 oligonucleotides representing all open reading frame sequences of B. anthracis Ames A2012 (www.jcvi.org) were prehybridized in 5× SSC (1× SSC is 0.15 mol/L NaCl plus 0.015 mol/L sodium citrate) (Life Technologies), 0.1% sodiumdodecyl sulfate (SDS), and 1% bovine serum albumin at 42°C for 60 min. The slides then were washed at room temperature with distilled water, dipped in isopropanol, and allowed to dry. Equal volumes of the appropriate Cy3- and Cy5-labeled probes were combined, dried, and then resuspended in a solution of 40% formamide, 5× SSC, and 0.1% SDS. Resuspended probes were heated to 95ºC prior to hybridization. The probe mixture then was added to the microarray slide and allowed to hybridize overnight at 42°C. Hybridized slides were washed sequentially in solutions of 1× SSC-0.2% SDS, 0.1× SSC-0.2% SDS, and 0.1× SSC at room temperature, then dried in air, and scanned with an Axon GenePix 4000 scanner (Molecular Devices, Sunnyvale, CA). All wash buffers were supplemented with 1 mL of 0.1 mol/L dithiothreitol per liter of wash buffer. Individual TIFF images from each channel were analyzed with TIGR Spotfinder (available at [www.tm4.org]). Microarray data were normalized by LOWESS normalization and with in-slide replicate analysis using TM4 software MIDAS (available at www.tm4.org). The array design is available at Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) with accession number GPL10188.
Quantitative real-time polymerase chain reaction analysis
cDNA was prepared from each RNA sample using random primers and Invitrogen SuperScript III RT (Life Technologies) as per the manufacturer's protocol. Triple or quadruple technical replicates were performed for each quantitative real-time polymerase chain reaction (RT-qPCR) assay, from either two (for microarray validation) or three (for diamide treatment) independently isolated RNA samples, in a 96-well plate using an ABI Prism Step-One Plus with Step-One Plus (Life Technologies) Software version 2.0 sequence detection system, an annealing temperature of 58°C, and extension at 72°C for 1 min for 40 cycles. Primer sequences (Table S6) were designed to specifically amplify a 100–250-bp portion of each transcript of interest (see Table S6). The amplification efficiencies were roughly equivalent across all primer sets (E% = 80–110). Control reactions (cDNA reactions lacking RT) were performed to verify that no genomic DNA contamination was present (that is, the threshold cycle [CT] for detection in the control without RT was above 35). Normalization of CT values was done relative to the signal obtained from reactions amplifying a portion of the gatB/Yqey (BA4533) transcript; the expression level of this gene has been shown previously to remain stable across the entire B. anthracis life cycle (Reiter et al. 2011), and the microarray experiments described in this study suggest that its expression levels change less than twofold when increased amounts of SpxA1 or SpxA2 are present (see Tables S1, S2). Fold-changes for microarray validation studies were obtained by taking the ratio of +IPTG/−IPTG samples after normalization to gatB/Yqey. Transcripts (ng) were directly reported for RT-qPCR assays of diamide-treated cultures following normalization to the gatB/Yqey endogenous control.
Promoter-lacZ strain construction and β-galactosidase assay
Amplified promoter regions, generated by primers listed in Table S6, were digested with EcoR1 and BamHI, ligated into pDG793 (harboring the lacZ reporter construct, [Guerout-Fleury et al. 1996]) and propagated by transformation of competent DH5α cells. Plasmids containing the correct insert were introduced by transformation into ORB3834 (B. subtilis JH642 Δspx::neo) with selection for Erm resistance and screening for thrC (threonine auxotrophy). These resulting strains were transformed with pMMN818 (Pspank(hy)-spxA1DD) or pMMN819 (Pspank(hy)-spxA2DD) (see Table 1) to obtain the final strains (e.g., B. subtilis JH642 Δspx::neo [Neo] PexoA-lacZ [Erm] amyE::Pspank(hy)-spxA1DD [Spec]) used in β-galactosidase assays. Strains bearing various promoters (see Table 1) fused to a lacZ-reporter were grown at 37°C overnight on DSM supplemented with appropriate antibiotics. Assays of β-galactosidase in DSM cultures were performed as previously described (Nakano et al. 1998); activity was calculated in Miller units (Miller 1972).
Western blot analysis
Bacillus anthracis Sterne 7702, and derivatives ORB8170 (ΔspxA1), ORB8438 (ΔspxA2), and ORB8285 (ΔspxA1 ΔspxA2), were grown in 40–60 mL of LB liquid medium lacking antibiotic at 37°C with agitation (200 rpm) and culture samples were taken at various time points during vegetative growth (see Fig. 2B). Cell pellets were harvested by centrifugation (5180g, 4°C, 10 min.) and stored until use at −80°C. Cell lysate was prepared by suspending the cell pellet in 200 μL–1 mL phosphate-buffered saline and vortexing with 0.1 mm glass beads for a total of 10 min, resting on ice every 5 min. After brief centrifugation to pellet the beads, the supernatant was analyzed for protein amount (Coomassie protein assay, Pierce) and an equal amount (25 or 30 μg) of cell lysate was separated on a SDS-polyacrylamide gel (15%) and transferred to nitrocellulose membranes. Immunoblotting was carried out in Tris-buffered saline plus 0.05% (v/v) Tween-20 using preabsorbed anti-Spx derived from B. subtilis purified Spx protein (Nakano et al. 2005) and alkaline phosphatase-anti-rabbit conjugated secondary antibody (Sigma) at appropriate dilutions. A representative immunoblot is shown for each experiment in Figures 1, 2, with each experiment done twice. Densitometry obtained with ImageJ (Schneider et al. 2012).
Figure 2.
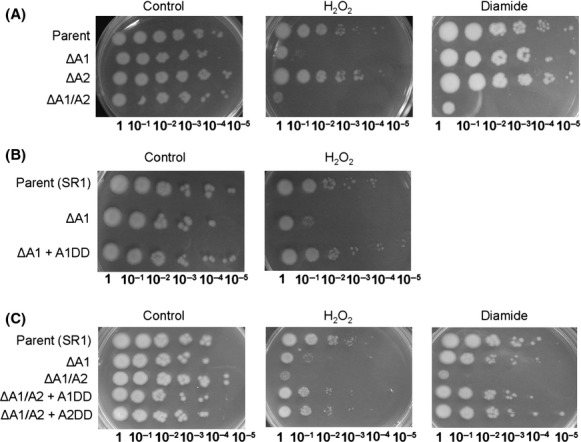
Spx mutants are sensitive to oxidative stress. Strains were grown on LB medium in the presence or absence of 100 μmol/L diamide or 0.44 mmol/L H2O2. Five microliter of the indicated dilutions were spotted onto LB agar (see Experimental Procedures). (A) Strains 7702 (Parent), ORB8170 (∆spxA1, ∆A1), ORB8438 (∆spxA2, ∆A2), ORB8285 (∆spxA1 ∆spxA2, ∆A1/A2). (B) 7702 StrR (Parent [SR1]), ORB8398 (∆spxA1 StrR, ∆A1), ORB8404 (∆spxA1 StrR ICEBs1::spxA1DD, ∆A1 +A1DD). (C) 7702 StrR (Parent [SR1]), ORB8398 (∆spxA1 StrR, ∆A1), ORB8481 (∆spxA1 ∆spxA2 StrR, ∆A1/A2), ORB8485 (∆spxA1 ∆spxA2 StrR ICEBs1::spxA1DD, ∆A1/A2 +A1DD), ORB8486 (∆spxA1 ∆spxA2 StrR ICEBs1::spxA2DD, ∆A1/A2 +A2DD). StrR, streptomycin resistance.
Phenotype testing
A fresh colony grown on LB agar was used to inoculate an overnight LB culture (37°C, 200 rpm). Ten microliter of the overnight cultures were subcultured into 2 mL LB and grown (37°C, 200 rpm) until OD600 reached around 1.0. Five microliter of 10-fold serial dilutions (in T-base) of the cultures were spotted onto LB agar unsupplemented or supplemented with either 100 μmol/L diamide, 1.5 × 10−3% (0.44 mmol/L) hydrogen peroxide, or 0.001% deoxycholate. Sensitivity was checked after overnight incubation (37°C) of the agar plates.
