Abstract
Agrobacterium-mediated transformation is a useful tool for the genetic modification in plants, although its efficiency is low for several plant species. Agrobacterium-mediated transformation has three major steps in laboratory-controlled experiments: the delivery of T-DNA into plant cells, the selection of transformed plant cells, and the regeneration of whole plants from the selected cells. Each of these steps must be optimized to improve the efficiency of Agrobacterium-mediated plant transformation. It has been reported that increasing the number of cells transformed by T-DNA delivery can improve the frequency of stable transformation. Previously, we demonstrated that a reduction in ethylene production by plant cells during cocultivation with A. tumefaciens-expressing 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase resulted in increased T-DNA delivery into the plant cells. In this study, to further improve T-DNA delivery by A. tumefaciens, we modified the expression cassette of the ACC deaminase gene using vir gene promoter sequences. The ACC deaminase gene driven by the virD1 promoter was expressed at a higher level, resulting in a higher ACC deaminase activity in this A. tumefaciens strain than in the strain with the lac promoter used in a previous study. The newly developed A. tumefaciens strain improves the delivery of T-DNA into Solanum lycopersicum (tomato) and Erianthus ravennae plants and thus may be a powerful tool for the Agrobacterium-mediated genetic engineering of plants.
Keywords: ACC deaminase, Agrobacterium, promoter, T-DNA delivery
Introduction
Agrobacterium tumefaciens is a phytopathogenic, soilborne, Gram-negative bacterium that is used to introduce transgenes into plant genomes. Agrobacterium-mediated transformation is an important technique in plant science research. The process of Agrobacterium-mediated transformation is divided into three steps: (1) the delivery of T-DNA into plant cells via A. tumefaciens, (2) the selection of transformed cells by antibiotics and the resistance marker genes, and (3) the regeneration of whole plants from the selected cells. Although this Agrobacterium-mediated transformation is well established for model plants, it remains ineffective in many plant species of practical importance. It is necessary to improve the efficiency of each step to generate sufficient numbers of transformed plants for evaluation.
Agrobacterium-mediated T-DNA delivery into plant cells occurs via the integration of T-DNA from a tumor-inducing (Ti) plasmid in A. tumefaciens into the host plant cells (reviewed in Tzfira et al. 2004). The virulence genes that are essential for T-DNA delivery are regulated by the two-component VirA/VirG system and a plant-derived phenolic compound such as acetosyringone (Stachel et al. 1985, reviewed in Pitzschke and Hirt 2010). It has been reported that certain plant hormones, such as salicylic acid, ethylene, cytokinin, auxin, and abscisic acid, induce defense responses against Agrobacterium-mediated plant transformation (Davis et al. 1992; Yuan et al. 2007; Lee et al. 2009; Hwang et al. 2010; Rico et al. 2010). For instance, salicylic acid interferes with the ability of A. tumefaciens to infect plants by suppressing the transcription of the vir genes, the repABC operon, and genes associated with quorum sensing (Anand et al. 2008). In addition, ethylene represses vir gene expression during transformation (Nonaka et al. 2008a). Consequently, ethylene functions as a repressor that inhibits Agrobacterium-mediated gene transfer (Ezura et al. 2000; Han et al. 2005).
We previously constructed an A. tumefaciens harboring a plasmid-encoding 1-aminocyclopropane-1 carboxylate, enzyme devoted to the degradation of the immediate precursor of ethylene (Nonaka et al. 2008b). The ACC deaminase-expressing A. tumefaciens strain suppressed ethylene synthesis and enhanced the gene transfer efficiency into melon cotyledon cells (Nonaka et al. 2008b). It has also been reported that the ACC deaminase-expressing A. tumefaciens strain allows for the efficient genetic transformation of the “Egusi” melon (Ntui et al. 2010). Furthermore, a similar A. tumefaciens strain with ACC deaminase activity was constructed and was also shown to be effective for the transformation of canola cultivars (Hao et al. 2010). Thus, the repression of ethylene synthesis in plant cells during T-DNA delivery is an effective method to efficiently generate transgenic plants using A. tumefaciens.
In this study, to improve the ACC deaminase-expressing A. tumefaciens strain, we modified the ACC deaminase expression cassette. Previously, the ACC deaminase gene in the pBBRacdS plasmid was expressed under the control of the lac promoter of Escherichia coli (Nonaka et al. 2008b), which exhibits transcriptional activity in acidic media (Chen and Winans 1991). Klüsener et al. (2010) showed that the expression levels of several A. tumefaciens virulence genes were the highest in acidic media (AB medium at pH 5.5) containing 100-μmol/L acetosyringone (Klüsener et al. 2010). It is speculated that the promoters of the virulence genes would exhibit higher transcriptional activity than the lac promoter in A. tumefaciens upon cocultivation with plants. Therefore, to express high levels of ACC deaminase in A. tumefaciens during cocultivation with plants, the lac promoter sequence should be substituted with virulence gene promoter sequences that permit a high level of gene expression under these particular culture conditions. In this study, we demonstrated that an A. tumefaciens strain harboring a plasmid with a virulence gene promoter for the expression of ACC deaminase resulted in an increased efficiency of Agrobacterium-mediated transient transformation in Solanum lycopersicum, and Erianthus ravennae plants relative to that for A. tumefaciens harboring the pBBRacdS plasmid with a lac promoter.