Microscopy
Two microliter aliquots of each spore culture grown for 18 h in SM broth were added to a precleaned glass slide, a coverslip was added, and cells were subsequently examined using a Leica DMIL Inverted Contrasting Microscope (Leica, Buffalo Grove, IL) equipped with a Leica HI Plan I 40× (numerical aperture, 0.5). Images were captured using a Leica DFC295 digital color camera and processed with Leica Software Application Suite V3.8.0.
Results and Discussion
Phenotypic analyses
Construction of Spx protease resistant and Δspx mutants
Low levels of Spx are normally maintained in B. subtilis in part through posttranslational control involving degradation catalyzed by ClpXP and mediated by an adaptor protein, YjbH (Larsson et al. 2007; Garg et al. 2009). This mechanism can be averted in B. subtilis by substituting the last two C-terminal residues of SpxA to DD (see sequence alignment Fig. 1A, [Nakano et al. 2003]). We utilized the same method in B. anthracis. The SpxDD-producing strains used in this study were constructed by interspecies conjugation using a recombinant form of the integrative conjugative element ICEBs1 of B. subtilis ([Auchtung et al. 2005], Experimental Procedures) and a streptomycin-resistant variant of B. anthracis 7702 as recipient (7702 SR1). Mutation of SpxA1 and SpxA2 in this manner (SpxA1DD and SpxA2DD) caused accumulation of either protein compared to levels observed in the parental strain (see below in the case of SpxA1). In-frame deletions of both spx genes, ΔspxA1 and ΔspxA2, were also constructed in order to study the phenotypic effects of these two paralogs in B. anthracis. These mutations were constructed using a markerless allelic replacement method developed by Janes and Stibitz ([Janes and Stibitz 2006], Experimental Procedures).
spxA1 is essential for peroxide resistance; both spxA1 and spxA2 participate in disulfide stress tolerance
In B. subtilis, a ΔspxA mutant is sensitive to oxidant stress (Nakano et al. 2003). Therefore, experiments were conducted to determine if a similar phenotype was conferred by B. anthracis spx null mutations. ΔspxA1 (ORB8170), ΔspxA2 (ORB8438), and ΔspxA1 ΔspxA2 (ORB8285) mutant cells were treated with several oxidants that had been shown to trigger a stress response in related bacteria ([Kristoffersen et al. 2007; Rukmana et al. 2009; McLean et al. 2010; Antelmann and Helmann 2011; Tu et al. 2012b]; Experimental Procedures). Either SpxA1 or SpxA2 can function in disulfide-stress tolerance, as only the ΔspxA1 ΔspxA2 (ΔA1/A2 in Fig. 2A and C) double mutant shows growth impairment upon diamide treatment. However, SpxA1 is essential for peroxide resistance, as ΔspxA1 (ΔA1 in Fig. 2A, B, and C) showed significant peroxide sensitivity. Defects in diamide and peroxide resistance could be corrected by introducing either spxA1DD or spxA2DD constructs in trans (+A1DD or +A2DD in Fig. 2C), suggesting that either Spx protein can potentially function in peroxide-induced stress.
Bile salt can induce oxidative stress and disulfide formation in bacteria (Rodriguez-Beltran et al. 2012; Yang et al. 2013). ΔspxA1 and ΔspxA1 ΔspxA2, but not ΔspxA2, mutants were sensitive to the bile salt deoxycholate (Fig. S1).
SpxA1 and SpxA2 differ in transcriptional regulation during disulfide stress
Experiments were conducted to determine if the spxA paralogous genes were induced by disulfide stress, as part of the oxidative stress response in B. anthracis. cDNA was synthesized using RNA harvested from cells that were untreated or treated with 1 mmol/L diamide for 20 min and analyzed by RT-qPCR to determine the relative amount of spxA1 or spxA2 transcript produced. spxA1 or spxA2 transcript amounts were normalized to a portion of the gatB/Yqey (BA4533) transcript, which served as an endogenous control. The gatB/Yqey transcript is stable during the B. anthracis cell cycle (Reiter et al. 2011), and we observed no increase in this transcript in response to elevated SpxA1 or SpxA2 levels (Table 2); however, in the presence of diamide the gatB/Yqey transcript increased more than threefold (1.65 CT). We also found a similar change using the16S gene transcript when used as an endogenous control (data not shown). Although fold-changes are not absolutely comparable in this experiment because of the severe stress applied to the cell by diamide treatment, we believe that general trends in transcript amount could be discerned for both spx paralogs. The level of spxA2 transcript increased 150- to 350-fold after diamide treatment; conversely, the spxA1 transcript amount remained nearly unchanged or decreased slightly (data not shown), suggesting that the observed repression of spxA1 exerted by SpxA2 that was uncovered from transcriptomic analysis (see below) was ongoing during disulfide stress.
Comparison of select transcript fold-changes by microarray and RT-qPCR
| Gene tag | Description | Microarray results1 | RT-qPCR validation2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SpxA1DD | SpxA2DD | SpxA1DD | SpxA2DD | ||||||||
| BA4533 | gatB/Yqey | No change under conditions studied | – | ||||||||
| BA1200 | spxA1 | 3.8 | −3.5 | Not determined | −4.4 | ||||||
| BA1118 | yvrG, sensor histidine kinase of YvrHG two-component system involved in peptidoglycan biosynthesis | 17.9 | No change3 | 2.9 | 1.4 | ||||||
| BA1263 | CoADR homolog | 12.9 | 5.9 | 4.2 | 3.4 | ||||||
| BA0774 | CoADR-RHD (rhodanese homology domain) | 2.3 | 12.8 | 3.7 | 7.9 | ||||||
| BA1951 | Putative oxidoreductase, conserved only in B. cereus/thuringiensis group | 32.6 | 2.9 | 5.1 | 3.5 | ||||||
| BA0847 | racE-1, Glu racemase | No change3 | 15.9 | 4.6 | 7.3 | ||||||
| BA1208 | yjbH, adaptor protein for SpxBsu proteolysis | 8.1 | 6.9 | 10.7 | 6.4 | ||||||
| BA5387 | trxB, thioredoxin reductase | 3.1 | 4.3 | 4.1 | 4.0 | ||||||
Averaged fold-change of three biological replicates (see Experimental Procedures).
Averaged fold-change of two biological replicates, one biological replicate used RNA extracted for microarray experiments and one replicate independently isolated under similar growth conditions (see Experimental Procedures).
No change is a fold-change <2.
Although spxA1 transcription was not stimulated by diamide treatment, the protein concentration might increase if a posttranslational control mechanism similar to that governing Spx in B. subtilis is operating in B. anthracis. The presence of both yjbH and clpXP orthologs in B. anthracis suggests that SpxA1 and SpxA2 might be subject to a similar mechanism of proteolytic control (Larsson et al. 2007; Garg et al. 2009). Therefore, the amount of SpxA1 protein after diamide-induced stress in B. anthracis was examined. The culture extracts of the B. anthracis parent (7702), ΔspxA1 (ORB8170), and ΔspxA2 (ORB8438) grown in LB medium were used in Western blotting experiments with preabsorbed rabbit polyclonal anti-serum raised against B. subtilis SpxA, which cross-reacts only with SpxA1. Although very low levels of SpxA1 protein were observed in the parent strain grown without stress, the amount of SpxA1 increased at 20 min after the addition of 1 mmol/L diamide (57-fold), suggesting that posttranslational, rather than transcriptional, control of spxA1 governs SpxA1 concentration in response to oxidative stress (Fig. 3A). Interestingly, SpxA1 protein levels also increased in the ΔspxA2 (ORB8438) mutant without stress (7.4-fold), but remained unchanged in the ΔspxA2 mutant after diamide treatment (1.2-fold), suggesting that SpxA2-dependent dampening of spxA1 expression likely occurs at the transcriptional level (Fig. 3A).
Figure 3.
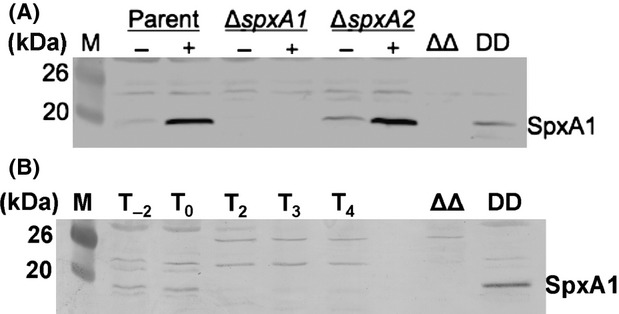
Protein levels of SpxA1 measured by Western blot. (A) SpxA1 levels in cultures grown with and without 1 mmol/L diamide (25 μg total protein applied; ΔΔ, ΔspxA1 ΔspxA2 (ORB8285); DD, Pspank(hy)-spxA1DD (ORB7863) +IPTG 45 min.). (B) Expression profile of SpxA1 during the B. anthracis vegetative life cycle (30 μg total protein loaded; Lanes: T−2 through T4, hours during vegetative growth; T0, transition to stationary phase; ΔΔ, ΔspxA1 ΔspxA2 [ORB8285]; DD, Pspank(hy)-spxA1DD [ORB7863] +IPTG 45 min.). Representative gels are shown in (A) or (B), each experiment was done twice. Growth conditions for obtaining cells for whole cell extracts are presented in Experimental Procedures.
One likely scenario for the increase in B. anthracis SpxA1 and SpxA2 concentration may be during germination and outgrowth, where newly formed vegetative cells would presumably encounter higher levels of ROS. This regulation could extend to spore germination and outgrowth in the oxidizing environment of the lysosome within the host macrophage. In line with this argument, previous transcriptomic studies identified increased spxA1 transcript during spore outgrowth, increased spxA2 transcript during sporulation, and both paralogous transcripts increased inside murine macrophage cells (Bergman et al. 2006, 2007). In keeping with the previous transcriptomic results, we found that SpxA1 was produced during exponential growth and that its concentration declined during early stationary phase (Fig. 3B).
The spike in spxA2 transcriptional activity upon diamide treatment does not correlate well with the contribution of its product toward diamide resistance, as this function is served primarily by SpxA1 based on sensitivity tests (Fig. 2). There are other factors, aside from transcriptional control that can figure prominently in the final level of SpxA2 activity, notably protein stability and redox activation of the Spx protein through its redox disulfide center. How these features of Spx control might differ between the two paralogs is currently not known. Differences in affinity for RNA polymerase binding surface that engages Spx protein, or holoenzyme preference exhibited by the two paralogs might also account for the disparate contributions of SpxA1 and A2 to oxidant resistance. Evidence from studies of B. subtilis suggest that the spx paralog, mgsR is transcriptionally activated by the σB form of RNA polymerase, after which the MgsR protein associates with σB holoenzyme (Reder et al. 2012). However, the finding that each of the B. anthracis spx paralogous genes can complement a ΔspxA mutation in B. subtilis suggests that each product can engage the σA form of RNA polymerase (data not shown).
Together our data suggest that both SpxA1 and spxA2 are upregulated when grown in the presence of the oxidant diamide; but different mechanisms exist for maintaining the level of each paralog in B. anthracis. SpxA1 is likely governed posttranslationally by a mechanism similar to that in B. subtilis; whereas another unknown control mechanism responsive to oxidative stress functions in elevating spxA2 transcript levels.
Increased SpxA1 affects sporulation efficiency in B. anthracis
The requirement for SpxA1 and SpxA2 in germination/outgrowth and sporulation was also examined (Data S1). The results showed that there was no difference in the sporulation, germination, and outgrowth efficiency between the Δspx mutants and the parent strain (7702 SR1), indicating that SpxA1 or SpxA2 are not required for the development or outgrowth from the spore state in B. anthracis (data not shown).
Similar to overexpressing Spx in B. subtilis (Nakano et al. 2001), overexpression of SpxA1DD, but not SpxA2DD, inhibited sporulation to a large extent. Cells containing ICEBs1::Pspank(hy)-spxA1DD (ORB8404) grown in SM the presence of 100 μmol/L IPTG exhibited aberrant cell morphology, compared with the single ΔspxA1 (ORB8170) or parental 7702 strains, at 18 h after inoculation with no visible endospores (Fig. 4), although some colony-forming units (CFUs) were obtained after heat treatment and plating onto LB-agar plates (postheat plated CFU/mL: ORB8404 −IPTG = 2.9 × 107, +IPTG = 2.0 × 106; compared with ORB8170 [7702 ΔspxA1] = 4.4 × 107 and Parent 7702 = 4.9 × 107). At 42 h after inoculation, the aberrant cells had completely lysed, no phase bright spores could be detected by microscopy, and plated CFUs resulted in (5% of the CFUs after heat treatment obtained on minus IPTG culture plates (preheat plated CFU/mL: ORB8404 −IPTG = 1.62 × 108, +IPTG = 3.8 × 106; postheat plated CFU/mL: ORB8404 −IPTG = 1.62 × 108, +IPTG = 2.8 × 106). These results suggest a role for activated Spx in delaying complex developmental process during periods of oxidative stress, a role previously proposed for B. subtilis SpxA (Nakano et al. 2003).
Figure 4.
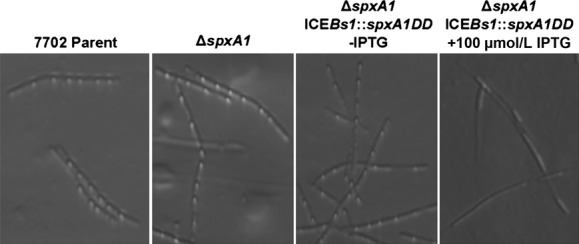
Micrographs of endospore-forming cells containing varying levels of SpxA1 protein after 24 h of growth in SM at 30°C. The phase-bright endospores are readily visualized in the Bacillus anthracis Sterne 7702 Parent and ΔspxA1 mutant (1st and 2nd Panels). In the presence of 100 μmol/L IPTG, the ΔspxA1 ICEBs1::Pspank(hy)-spxA1DD mutant (4th Panel) produced somewhat phase-bright bodies/filaments instead of forming endospores, whereas this strain readily produced endospores similar to the 7702 Parent strain in the absence of IPTG (3rd Panel).
Transcriptomic analyses
Transcriptomic changes in the presence of protease-resistant SpxA1DD or SpxA2DD in B. anthracis
To identify genes potentially regulated by SpxA1 or SpxA2, we compared gene expression profiles between the parent 7702 strain and its isogenic derivatives expressing protease-resistant forms of either Spx paralog (parent vs. ICEBs1::Pspank(hy)-spxA1DD or parent vs. ICEBs1::Pspank(hy)-spxA2DD) using microarray hybridization array analysis. The strains were grown in LB broth and induced by the addition of IPTG for 15 or 45 min prior to culture sampling. Figures 5A and B summarize the overall transcriptional trends from the microarray experiments, and lists of all transcript changes can be found in Tables S1, S2.
Figure 5.

Transcriptomic changes in the presence of SpxA paralogs in Bacillus anthracis. (A) The total number of transcripts that change after induction of SpxA1DD (dark gray bars: A1DD[+]/A1D[−]) or SpxA2DD (light gray bars: A2DD[+]/A2DD[−]) for either 15 or 45 min. Total number of transcripts in each category listed above each bar. (B) Venn diagrams representing the total common transcript changes (≥twofold) observed when either SpxA1DD or SpxA2DD is induced for 45 min. Genes that overlap between the 15- and 45-min time points are considered common genes. A list of up- or downregulated transcripts, grouped by cellular function, can be found in Tables S1, S2. Transcripts were grouped by cellular function based on annotations from Genolist (http://genolist.pasteur.fr) and primary literature sources (Moszer et al. 2002; Pohl et al. 2011). Growth conditions for obtaining total cellular RNA and subsequent microarray experiments are presented in Experimental Procedures.