Materials and Methods
Bacterial strains and culture conditions
The bacterial strains used in this study are listed in Table 1. E. coli strains were grown at 37°C in LB medium (1% Bacto-Tryptone, 0.5% yeast extract, 0.5% NaCl). A. tumefaciens strains were grown at 28°C in LB medium or Murashige and Skoog (MS) medium (Murashige and Skoog 1962). E. coli (pQE60) was cultured in LB medium containing 100 mg/L ampicillin and 20 mg/L kanamycin. A. tumefaciens (pBBR1MCS5 and the derivative strain pIG121-Hm) was selected with 100 mg/L ampicillin, 50 mg/L gentamicin, and 100 mg/L kanamycin. Gentamicin (50 mg/L), ampicillin (100 mg/L), and spectinomycin (50 mg/L) were used for the selection of A. tumefaciens (pBBR1MCS5 and the derivative strain pEKH2).
Table 1.
Plasmids and strains used in this study
| Description | Reference | |
|---|---|---|
| Strain | ||
| GV2260 | Nononcogenic A. tumefaciens strain | Deblaere et al. (1985) |
| MCS | GV2260 containing the pBBR1MCS5 plasmid | Nonaka et al. (2008a,b) |
| acdS | GV2260 containing the pBBRacdS plasmid | Nonaka et al. (2008a,b) |
| virB1acdS | GV2260 containing the pvirB1acdS plasmid | This study |
| virD1acdS | GV2260 containing the pvirD1acdS plasmid | This study |
| virE1acdS | GV2260 containing the pvirE1acdS plasmid | This study |
| Plasmid | ||
| pQEAcdS | Overexpression vector for ACC deaminase; AmpR | This study |
| pBBR1MCS-5 | Broad host-range shuttle vector; GenR | Kovach et al. (1995) |
| pBBRacdS | Overexpression vector for ACC deaminase under the control of the lac promoter; GmR | Nonaka et al. (2008a,b) |
| pvirB1acdS | Overexpression vector for ACC deaminase under the control of the virB1 promoter; GmR | This study |
| pvirD1acdS | Overexpression vector for ACC deaminase under the control of the virD1 promoter; GmR | This study |
| pvirE1acdS | Overexpression vector for ACC deaminase under the control of the virE1 promoter; GmR | This study |
| pIG121-Hm | Binary vector plasmid carrying the b-glucuronidase gene (gusA) between the T-borders; KmR | Ohta et al. (1990) |
| pEKH2 | Binary vector plasmid carrying the b-glucuronidase gene (gusA) between the T-borders; SpR | Hoshikawa et al. (2012) |
Construction of plasmids
The primers and plasmids used in this study are listed in Table S1 and Table 1, respectively. The transcriptional fusion plasmids were constructed as follows: to delete the restriction site downstream of the acdS gene in the pBBRacdS plasmid, an acdS fragment was amplified with the primers acdS-NcoI and acdS-SpeI. The resulting product was digested with NcoI and SpeI and then ligated to an NcoI- and XbaI-digested pBBR1MCS-5 vector. The lacZ-acdS ORF fragment was then amplified by the primers plac-ATG and acdS_EcoRI. The DNA fragments of various vir promoter regions, including the vir box, were amplified by PCR using the genomic DNA of A. tumefaciens as the template. Each of the amplified promoter fragments and the lacZ-acdS ORF fragment was ligated by fusion PCR, and these fusion fragments were then digested with NcoI and ScaI and cloned into the pBBR1MCS-5 vector. The overexpression vector, designated pQE60AcdS, for the C-terminal 6× His AcdS fusion protein (AcdS-His) was constructed by inserting a PCR-amplified acdS gene into the NcoI-BglII-digested pQE60 expression vector (Qiagen, Hamburg, Germany). The PCR product was obtained using the pQEacdS-F and pQEacdS-R primers.