The expression trends from the microarray transcriptomic analysis underwent validation by (RT-qPCR, Table 2) and by assaying β-galactosidase activity in B. subtilis strains harboring promoter–lacZ transcriptional fusions (Fig. 6). For RT-qPCR, the transcripts were normalized against a portion of the gatB/Yqey (BA4533) transcript (Reiter et al. 2011). The individual transcript values were reported as the averaged fold-change for two biological replicates (Table 2). β-galactosidase assays were performed by constructing lacZ transcriptional fusions to several gene promoters (intergenic regions roughly 200–600 nucleotides upstream of the start codon, extending downstream into the 5′ end of the coding sequence), and integrating these alleles into the chromosome of B. subtilis strains harboring a deletion of the native spxA gene as well as IPTG-inducible versions of B. anthracis spxA1DD or spxA2DD. β-galactosidase activity in the presence or absence of IPTG was then measured in these strains and the ratio of β-galactosidase activity (highest activity/starting time point activity) was plotted for each strain when grown in the presence or absence of IPTG (Fig. 6, see Experimental Procedures).
Figure 6.
SpxA1DD and SpxA2DD activate several Bacillus anthracis genes in B. subtilis. Promoters of selected genes (horizontal axis) controlled by either SpxA1DD or SpxA2DD, as identified by microarray, were transcriptionally fused to a promoterless lacZ and integrated into the chromosome at the thrC locus of a B. subtilis JH642 Δspx strain harboring an IPTG-inducible copy of either spxA1DD or spxA2DD integrated at the amyE locus. β-galactosidase activity was measured in these strains at 30 min. intervals after the addition of 1 mmol/L IPTG. The maximal activity, which was usually observed at 1–2 h after IPTG induction, was divided by the β-galactosidase activity when IPTG was added; except for PspxA1-lacZ, where the minimal activity was divided by the β-galactosidase activity when IPTG was added (SpxA1DD: minus IPTG 0.64 ± 0.16, plus IPTG 0.49 ± 0.2; SpxA2DD: minus IPTG 0.52 ± 0.27, plus IPTG 0.44 ± 0.18). The ratio was shown as the average of three biological triplicates with standard deviation. Significance was determined by a two-tailed T-test comparing plus IPTG to minus IPTG ratios; *P < 0.05 and **P < 0.005 (SpxA1DD: Control [empty vector] P = 0.272, BA0847 [racE-1] P = 0.026, BA1119 [yvrH] P = 0.055, BA5387 [trxB] P = 0.020, BA3868 [exoA] P = 0.0057, BA1951 P = 0.036, BA1263 [coADR] P = 0.005, BA0774 [coADR-RHD] P = 0.108, BA1200 [spxA1] P = 0.463; SpxA2DD: Control [empty vector] P = 0.078, BA0847 [racE-1] P = 0.013, BA1119 [yvrH] P = 0.179, BA5387 [trxB] P = 0.001, BA3868 [exoA] P = 0.015, BA1951 P = 0.032, BA1263 [coADR] P = 0.027, BA0774 [coADR-RHD] P = 0.009, spxA1 P = 0.020).
There were 773 and 699 transcript changes (≥twofold), respectively, when either spxA1DD or spxA2DD was expressed for 15 min. After 15 min of induction, transcript levels of 377 genes were higher and those of 396 genes were lower in the SpxA1DD-producing strain; and transcript levels of 367 genes were higher and 332 genes were lower when SpxA2DD production was induced (Fig. 5A). After 45 min of induction, only 363 transcript changes (≥twofold; 211 up- and 152 downregulated) were noted in SpxA1DD with 306 of those transcripts also found at higher levels after 15 min of induction (Fig. 5A and B). Far more transcripts changed after prolonged SpxA2DD induction (45 min) with 509 total transcript changes (≥twofold; 280 up- and 229 downregulated), while 358 of these also exhibited a change in expression after 15 min of induction (Fig. 5A and B). The total number of upregulated genes between SpxA1DD and SpxA2DD after 45 min of induction was 163, with 110 upregulated and 53 downregulated (Fig. 5B). A list of all transcriptomic changes can be found in Tables S1, S2. Due to the large number of transcripts that showed changes in abundance when either SpxA1DD or SpxA2DD was present, we chose to focus on upregulated transcripts after 15 min of SpxA1DD or SpxA2DD induction for further analysis. Lists of all transcript changes ≥twofold can be found in Tables S1, S2; and lists of genes whose transcripts changed by ≥threefold, grouped by function, can be found in Tables S3, S4.
Many genes having elevated transcript amounts were uncovered in analysis of spxA1DD and spxA2DD transcriptomes, suggesting some overlap in the composition of SpxA1 and SpxA2 regulons. In total, several upregulated genes unique to either SpxA1 or SpxA2 regulons, 184 and 222, respectively, were also identified (Fig. 5B), supporting the hypothesis that each Spx protein has some unique sequence recognition properties (Tables S1, S2) and/or perhaps holoenzyme specificity.
Redox homeostasis
In Bacilli, as in many bacteria, low-molecular-weight thiols are important for maintaining redox homeostasis and alleviating stress caused by toxic oxidants (Newton et al. 1996, 2009). As regulated by orthologous spx in B. subtilis, trxA, trxB, and thioredoxin family protein, ytpP, among others, were also upregulated in the spxA1DD and spxA2DD B. anthracis strains (Nakano et al. 2003; Zuber et al. 2011; Rochat et al. 2012). RT-qPCR and β-galactosidase assays further confirmed a role for both paralogs in the transcriptional control of trxB (Table 2 and Fig. 6).
In several organisms that do not produce glutathione (GSH), coenzyme A (CoASH) can become an important molecule involved in intracellular redox functions (Delcardayre and Davies 1998; Delcardayre et al. 1998). In the presence of SpxA1DD or SpxA2DD, the transcripts specifying two CoASH disulfide reductases, BA1263 (CoADR) and BA0774, which encodes an isoform of CoADR that bears a rhodanese domain (CoADR-RHD), increased >threefold, suggesting that both genes are under the control of the spx paralogs in B. anthracis (Table 2). By RT-qPCR, we observed an increase in BA1263 and BA0774 transcripts with either SpxA1DD or SpxA2DD induction (Table 2); however, in β-galactosidase assays only SpxA2 could activate PBA0774-lacZ, while either paralog activated PBA1263-lacZ (Fig. 6). It may be possible that an activator (missing in B. subtilis) is necessary, in conjunction with SpxA1, to stimulate BA0774 transcription. B. subtilis lacks CoADR and CoADR-RHD isoforms, perhaps utilizing bacillithiol and cysteine instead of CoA in the cytoplasmic redox buffer (Helmann 2011).
The synthesis of bacillithiol has been linked to diamide stress and the spx regulon (Gaballa et al. 2010). Similarly, we found several orthologous genes encoding proteins functioning in the sequential synthesis of UDP-GlcNAc to bacillithiol that are upregulated upon induction of SpxA1DD and SpxA2DD ([Gaballa et al. 2010]; Tables S1, S2). In particular, the transcript for the glycosyltransferase gene, bshA, increased 4.3-fold and 2.5-fold in the SpxA1DD- and SpxA2DD-producing strains, respectively (Tables S1, S2); and both orthologous genes encoding N-acetylhydrolases, bshB1 and yojG, as well as the Cys-adding enzyme, yllA, were also upregulated >twofold in the presence of either SpxA1DD or SpxA2DD (Tables S1, S2), suggesting expression that is governed by SpxA1 and SpxA2 resulting in increased production of bacillithiol in B. anthracis. Additionally, an ortholog of a B. subtilis gene putatively involved in the reduction of oxidized bacillithiol, namely ypdA, encoding a pyridine nucleotide-dependent disulfide oxidoreductase, was upregulated >twofold by either SpxA1DD or SpxA2DD (Tables S1, S2). Two putative bacilliredoxin genes, yphP and ytxJ, also showed elevated transcript levels after SpxA1DD or SpxA2DD production. ytxJ transcript level was increased 2.5-fold by SpxA1DD and yphP 2.3-fold by SpxA2DD (Tables S1, S2).
Several SpxA1DD- and SpxA2DD-activated genes were also observed to be transcriptionally upregulated in peroxide-treated cells (Table S5; [Pohl et al. 2011]), and likely include genes required for peroxide-induced stress tolerance, including (trxB), DNA repair (uvrC), detoxification (nitroreductase, aldehyde dehydrogenase), and export (MATE efflux family protein).
Bacillus anthracis Spx proteins also stimulated the transcription of genes whose products function in overcoming DNA damage, which studies have shown is a direct consequence of oxidative stress brought about by encounters with oxidizing agents such as H2O2 (Imlay and Linn 1988; Imlay et al. 1988). We found that SpxA2DD upregulated BA3868, the ortholog of B. subtilis exoA, an apurinic-apyrimidinic endonuclease important in base excision repair (Ibarra et al. 2008) and genes encoding components of the ABC excinuclease (uvrC and uvrD) (Tables S1, S2) are also upregulated by SpxA1DD and SpxA2DD. Both Spx paralogs were able to activate a PBA3868-lacZ reporter construct in B. subtilis, suggesting that BA3868 is a member of SpxA1 and SpxA2 regulons (Fig. 6). ExoABsu is an important component of spore outgrowth and germination when oxidative DNA damage occurs, presumably when the once dormant spore is suddenly exposed to oxygen upon germinated spore hydration (Ibarra et al. 2008). Bacillus subtilis cells deficient in exoA were significantly delayed in spore outgrowth and increasingly sensitive to H2O2 (Ibarra et al. 2008).
Several genes encoding hypothetical products, many having no homology outside of the B. cereus/B. thuringiensis group, were also upregulated when SpxA1DD or SpxA2DD were produced (Tables S1, S2). We focused on one of these genes, a putative oxidoreductase (BA1951), which in the microarray analysis was induced 32.6-fold in cells expressing SpxA1DD. Cells expressing SpxA2DD had a smaller increase in BA1951 transcript (2.9-fold) (Table 2). The microarray trends were mirrored in β-galactosidase (activation of PBA1951-lacZ by SpxA1DD [P = 0.036] or SpxA2DD [P = 0.032]) and RT-qPCR (5.1-fold, SpxA1DD, 3.5-fold SpxA2DD) assays, although the transcript changes determined by RT-qPCR were lower than transcript changes seen by microarray (Table 2).
Several genes functioning in NAD(P)H-dependent interconversions were also highly upregulated by SpxA1 and SpxA2. These include genes encoding putative alcohol, aldehyde, and quinone dehydrogenases (BA0838, BA2647, and BA3438) for which orthologs exist in B. subtilis (YogA and YdeQ). Interestingly, the B. subtilis orthologous genes are not members of the Spx regulon, suggesting that the SpxA1 and SpxA2 regulons of B. anthracis include members with different metabolic capabilities than those belonging to the B. subtilis Spx regulon. The products of the Spx-activated genes could function in detoxification or generation of reduced NAD(P)H for coping with oxidative stress (Rochat et al. 2012). Taken together these results reinforce the view that SpxA1 and SpxA2 act as transcriptional regulators to activate genes involved in the reduction of oxidized thiols in B. anthracis.
SpxA2 repression of spxA1
Increasing the amount of SpxA2DD led to a 3.5-fold decrease in the spxA1 transcript, suggesting a relationship between SpxA2 and the transcriptional control of spxA1 expression (Table 2). By microarray, the induction of spxA2 was not dependent on SpxA1DD, suggesting that another regulatory mechanism functions in elevating spxA2 transcript levels during oxidative stress.
The spxA1 repression by SpxA2DD was confirmed in three ways: (1) a roughly fourfold decrease in transcript amount was found by RT-qPCR when SpxA2DD was present (Table 2); (2) a roughly twofold increase in spxA1 transcript was detected in a ΔspxA2 background (data not shown); and (3) expression of a PspxA1-lacZ reporter fusion was repressed when SpxA2DD was produced in B. subtilis (Fig. 6). These transcriptomic results parallel the expression results reported above, in which an increase in the SpxA1 protein amount was detected in the ΔspxA2 mutant (Fig. 3A). In conjunction with the large increase in the spxA2 transcript in the presence of diamide stress, it may be that the muted transcriptional response of spxA1 in B. anthracis Sterne 7702 is a consequence of SpxA2-dependent repression, which seems to persist even in the presence of disulfide stress. However, as shown by Western blot experiments, the SpxA1 protein amount increased during diamide stress (Fig. 3), suggesting that the oxidative stress response in B. anthracis includes a posttranscriptional mechanism that oversees SpxA1 accumulation instead of a surge in transcriptional activity that characterizes spxA2 induction. Together, these experiments indicate that SpxA2 negatively affects spxA1 expression.
Cell wall biosynthesis and sporulation
The involvement of SpxA orthologs in control of genes that function in cell wall metabolism has been observed in other Gram-positive bacteria (Prudhomme et al. 2006; Veiga et al. 2007; Suntharalingam et al. 2009; Turlan et al. 2009; Eldholm et al. 2010; Kajfasz et al. 2010). The microarray data suggest that both B. anthracis paralogs can increase expression of genes involved in cell wall or membrane synthesis, remodeling, and maintenance (Tables S1, S2).
Two two-component signal transduction systems specified by the yycFG and BA1119-BA1118 (yvrHG) operons were upregulated in response to increased amounts of SpxA1DD, but not SpxA2DD. Transcription of yycFG increased more than twofold (2.2-fold yycF, 3.5-fold yycG), while yvrGH, showed an even greater transcript increase (17.9-fold yvrG, 12.9-fold yvrH). Upon induction of SpxA1DD, PBA1119-lacZ, encompassing the promoter for the gene encoding the YvrH DNA-binding protein, was activated to nearly significant levels (P = 0.055; Fig. 6). Similarly in RT-qPCR experiments, SpxA1DD increased the BA1118 (yvrG) transcript (2.9-fold), although to lesser amounts than found by microarray. Neither BA1119 promoter activation (Fig. 6) or BA1118 transcript increase (1.4-fold, Table 2) were detected after SpxA2DD induction, suggesting that the yvrHG operon is regulated by SpxA1DD only. Both YycFG and YvrGH are responsible for peptidoglycan biosynthesis and cell wall homeostasis in B. subtilis (Serizawa et al. 2005; Szurmant et al. 2007a). The essential YycFG system of B. subtilis also has two accessory proteins YycH and YycI, which are transcribed as part of the same operon (Szurmant et al. 2005, 2007b) and both yycHI orthologous transcripts in B. anthracis were elevated only when SpxA1DD was produced (3.3-fold yycH, 2.1-fold yycI).
Bacillus anthracis possesses two glutamate racemases, which catalyze the conversion of L-glu to D-glu for the synthesis of poly-γ-glutamate capsule and peptidoglycan (Dodd et al. 2007). One of these glutamate racemase-encoding genes, BA0847 (racE-1), was upregulated in the SpxA2DD-producing strain almost 16-fold by microarray analysis, while its expression did not change when SpxA1DD was overproduced. However, using RT-qPCR, we were able to detect an elevated BA0847 (racE-1) transcript concentration in the SpxA1DD-producing strain (4.6-fold) as well as the expected transcript increase (7.3-fold) by SpxA2DD (Table 2), suggesting that there may be an overestimation of genes uniquely controlled by only one paralogous Spx, as indicated by microarray. A PBA0847-lacZ transcriptional fusion was also activated in the presence of SpxA2DD or SpxA1DD (Fig. 6), confirming our RT-qPCR results.
Production of SpxA2DD also increased the level of transcripts encoded by three sporulation genes, spoIIID, spoIIIE, and rsfA, whose gene products are involved in sporulation-specific transcriptional regulation and DNA translocation (Kunkel et al. 1989; Bath et al. 2000; Wang et al. 2006). As the ΔspxA2 mutant does not seem to have altered sporulation efficiency or outgrowth patterns, the increased transcript amount of these sporulation-specific genes after SpxA2DD induction may be an indirect effect. A more detailed study of spxA2 is underway.