Antibody preparation
The expression of AcdS-His in E. coli M15/pREP4 was induced for 5 h in the presence of 1 mmol/L IPTG. The cells were harvested by centrifugation at 5000 g at 4°C for 15 min. The wet cells were suspended in 10 mL of lysis buffer (50-mmol/L NaH2PO4, 300-mmol/L NaCl, pH 8.0), lysed on ice by sonication, and centrifuged at 5000 × g at 4°C for 15 min. The supernatant was incubated with His-select™ Nickel Affinity gel (Sigma, St. Louis, MO) at 4°C for 1 h and eluted with elution buffer (50-mmol/L NaH2PO4, 300-mmol/L NaCl, 250-mmol/L imidazole, pH 8.0). The purity of the AcdS-His protein was analyzed by 15% SDS-PAGE. The eluted sample was dialyzed against 50-mmol/L Tris-HCl buffer (pH 8.0) containing 100-mmol/L NaCl and 50% glycerol. The AcdS-His fusion protein was injected into rabbits to prepare polyclonal antibodies.
Western blots
The samples were prepared from whole-cell extracts of strains grown in MS medium containing 200-μmol/L acetosyringone. The cells were harvested by centrifugation at 5000 × g at 4°C for 15 min. Wet cells were suspended in lysis buffer (100-mmol/L Tris-HCl, 0.1% SDS, 0.1% Triton X-100, pH 8.5), lysed on ice by sonication, and centrifuged at 5000 × g at 4°C for 15 min. Equivalent volumes containing 0.03 OD600 units of protein were resolved by SDS-PAGE and then electroblotted onto polyvinylidene difluoride membranes. The membranes were blocked with 5% skim milk in TBS-containing 0.2% Tween 80. Primary and secondary antibodies were diluted in TBS-containing 0.2% Tween 80. The primary antibody was a rabbit polyclonal anti-ACC deaminase antibody, and the secondary antibody was an HRP-conjugated donkey anti-rabbit IgG. Bound antibodies were then detected using ImmunoSar LD (Wako, Tokyo, Japan).
ACC deaminase activity assay
Cells were collected and washed twice with 100-mmol/L Tris-HCl (pH 8.5) and resuspended in 1.5 mL of lysate buffer. The cells were lysed on ice by sonication and centrifuged at 5000 × g at 4°C for 15 min. The ACC deaminase activity was measured according to a modified protocol based on that of Honma and Shimomura (1978). The ACC deaminase activity was measured spectrophotometrically at 340 nm. The protein content of the extracts was determined using the Bradford method (Bradford 1976).
Plant material
Seeds of S. lycopersicum cv. Moneymaker were washed with 70% ethanol for 10 sec, sterilized with 5% hypochlorous acid containing 10% Triton X-100 for 45 min, and washed three times with sterilized water. After the third wash, the seeds were kept in water overnight. The sterilized S. lycopersicum seeds were sown on MS medium containing 15 g/L sucrose (Wako) and 3% gellan gum (Wako) and then grown for 7 days. E. ravennae calli, which were induced from seeds on MS medium-containing 1 g/L casamino acids, 2 mg/L 2,4-D, 0.2 mg/L 6-benzylaminopurine (BAP), 30 g/L maltose H (Wako), and 3% gellan gum, were kindly provided by Prof. Masashiro Mii of Chiba University in Japan. The calli were subcultured for 2 weeks before A. tumefaciens inoculation.
Agrobacterium-mediated T-DNA transfer
Preparation of A. tumefaciens
Agrobacterium tumefaciens was cultured on solid LB medium at 28°C for 2 days. A single colony was picked and cultured in 2 mL of LB medium at 28°C and 200 rpm for 2 days until the culture reached the stationary phase. From this culture, 15 μL was harvested and added to 15 mL of LB medium, which was then cultured at 28°C and 200 rpm for 20 h. When the optical density of the culture reached 0.8-1.0, the cells were centrifuged, and the pelleted bacterial cells were resuspended in liquid MS medium containing 30 g/L glucose and 200 μmol/L acetosyringone at pH 5.2. The optical density was adjusted to 0.4-0.5.
Solanum lycopersicum cv. Moneymaker
Cotyledons from 7-day-old S. lycopersicum seedlings were cut into four explants and used to generate two locations for inoculation with A. tumefaciens. Thirty explants were subjected to each treatment. The inoculated explants were cultured on cocultivation medium, which contained MS salts, 30 g/L glucose, 200 μmol/L acetosyringone (pH 5.2), and 3% gellan gum, at 25°C for 3 days in the dark. After 3 days of cocultivation, the S. lycopersicum explants were assayed histochemically for GUS activity with X-Gluc buffer.
Erianthus ravennae
Erianthus ravennae calli were inoculated with A. tumefaciens and cultured on MS medium containing 1 g/L casamino acids, 30 g/L glucose, 3% gellan gum, 2 mg/L 2,4-D, and 200-μmol/L acetosyringone at pH 5.2 for 3 days. After 3 days of cocultivation, the calli were washed with sterilized water and transferred to MS medium containing 1 g/L casamino acids, 2 mg/L 2,4-D, 0.2 mg/L BAP, 30 g/L glucose, 3% gellan gum, and 12.5 mg/L meropenem trihydrate (Dainippon Sumitomo Pharma) for 1 day to eliminate A. tumefaciens. The inoculated calli were assayed using histochemistry and quantitative activity assays.
Estimation of the T-DNA transfer efficiency
Solanum lycopersicum cv. Moneymaker
After 3 days of cocultivation, S. lycopersicum segments were placed in GUS staining buffer-containing 100-mmol/L phosphate buffer, 10-mmol/L EDTA, 2.5-mmol/L potassium ferricyanide, 2.5-mmol/L potassium ferrocyanide, 0.1% Triton X-100, and 0.5 mg/L X-glucuronide. GUS-stained S. lycopersicum cotyledon explants were observed, and images were taken using a stereoscopic microscope system (Leica: MX FLIII, DFC300 FX, Application Suite, Leica, Germany). The GUS-stained area was converted into a numerical value using Image J (National Institutes of Health: http://rsbweb.nih.gov/ij/), and the percentage of GUS-stained area was calculated for each explant. According to these results, the GUS-stained S. lycopersicum explants were categorized into six classes (less than 1%, 1% to 3%, 3% to 5%, 5% to 10%, 10% to 20%, and more than 20%). To estimate the T-DNA transformation efficiency, the frequency of each class was calculated.
Erianthus ravennae
After cocultivation, the GUS activity of E. ravennae calli was assayed histochemically with GUS staining buffer as described above. The GUS-stained calli were observed using a stereoscopic microscope, and the number of GUS-stained spots was counted. After counting, the number of GUS-stained spots per 1 g of calli was calculated. The T-DNA transfer efficiency was estimated based on the relative number of GUS spots.
The T-DNA transfer efficiency was also estimated based on the quantitative GUS activity measured using the fluorometric assay previously described by Nonaka et al. (2008b) with a Perkin-Elmer ARYO MX-FL 1420 Multilabel Counter fluorometer (Perkin-Elmer, Waltham, MA) and 4-methylumbelliferyl beta-d-glucuronide (MUG) as the substrate.
Results and Discussion
Effects of different promoters on the expression of ACC deaminase
The ACC deaminase gene in the pBBRacdS plasmid was expressed under the control of the lac promoter from E. coli (Nonaka et al. 2008b). To increase the expression of ACC deaminase, we constructed transcriptional fusion plasmids. The promoter sequences originated from A. tumefaciens and were selected due to their high expression of genes in A. tumefaciens during cocultivation with plant cells. Klüsener et al. (2010) reported that the virB1 operon, virD1 operon, and virE1 operon, which vir genes were induced in response to acetosyringone, and therefore we attempted to utilize the vir gene promoters to increase compared to ACC deaminase expression and the activity in A. tumefaciens cells. We constructed three plasmids with these promoters driving ACC deaminase expression. These plasmids included the VirG-binding site upstream of the promoter sequence (Steck et al. 1988). A. tumefaciens strains harboring these expression plasmids were cultured in liquid MS medium-containing 200-μmol/L acetosyringone for 14 h, and the expression of ACC deaminase in the whole-cell lysates was measured by Western blotting (Fig. 1A). The expression of ACC deaminase driven by the virulence gene promoters was greater than that from the pBBRacdS plasmid with the lac promoter (Fig. 1A,B). The expression of ACC deaminase increased by fourfold in the A. tumefaciens GV2260 (pvirD1acdS) strain (Fig. 1A,B). Next, we analyzed the time course (0–72 h) of acetosyringone-induced ACC deaminase accumulation in MS medium. In the A. tumefaciens GV2260 (pvirD1acdS) strain, ACC deaminase expression was induced at 6 h by adding acetosyringone, and high expression was still observed at 72 h (Fig. 1C). In contrast, the expression of ACC deaminase decreased in the A. tumefaciens GV2260 (pBBRacdS) strain (Fig. 1C). The expressed protein in both strains containing either plasmid was degraded by endogenous proteases.
Figure 1.