SpxA-activated gene expression during disulfide stress
We chose to further characterize two genes regulated by both Spx paralogs and putatively involved in thiol homeostasis, BA5387 (trxB) and BA1951 (a putative oxidoreductase), by measuring their transcript levels after diamide stress (1 mmol/L) for 20 min (Fig. 7; Experimental Procedures). The levels of the two transcripts were normalized to the gatB/Yqey (BA4533) transcript as described above. Without diamide treatment, gatB/Yqey transcript levels remained unchanged. However, in the presence of diamide the gatB/Yqey transcript increased more than threefold in the parent and roughly eightfold in the ΔspxA1 ΔspxA2 mutant. As BA1951 and BA5387 transcripts increased by at least 80-fold when diamide was applied, we believe that general comparisons between untreated and treated parent and ΔspxA1 ΔspxA2 transcripts could reasonably be made.
Figure 7.
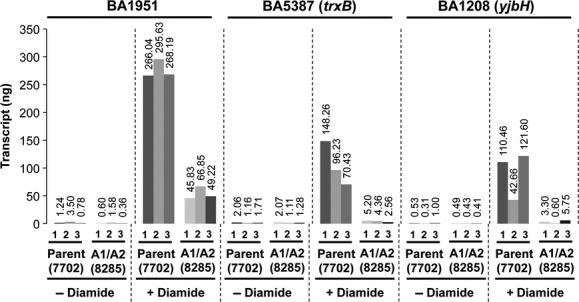
SpxA1 and SpxA2 activate genes that potentially function in thiol homeostasis. RT-qPCR of BA1951, BA5387, and BA1208 transcripts of the parent (Bacillus anthracis Sterne 7702) and ΔspxA1 ΔspxA2 mutant (A1/A2, ORB8285) from triplicate cultures with and without 1 mmol/L diamide for 20 min (see Experimental Procedures). Each biological replicate is graphed individually (denoted as 1, 2, or 3) to show the variation in the transcript amount (values listed above each bar).
In the absence of oxidative stress, BA1951 and BA5387 transcript levels remained low, but in the presence of diamide, both transcripts increased dramatically in the parent (7702), suggesting a role for both genes in SpxA-dependent disulfide stress tolerance (Fig. 7). In the double spxA mutant, BA1951 transcript levels decreased, but not to untreated levels, signifying that BA1951 is also regulated by a Spx-independent mechanism during oxidative stress (Fig. 7). The level of BA5837 (trxB) transcript in the double spxA mutant was only slightly induced by diamide, indicating a requirement for SpxA1 and/or A2 in activating BA5387 transcription during diamide-induced stress (Fig. 7). Taken together, we believe that the B. anthracis (cereus/thuringiensis)-specific BA1951 gene product is an oxidoreductase, and along with BA5387, have potential roles in oxidative stress defense.
In microarray experiments, the transcript levels of the B.subtilis yjbH ortholog, BA1208, was elevated when either SpxA1DD or SpxA2DD was present (Table 2, Tables S1, S2), indicating a requirement for one or both Spx paralogs in the activation of yjbH transcription. RT-qPCR experiments confirmed SpxA1DD and SpxA2DD transcriptional control of BA1208, even during disulfide stress where the transcript amount of BA1208 increased in the parent (7702) but not in the ΔspxA1 ΔspxA2 mutant (Table 2 and Fig. 7). Elevating the concentrations of YjbH and ClpX through a Spx-dependent mechanism of control might ensure that the means of Spx elimination is in place for when thiol homeostasis is restored following stress. Recent evidence from chromatin immune precipitation and transcription analysis indicated that the clpX gene is under positive transcriptional control by SpxA in B. subtilis (Rochat et al. 2012). However, we were unable to detect upregulation of the B. anthracis clpX (BA4074) gene in SpxA1DD- or SpxA2DD-producing cells.
In summary, four lines of evidence indicate that SpxA proteins of B. anthracis participate in the control of the oxidative stress response: (1) The spxA1 null mutant shows heightened sensitivity to H2O2 treatment, while the spxA1/spxA2 double mutant shows a severe growth defect when cells are treated with diamide; (2) SpxA1 protein is elevated upon diamide-induced stress, in parallel with elevated SpxA-dependent transcriptional induction; (3) SpxA1 or SpxA2, in protease-resistant form, activate the transcription of many genes that have been implicated in disulfide stress resistance and thiol homeostasis; (4) Genes likely involved in thiol homeostasis are induced by diamide treatment through an SpxA-dependent mechanism. Similar to the role of spx in B. subtilis, the B. anthracis spx paralogs are likely to be important transcriptional regulators during disulfide stress. Each paralog may differentially regulate genes involved in other stress-alleviating processes. Additional experiments are underway to further examine the function and regulation of these two paralogous genes and their products.
Acknowledgments
We thank Scott Stibitz for the gift of plasmids pDR1028, pSS1827, and pSS4332; C. Lee and A. D. Grossman for providing plasmid pJMA402. Theresa Koehler for B. anthracis Sterne 7702; and Cole Zuber for strain construction. This research was supported by National Institutes of Health Grant AI092313 to P. Z.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Spx mutants sensitive to oxidative stress. Strains were grown in LB medium in the presence or absence of 0.01% deoxycholate. Five microliter of the indicated dilutions were spotted onto LB agar (see Experimental Procedures). 7702 StrR(Parent [SR1]), ORB8398 (ΔspxA1 StrR, ΔA1), ORB8481 (ΔspxA1 ΔspxA2 StrR, ΔA1/A2), ORB8485 (ΔspxA1 ΔspxA2 StrR ICEBs1::spxA1DD, ΔA1/A2 +A1DD), ORB8486 (ΔspxA1 ΔspxA2 StrR ICEBs1::spxA2DD, ΔA1/A2 +A2DD). StrR, streptomycin resistance.
Data S1. Sporulation and germination assays.
Table S1. SpxA1DD-regulated genes
Table S2. SpxA2DD-regulated genes.
Table S3. SpxA1DD-activated genes. Fifteen minutes after induction. .threefold increase.
Table S4. SpxA2DD-activated. Fifteen minutes after induction. .threefold increase.
Table S5. SpxA1- and SpxA2- controlled genes shown to be induced by peroxide treatment (Pohl et al. 2011).
Table S6. Oligonuceotide primers.