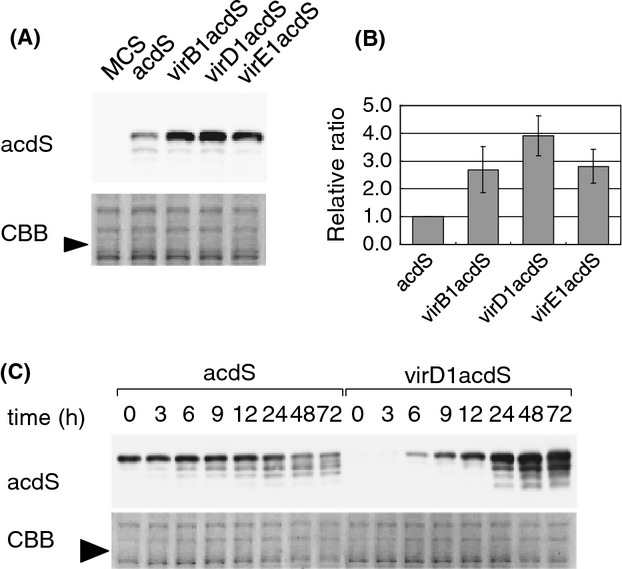
Semiquantitative analysis of ACC deaminase expression in A. tumefaciens strains. (A) Western blot analysis of ACC deaminase expression was performed using cell extracts. ACC deaminase was probed with an anti-ACC deaminase antibody. Coomassie Brilliant Blue staining (bottom panel) is shown as an internal control. Arrow head indicates 57kDa. (B) Relative intensities of the immunoreactive signals from the Western blots in (A). The ACC deaminase expression level in the A. tumefaciens GV2260 (pBBRacdS) strain was set to 1.0. The error bars show the standard deviation (n = 3). (C) Western blot analysis of ACC deaminase after 0–72 h of subculture on MS medium with acetosyringone [left panel: A. tumefaciens GV2260 (pBBRacdS); right panel: A. tumefaciens GV2260 (pvirD1acdS)]. MCS: A. tumefaciens GV2260 (pBBRMCS1-5); acdS: A. tumefaciens GV2260 (pBBRacdS); virB1acdS: A. tumefaciens GV2260 (pvirB1acdS); virD1acdS: A. tumefaciens GV2260 (pvirD1acdS); virE1acdS: A. tumefaciens GV2260 (pvirE1acdS). Arrow head means 57kDa.
In addition, we determined whether the expressed ACC deaminase was active. ACC deaminase catalyzes the degradation of ACC to α-ketobutyric acid and ammonia. We determined the level of α-ketobutyric acid using the whole-cell lysates from cells that had been cultured in MS medium containing 200-μmol/L acetosyringone. The amounts of α-ketobutyric acid after a 1-h reaction time for A. tumefaciens GV2260 (pBBRacdS) and A. tumefaciens GV2260 (pvirD1acdS) were 21.3 ± 3.3 and 41.1 ± 5.3 μmol/mg protein, respectively (Fig. 2). This result indicates that the ACC deaminase expressed from the pvirD1acdS plasmid was enzymatically active in the cells, and the A. tumefaciens GV2260 (pvirD1acdS) strain showed higher ACC deaminase activity than the A. tumefaciens GV2260 (pBBRacdS) strain.
Figure 2.
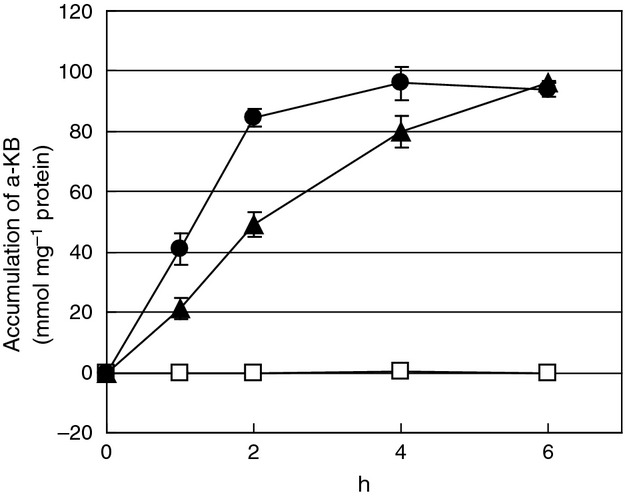
1-aminocyclopropane-1-carboxylic acid deaminase activity for various expression cassettes in A. tumefaciens. The accumulation of α-ketobutyrate in cell lysates was determined as described in the Materials and Methods section. The open squares, closed triangles, and closed circles indicate the A. tumefaciens GV2260 (pBBR1MCS-5), A. tumefaciens GV2260 (pBBRacdS), and A. tumefaciens GV2260 (pvirD1acdS) strains, respectively. The values were standardized to the value at 0 h. The error bars show the standard deviation (n = 3).
Efficiency of T-DNA delivery
Solanum lycopersicum cv. Moneymaker
To evaluate Agrobacterium-mediated T-DNA delivery, 60–80 S. lycopersicum explants from 7-day-old seedlings were prepared and inoculated with A. tumefaciens. Three types of A. tumefaciens strains [A. tumefaciens GV2260 (pBBR1MCS-5, pIG121-Hm), A. tumefaciens GV2260 (pBBRacdS, pIG121-Hm), and A. tumefaciens GV2260 (pvirD1acdS, pIG121-Hm)] were used in this experiment. The uidA gene was used as an indicator of T-DNA delivery, and the efficiency of T-DNA delivery was estimated by GUS staining. We determined the GUS-stained area in each of the explants with Image J, as described in the Materials and Methods. The degree of staining was categorized into six classes (Fig. 3A), and the frequency of each class was calculated (Fig. 3). This experiment was repeated three times. A. tumefaciens GV2260 (pBBRacdS, pIG121-Hm) decreased the frequency of S. lycopersicum explants with a low degree of staining (less than 5%) relative to the frequency for inoculation with A. tumefaciens GV2260 (pBBRMCS, pIG121-Hm) (Fig. 3B). Compared with A. tumefaciens GV2260 (pBBRacdS, pIG121-Hm), A. tumefaciens GV2260 (pvirD1acdS, pIG121-Hm) increased the frequency of high staining (more than 10%) and decreased the frequency of low staining (less than 5%) (Fig. 3B). All these results showed the same tendency in three repetitions. Therefore, the increases in ACC deaminase gene expression and activity improved the delivery of T-DNA by A. tumefaciens, and we succeeded in developing an A. tumefaciens strain capable of effectively transforming S. lycopersicum cells.
Figure 3.
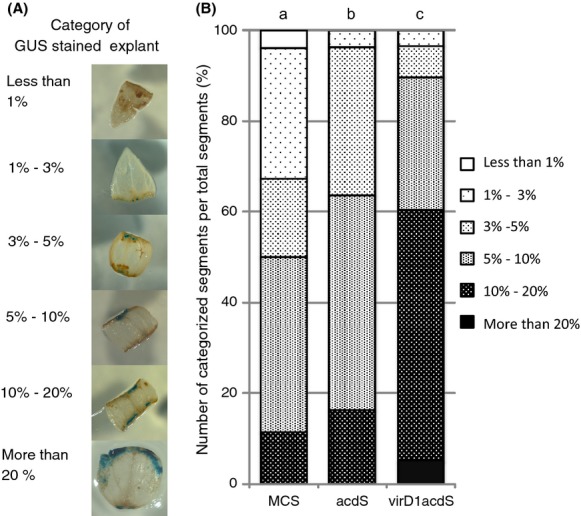
Estimation of the Agrobacterium-mediated T-DNA transfer efficiency in S. lycopersicum plants. (A) Classification of GUS-stained cotyledon explants. GUS-stained S. lycopersicum cotyledons were categorized based on the stained area: less than 1%, 1% to 3%, 3% to 5%, 5% to 10%, 10% to 20%, and more than 20%. (B) The frequency of each GUS staining category in S. lycopersicum explants. Bacterial strains resulting in significant differences (Student's T-test and Kruskal–Wallis test; P < 0.01) are indicated by different letters. MCS: A. tumefaciens GV2260 (pBBRMCS1-5, pIG121-Hm); acdS: A. tumefaciens GV2260 (pBBRacdS, pIG121-Hm); virD1acdS: A. tumefaciens GV2260 (pvirD1acdS, pIG121-Hm).
Erianthus ravennae
Erianthus ravennae calli subcultured every 2 weeks were inoculated with A. tumefaciens GV2260 (pBBR1MCS-5, pEKH2), A. tumefaciens GV2260 (pBBRacdS, pEKH2), or A. tumefaciens GV2260 (pvirD1acdS, pEKH2). The calli were cocultured in the dark for 3 days. The calli were then subcultured in callus induction medium containing 12.5 mg/L meropenem trihydrate for 1 day to eliminate A. tumefaciens. After the 1-day A. tumefaciens GV2260 elimination step, the calli were stained with GUS staining solution. Blue GUS-stained spots indicated T-DNA delivery into E. ravennae cells (Fig. 4A–C). To estimate the efficiency of Agrobacterium-mediated T-DNA delivery, the blue spots were counted. The number of blue spots per 1 g of calli was threefold higher with A. tumefaciens GV2260 (pBBRacdS, pEKH2) than with A. tumefaciens GV2260 (pBBR1MCS-5, pEKH2). ACC deaminase expression driven by the virD1 promoter resulted in the highest number of blue spots (Fig. 4D). A. tumefaciens GV2260 (pvirD1acdS, pEKH2) resulted in 1.5-fold blue spots compared with A. tumefaciens GV2260 (pBBRacdS, pEKH2), and 4.5-fold more blue spots than the control. The effect of increasing ACC deaminase gene expression in A. tumefaciens on the transformation efficiency was also evaluated by measuring the GUS activity fluorometrically (Fig. 4E). Inoculation with A. tumefaciens GV2260 (pvirD1acdS, pEKH2) increased the transient GUS activity by 1.5-fold relative to that for inoculation with A. tumefaciens GV2260 (pBBRacdS, pEKH2). This result was consistent with the number of GUS-stained spots (Fig. 4D).