References
- Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid. Redox Signal. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. USA. 2005;102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua S, McKevitt M, Degiusti K, Hamm EE, Larabee J, Shakir S, et al. The mechanism of Bacillus anthracis intracellular germination requires multiple and highly diverse genetic loci. Infect. Immun. 2009;77:23–31. doi: 10.1128/IAI.00801-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J, Wu LJ, Errington J, Wang JC. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science. 2000;290:995–997. doi: 10.1126/science.290.5493.995. [DOI] [PubMed] [Google Scholar]
- Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J. Bacteriol. 2006;188:6092–6100. doi: 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman NH, Anderson EC, Swenson EE, Janes BK, Fisher N, Niemeyer MM, et al. Transcriptional profiling of Bacillus anthracis during infection of host macrophages. Infect. Immun. 2007;75:3434–3444. doi: 10.1128/IAI.01345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer W, Turnbull PC. Anthrax in animals. Mol. Aspects Med. 2009;30:481–489. doi: 10.1016/j.mam.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, et al. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi A, Labruyere E, Mock M. Construction and characterization of a protective antigen-deficient Bacillus anthracis strain. Mol. Microbiol. 1990;4:1111–1117. doi: 10.1111/j.1365-2958.1990.tb00685.x. [DOI] [PubMed] [Google Scholar]
- Chen L, Ge X, Wang X, Patel JR, Xu P. SpxA1 involved in hydrogen peroxide production, stress tolerance and endocarditis virulence in Streptococcus sanguinis. PLoS One. 2012;7:e40034. doi: 10.1371/journal.pone.0040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi BK, Gronau K, Maeder U, Hessling B, Becher D, Antelmann H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell. Proteomics. 2011;10:M111.009506. doi: 10.1074/mcp.M111.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Reyes D, Leelakriangsak M, Zuber P. The global regulator Spx functions in the control of organosulfur metabolism in Bacillus subtilis. J. Bacteriol. 2006;188:5741–5751. doi: 10.1128/JB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson RM, Liu CQ. Properties and applications of antimicrobial peptides in biodefense against biological warfare threat agents. Crit. Rev. Microbiol. 2008;34:89–107. doi: 10.1080/10408410802143808. [DOI] [PubMed] [Google Scholar]
- Delcar SB, Stock KP, Newton GL, Fahey RC, Davies JE. Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. J. Biol. Chem. 1998;273:5744–5751. doi: 10.1074/jbc.273.10.5744. [DOI] [PubMed] [Google Scholar]
- Delcardayre SB, Davies JE. Staphylococcus aureus coenzyme A disulfide reductase, a new subfamily of pyridine nucleotide-disulfide oxidoreductase. Sequence, expression, and analysis of cdr. J. Biol. Chem. 1998;273:5752–5757. doi: 10.1074/jbc.273.10.5752. [DOI] [PubMed] [Google Scholar]
- Dodd D, Reese JG, Louer CR, Ballard JD, Spies MA, Blanke SR. Functional comparison of the two Bacillus anthracis glutamate racemases. J. Bacteriol. 2007;189:5265–5275. doi: 10.1128/JB.00352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W, Helmann JD. The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 2008;67:830–848. doi: 10.1111/j.1365-2958.2007.06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldholm V, Gutt B, Johnsborg O, Bruckner R, Maurer P, Hakenbeck R, et al. The pneumococcal cell envelope stress-sensing system LiaFSR is activated by murein hydrolases and lipid II-interacting antibiotics. J. Bacteriol. 2010;192:1761–1773. doi: 10.1128/JB.01489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher N, Hanna P. Characterization of Bacillus anthracis germinant receptors in vitro. J. Bacteriol. 2005;187:8055–8062. doi: 10.1128/JB.187.23.8055-8062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet A, Mesnage S, Tosi-Couture E, Gounon P, Mock M. Bacillus anthracis surface: capsule and S-layer. J. Appl. Microbiol. 1999;87:251–255. doi: 10.1046/j.1365-2672.1999.00882.x. [DOI] [PubMed] [Google Scholar]
- Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 2001;183:4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. Bacillus subtilis Fur represses one of two paralogous haem-degrading monooxygenases. Microbiology. 2011;157:3221–3231. doi: 10.1099/mic.0.053579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, et al. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. USA. 2010;107:6482–6486. doi: 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg SK, Kommineni S, Henslee L, Zhang Y, Zuber P. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J. Bacteriol. 2009;191:1268–1277. doi: 10.1128/JB.01289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Pane-Farre J, Volker U. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 2007;61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- Helmann JD. Bacillithiol, a new player in bacterial redox homeostasis. Antioxid. Redox Signal. 2011;15:123–133. doi: 10.1089/ars.2010.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD, Wu MF, Kobel PA, Gamo FJ, Wilson M, Morshedi MM, et al. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 2001;183:7318–7328. doi: 10.1128/JB.183.24.7318-7328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra JR, Orozco AD, Rojas JA, Lopez K, Setlow P, Yasbin RE, et al. Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in repair of DNA damage during outgrowth of Bacillus subtilis spores. J. Bacteriol. 2008;190:2031–2038. doi: 10.1128/JB.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo MM, Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J. Mol. Biol. 1986;191:615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- Janes BK, Stibitz S. Routine markerless gene replacement in Bacillus anthracis. Infect. Immun. 2006;74:1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jervis AJ, Thackray PD, Houston CW, Horsburgh MJ, Moir A. SigM-responsive genes of Bacillus subtilis and their promoters. J. Bacteriol. 2007;189:4534–4538. doi: 10.1128/JB.00130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, et al. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J. Bacteriol. 2010;192:2546–2556. doi: 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz JK, Mendoza JE, Gaca AO, Miller JH, Koselny KA, Giambiagi-Demarval M, et al. The Spx regulator modulates stress responses and virulence in Enterococcus faecalis. Infect. Immun. 2012;80:2265–2275. doi: 10.1128/IAI.00026-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler TM. Bacillus anthracis physiology and genetics. Mol. Aspects Med. 2009;30:386–396. doi: 10.1016/j.mam.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristoffersen SM, Ravnum S, Tourasse NJ, Okstad OA, Kolsto AB, Davies W. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14579 [corrected] J. Bacteriol. 2007;189:5302–5313. doi: 10.1128/JB.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel B, Kroos L, Poth H, Youngman P, Losick R. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes Dev. 1989;3:1735–1744. doi: 10.1101/gad.3.11.1735. [DOI] [PubMed] [Google Scholar]
- Larsson JT, Rogstam A, von Wachenfeldt C. YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol. Microbiol. 2007;66:669–684. doi: 10.1111/j.1365-2958.2007.05949.x. [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- Leelakriangsak M, Kobayashi K, Zuber P. Dual negative control of spx transcription initiation from the P3 promoter by repressors PerR and YodB in Bacillus subtilis. J. Bacteriol. 2007;189:1736–1744. doi: 10.1128/JB.01520-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liochev SI, Fridovich I. Superoxide and iron: partners in crime. IUBMB Life. 1999;48:157–161. doi: 10.1080/713803492. [DOI] [PubMed] [Google Scholar]
- McLean S, Bowman LA, Sanguinetti G, Read RC, Poole RK. Peroxynitrite toxicity in Escherichia coli K12 elicits expression of oxidative stress responses and protein nitration and nitrosylation. J. Biol. Chem. 2010;285:20724–20731. doi: 10.1074/jbc.M109.085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Moayeri M, Leppla SH. Cellular and systemic effects of anthrax lethal toxin and edema toxin. Mol. Aspects Med. 2009;30:439–455. doi: 10.1016/j.mam.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mols M, Abee T. Primary and secondary oxidative stress in Bacillus. Environ. Microbiol. 2011;13:1387–1394. doi: 10.1111/j.1462-2920.2011.02433.x. [DOI] [PubMed] [Google Scholar]
- Mostertz J, Scharf C, Hecker M, Homuth G. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology. 2004;150:497–512. doi: 10.1099/mic.0.26665-0. [DOI] [PubMed] [Google Scholar]
- Moszer I, Jones LM, Moreira S, Fabry C, Danchin A. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res. 2002;30:62–65. doi: 10.1093/nar/30.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Hoffmann T, Zhu Y, Jahn D. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J. Bacteriol. 1998;180:5344–5350. doi: 10.1128/jb.180.20.5344-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 2001;42:383–394. doi: 10.1046/j.1365-2958.2001.02639.x. [DOI] [PubMed] [Google Scholar]