Figure 4.
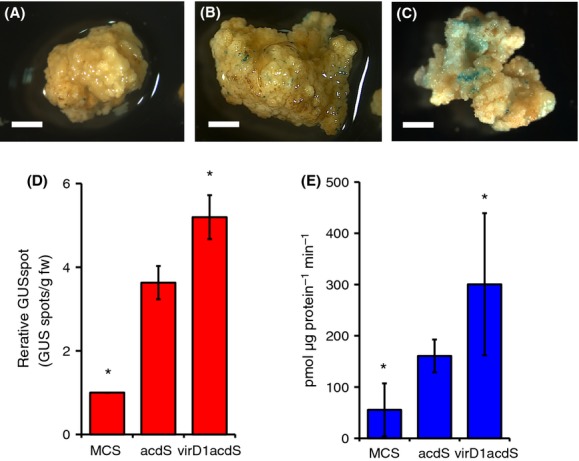
T-DNA delivery efficiency in E. ravennae. GUS-stained calli inoculated with A. tumefaciens (pBBRMCS1-5, pEKH2) (A), A. tumefaciens (pBBRacdS, pEKH2) (B), and A. tumefaciens (pviracdS, pEKH2) (C). Blue spots indicate transformed cells. The bar indicates 5 mm. (D) Occurrence of T-DNA transformation in E. ravennae. This graph shows the relative number of GUS spots. The number of GUS-stained spots per 1 g of E. ravennae calli was counted for each treatment. The bars indicate the standard deviation (n = 3). Asterisks indicate values that were significantly different from A. tumefaciens (pEKH2, pBBRacdS) inoculation according to the Student's T-test (P < 0.05). (E) Estimation of the transformation efficiency using the fluorometric GUS assay. GUS activity was measured immediately after cocultivation. The mean GUS activity ± SD was calculated from the results of three experiments. Approximately 2 g of E. ravennae calli was used in each experiment. Asterisks indicate statistically significant differences according to Student's T-test (P < 0.05). MCS: A. tumefaciens GV2260 (pBBRMCS1-5, pEKH2); acdS: A. tumefaciens GV2260 (pBBRacdS, pEKH2); virD1acdS: A. tumefaciens GV2260 (pvirD1acdS, pEKH2).
In this study, we increased the ACC deaminase gene expression and activity in A. tumefaciens GV2260 using the virD1 promoter, which, in turn, enhanced the ability of A. tumefaciens to deliver T-DNA into S. lycopersicum and E. ravennae cells. By enhancing Agrobacterium-mediated T-DNA delivery into plant cells, the number of stable transgenic plants might be increased. Ntui et al. (2010) and Hao et al. (2010) showed that the enhancement of Agrobacterium-mediated T-DNA delivery into plants by the addition of ACC deaminase driven by the lac promoter in A. tumefaciens increased the stable transformation efficiency in “Egusi” melon and canola cultivars, respectively. Therefore, A. tumefaciens GV2260, which harbors the ACC deaminase gene driven by the virD1 promoter and was created in this study, has the potential to increase the number of stable transgenic crop plants.
In this study, a newly produced strain of A. tumefaciens GV2260 (pvirD1acdS, pEKH2), was shown to deliver T-DNA to E. ravennae, which is an important biomass-producing plant, with improved efficiency. The effective production of plants for biomass that does not compete with food production is important for the utilization of biomass resources. In typical land usage patterns, the best land is used for food crop production and poor soil is used for biomass production. It is therefore necessary to make biomass-producing plants such as Erianthus and Sorghum tolerant to nutrient-deficient conditions through Agrobacterium-mediated transformation. However, reliable regeneration and transformation systems for Erianthus and Sorghum have not been established. A. tumefaciens GV2260, which harbors the ACC deaminase gene driven by the virD1 promoter and was established in this study, might contribute to the establishment of new reliable transformation systems for biomass-producing plants.
Acknowledgments
We would like to thank Prof. M. Mii (Chiba University, Japan) for kindly providing the E. ravennae calli. We also thank Dr. I. Nakamura (Chiba University, Japan) for the gift of the pEKH2 plasmid. This research was supported in part by the New Energy and Industrial Technology Development Organization (NEDO).
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. DNA primers used in this study.
References
- Anand A, Uppalapati SR, Ryu CM, Allen SN, Kang L, Tang Y, et al. Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol. 2008;146:703–715. doi: 10.1104/pp.107.111302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen CY, Winans SC. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J. Bacteriol. 1991;173:1139–1144. doi: 10.1128/jb.173.3.1139-1144.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME, Miller AR, Lineberger RD. Studies on the Effects of Ethylene on Transformation of Tomato Cotyledons (Lycopersicon esculentum Mill.) by Agrobacterium tumefaciens. J. Plant Pysiol. 1992;139:309–312. [Google Scholar]
- Deblaere R, Bytebier B, Greve HD, Deboeck F, Schell J, Montagu MV, et al. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 1985;13:4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezura H, Yuhashi K, Yasuta T, Minamisawa K. Effect of ethylene on Agrobacterium tumefaciens-mediated gene transfer to melon. Plant Breed. 2000;119:75–79. [Google Scholar]
- Han JS, Kim CK, Park SH, Hirschi KD, Mok I. Agrobacterium-mediated transformation of bottle gourd (Lagenaria siceraria Standl.) Plant Cell Rep. 2005;23:692–698. doi: 10.1007/s00299-004-0874-z. [DOI] [PubMed] [Google Scholar]
- Hao Y, Charles TC, Glick BR. ACC deaminase increases the Agrobacterium tumefaciens-mediated transformation frequency of commercial canola cultivars. FEMS Microbiol. Lett. 2010;307:185–190. doi: 10.1111/j.1574-6968.2010.01977.x. [DOI] [PubMed] [Google Scholar]
- Honma S, Shimomura T. Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 1978;42:1825–1831. [Google Scholar]
- Hoshikawa K, Ishihara G, Takahashi H, Nakamura I. Enhanced resistance to gray mold (Botrytis cinerea) in transgenic potato plants expressing thionin genes isolated from Brassicaceae species. Plant Biotechnol. 2012;29:87–93. [Google Scholar]
- Hwang HH, Wang MH, Lee YL, Tsai YL, Li YH, Yang FJ, et al. Agrobacterium-produced and exogenous cytokinin-modulated Agrobacterium-mediated plant transformation. Mol. Plant Pathol. 2010;11:677–690. doi: 10.1111/j.1364-3703.2010.00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klüsener S, Hacker S, Tsai YL, Bandow JE, Gust R, Lai EM, et al. Proteomic and transcriptomic characterization of a virulence-deficient phosphatidylcholine-negative Agrobacterium tumefaciens mutant. Mol. Genet. Genomics. 2010;283:575–589. doi: 10.1007/s00438-010-0542-7. [DOI] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Lee CW, Efetova M, Engelmann JC, Kramell R, Wasternack C, Ludwig-Müller J, et al. Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell. 2009;21:2948–2962. doi: 10.1105/tpc.108.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- Nonaka S, Yuhashi K, Takada K, Sugaware M, Minamisawa K, Ezura H. Ethylene production in plants during transformation suppresses vir gene expression in Agrobacterium tumefaciens. New Phytol. 2008a;178:647–656. doi: 10.1111/j.1469-8137.2008.02400.x. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Sugawara M, Minamisawa K, Yuhashi K, Ezura H. 1-Aminocyclopropane-1-carboxylate deaminase enhances Agrobacterium tumefaciens-mediated gene transfer into plant cells. Appl. Environ. Microbiol. 2008b;74:2526–2528. doi: 10.1128/AEM.02253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntui VO, Khan RS, Chin DP, Nakamura I, Mii M. An efficient Agrobacterium tumefaciens-mediated genetic transformation of “Egusi” melon (Colocynthis citrullus L.) Plant Cell Organ Cult. 2010;103:15–22. [Google Scholar]
- Ohta S, Mita S, Hattori T, Nakamura K. Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol. 1990;31:805–813. [Google Scholar]
- Pitzschke A, Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 2010;29:1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico A, Bennett MH, Forcat S, Huang WE, Preston GM. Agroinfiltration reduces ABA levels and suppresses Pseudomonas syringae-elicited salicylic acid production in Nicotiana tabacum. PLoS ONE. 2010;5:e8977. doi: 10.1371/journal.pone.0008977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel SE, Messens E, Montagu MV, Zambryski P. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium mumefaciens. Nature. 1985;318:624–629. [Google Scholar]
- Steck TR, Morel P, Kado CI. Vir box sequences in Agrobacterium tumefaciens pTiC58 and A6. Nucleic Acids Res. 1988;16:8736. doi: 10.1093/nar/16.17.8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Li J, Lacroix B, Citovsky V. Agrobacterium T-DNA integration: molecules and models. Trends Genet. 2004;20:375–383. doi: 10.1016/j.tig.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Yuan ZC, Edlind MP, Liu P, Saenkham P, Banta LM, Wise AA, et al. The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc. Natl. Acad. Sci. USA. 2007;104:11790–11795. doi: 10.1073/pnas.0704866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