- Nakano S, Kuster-Schock E, Grossman AD, Zuber P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2003;100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Erwin KN, Ralle M, Zuber P. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 2005;55:498–510. doi: 10.1111/j.1365-2958.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- Newberry KJ, Nakano S, Zuber P, Brennan RG. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. USA. 2005;102:15839–15844. doi: 10.1073/pnas.0506592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Arnold K, Price MS, Sherrill C, Delcardayre SB, Aharonowitz Y, et al. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Rawat M, Jothivasan JJ, la Clair VK, Budiarto T, Hamilton CJ, et al. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat. Chem. Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm GJ, Khanh Chi B, Waack P, Gronau K, Becher D, Albrecht D, et al. Structural insights into the redox-switch mechanism of the MarR/DUF24-type regulator HypR. Nucleic Acids Res. 2012;40:4178–4192. doi: 10.1093/nar/gkr1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passalacqua KD, Bergman NH. Bacillus anthracis: interactions with the host and establishment of inhalational anthrax. Future Microbiol. 2006;1:397–415. doi: 10.2217/17460913.1.4.397. [DOI] [PubMed] [Google Scholar]
- Passalacqua KD, Bergman NH, Herring-Palmer A, Hanna P. The superoxide dismutases of Bacillus anthracis do not cooperatively protect against endogenous superoxide stress. J. Bacteriol. 2006;188:3837–3848. doi: 10.1128/JB.00239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passalacqua KD, Bergman NH, Lee JY, Sherman DH, Hanna PC. The global transcriptional responses of Bacillus anthracis Sterne (34F2) and a Delta sodA1 mutant to paraquat reveal metal ion homeostasis imbalances during endogenous superoxide stress. J. Bacteriol. 2007;189:3996–4013. doi: 10.1128/JB.00185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersohn A, Brigulla M, Haas S, Hoheisel JD, Volker U, Hecker M. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 2001;183:5617–5631. doi: 10.1128/JB.183.19.5617-5631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piris-Gimenez A, Paya M, Lambeau G, Chignard M, Mock M, Touqui L, et al. In vivo protective role of human group IIa phospholipase A2 against experimental anthrax. J. Immunol. 2005;175:6786–6791. doi: 10.4049/jimmunol.175.10.6786. [DOI] [PubMed] [Google Scholar]
- Pohl S, Tu WY, Aldridge PD, Gillespie C, Hahne H, Mader U, et al. Combined proteomic and transcriptomic analysis of the response of Bacillus anthracis to oxidative stress. Proteomics. 2011;11:3036–3055. doi: 10.1002/pmic.201100085. [DOI] [PubMed] [Google Scholar]
- Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006;313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- Reder A, Pother DC, Gerth U, Hecker M. The modulator of the general stress response, MgsR, of Bacillus subtilis is subject to multiple and complex control mechanisms. Environ. Microbiol. 2012;14:2838–2850. doi: 10.1111/j.1462-2920.2012.02829.x. [DOI] [PubMed] [Google Scholar]
- Reiter L, Kolsto AB, Piehler AP. Reference genes for quantitative, reverse-transcription PCR in Bacillus cereus group strains throughout the bacterial life cycle. J. Microbiol. Methods. 2011;86:210–217. doi: 10.1016/j.mimet.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Rochat T, Nicolas P, Delumeau O, Rabatinova A, Korelusova J, Leduc A, et al. Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res. 2012;40:9571–9583. doi: 10.1093/nar/gks755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Beltran J, Rodriguez-Rojas A, Guelfo JR, Couce A, Blazquez J. The Escherichia coli SOS gene dinF protects against oxidative stress and bile salts. PLoS One. 2012;7:e34791. doi: 10.1371/journal.pone.0034791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukmana A, Morimoto T, Takahashi H, Giyanto, Ogasawara N. Assessment of transcriptional responses of Bacillus subtilis cells to the antibiotic enduracidin, which interferes with cell wall synthesis, using a high-density tiling chip. Genes Genet. Syst. 2009;84:253–267. doi: 10.1266/ggs.84.253. [DOI] [PubMed] [Google Scholar]
- van Schaik W, Prigent J, Fouet A. The stringent response of Bacillus anthracis contributes to sporulation but not to virulence. Microbiology. 2007;153:4234–4239. doi: 10.1099/mic.0.2007/010355-0. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa M, Kodama K, Yamamoto H, Kobayashi K, Ogasawara N, Sekiguchi J. Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and northern blot analyses. Biosci. Biotechnol. Biochem. 2005;69:2155–2169. doi: 10.1271/bbb.69.2155. [DOI] [PubMed] [Google Scholar]
- Shatalin K, Gusarov I, Avetissova E, Shatalina Y, McQuade LE, Lippard SJ, et al. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc. Natl. Acad. Sci. USA. 2008;105:1009–1013. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam P, Senadheera MD, Mair RW, Levesque CM, Cvitkovitch DG. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J. Bacteriol. 2009;191:2973–2984. doi: 10.1128/JB.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Nelson K, Kim EJ, Perego M, Hoch JA. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 2005;187:5419–5426. doi: 10.1128/JB.187.15.5419-5426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Fukushima T, Hoch JA. The essential YycFG two-component system of Bacillus subtilis. Methods Enzymol. 2007a;422:396–417. doi: 10.1016/S0076-6879(06)22020-2. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Mohan MA, Imus PM, Hoch JA. YycH and YycI interact to regulate the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 2007b;189:3280–3289. doi: 10.1128/JB.01936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier JN, Rossi Paccani S, Quesnel-Hellmann A, Baldari CT. Anthrax toxins: a weapon to systematically dismantle the host immune defenses. Mol. Aspects Med. 2009;30:456–466. doi: 10.1016/j.mam.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Tu WY, Pohl S, Gizynski K, Harwood CR. The iron-binding protein Dps2 confers peroxide stress resistance on Bacillus anthracis. J. Bacteriol. 2012a;194:925–931. doi: 10.1128/JB.06005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu WY, Pohl S, Summpunn P, Hering S, Kerstan S, Harwood CR. Comparative analysis of the responses of related pathogenic and environmental bacteria to oxidative stress. Microbiology. 2012b;158:636–647. doi: 10.1099/mic.0.057000-0. [DOI] [PubMed] [Google Scholar]
- Turlan C, Prudhomme M, Fichant G, Martin B, Gutierrez C. SpxA1, a novel transcriptional regulator involved in X-state (competence) development in Streptococcus pneumoniae. Mol. Microbiol. 2009;73:492–506. doi: 10.1111/j.1365-2958.2009.06789.x. [DOI] [PubMed] [Google Scholar]
- Veiga P, Bulbarela-Sampieri C, Furlan S, Maisons A, Chapot-Chartier MP, Erkelenz M, et al. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 2007;282:19342–19354. doi: 10.1074/jbc.M611308200. [DOI] [PubMed] [Google Scholar]
- Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, et al. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- Welkos S, Cote CK, Hahn U, Shastak O, Jedermann J, Bozue J, et al. Humanized theta-defensins (retrocyclins) enhance macrophage performance and protect mice from experimental anthrax infections. Antimicrob. Agents Chemother. 2011;55:4238–4250. doi: 10.1128/AAC.00267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Liu Z, Hughes C, Stern AM, Wang H, Zhong Z, et al. Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc. Natl. Acad. Sci. USA. 2013;110:2348–2353. doi: 10.1073/pnas.1218039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C, Sekowska A, Francetic O, Martin-Verstraete I, Wang Y, Danchin A. Spx mediates oxidative stress regulation of the methionine sulfoxide reductases operon in Bacillus subtilis. BMC Microbiol. 2008;8:128. doi: 10.1186/1471-2180-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 2004;186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P, Chauhan S, Pilaka P, Nakano MM, Gurumoorthy S, Lin AA, et al. Phenotype enhancement screen of a regulatory spx mutant unveils a role for the ytpQ gene in the control of iron homeostasis. PLoS One. 2011;6:e25066. doi: 10.1371/journal.pone.0025066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Spx mutants sensitive to oxidative stress. Strains were grown in LB medium in the presence or absence of 0.01% deoxycholate. Five microliter of the indicated dilutions were spotted onto LB agar (see Experimental Procedures). 7702 StrR(Parent [SR1]), ORB8398 (ΔspxA1 StrR, ΔA1), ORB8481 (ΔspxA1 ΔspxA2 StrR, ΔA1/A2), ORB8485 (ΔspxA1 ΔspxA2 StrR ICEBs1::spxA1DD, ΔA1/A2 +A1DD), ORB8486 (ΔspxA1 ΔspxA2 StrR ICEBs1::spxA2DD, ΔA1/A2 +A2DD). StrR, streptomycin resistance.
Data S1. Sporulation and germination assays.
Table S1. SpxA1DD-regulated genes
Table S2. SpxA2DD-regulated genes.
Table S3. SpxA1DD-activated genes. Fifteen minutes after induction. .threefold increase.
Table S4. SpxA2DD-activated. Fifteen minutes after induction. .threefold increase.
Table S5. SpxA1- and SpxA2- controlled genes shown to be induced by peroxide treatment (Pohl et al. 2011).
Table S6. Oligonuceotide primers.


