Abstract
In an attempt to develop a method to discriminate among isolates of Listeria monocytogenes, the sequences of all of the annotated genes from the fully sequenced strain L. monocytogenes EGD-e (serotype 1/2a) were compared by BLASTn to a file of the unfinished genomic sequence of L. monocytogenes ATCC 19115 (serotype 4b). Approximately 7% of the matching genes demonstrated 90% or lower identity between the two strains, and the lowest observed identity was 80%. Nine genes (hisJ, cbiE, truB, ribC, comEA, purM, aroE, hisC, and addB) in the 80 to 90% identity group and two genes (gyrB and rnhB) with approximately 97% identity were selected for multilocus sequence analysis in two sets of L. monocytogenes isolates (a 15-strain diversity set and a set of 19 isolates from a single food-processing plant). Based on concatenated sequences, a total of 33 allotypes were differentiated among the 34 isolates tested. Population genetics analyses revealed three lineages of L. monocytogenes that differed in their history of apparent recombination. Lineage I appeared to be completely clonal, whereas representatives of the other lineages demonstrated evidence of horizontal gene transfer and recombination. Although most of the gene sequences for lineage II strains were distinct from those of lineage I, a few strains with the majority of genes characteristic of lineage II had some that were characteristic of lineage I. Genes from lineage III organisms were mostly similar to lineage I genes, with instances of genes appearing to be mosaics with lineage II genes. Even though lineage I and lineage II generally demonstrated very distinct sequences, the sequences for the 11 selected genes demonstrated little discriminatory power within each lineage. In the L. monocytogenes isolate set obtained from one food-processing plant, lineage I and lineage II were found to be almost equally prevalent. While it appears that different lineages of L. monocytogenes can share habitats, they appear to differ in their histories of horizontal gene transfer.
Although disease caused by Listeria monocytogenes occurs at a low rate relative to those caused by other food-borne pathogens, the organism is second only to Salmonella spp. in the estimated number of food-related deaths in the United States (18). The majority of these deaths occur in individuals who are immunocompromised (6). With the continued high prevalence of AIDS, an increasingly elderly population, and numerous organ transplants, joint replacements, and other immunocompromising conditions, the number of people in the United States who are susceptible to listeriosis is growing.
Accurate tracing of Listeria strains is important for clinical epidemiology, food safety, and public health. Molecular tracing can help to manage and contain L. monocytogenes contamination in food-processing plants by giving a means to accurately evaluate the source of a specific contaminant. The last decade has seen a flurry of published studies in which inexpensive and rapid methods to type Listeria spp. have been designed and tested (5, 10, 11, 17, 29, 30, 32, 33). The overall goal has been to develop methods that are more discriminatory than existing serotyping and phage-typing methods. Ribotyping (10, 32) and pulsed-field gel electrophoresis of macrorestriction enzyme-digested chromosomal DNA (11) have demonstrated good discrimination of Listeria spp. However, the results are difficult to standardize between laboratories, making cooperative or retrospective studies difficult.
The current study began as an attempt to identify genes in L. monocytogenes with high levels of polymorphism that might serve as the basis for a discriminatory sequence-based typing method. Patterns of polymorphisms were evaluated in the partial DNA sequences of genes expected to be hypervariable, i.e., genes that demonstrated polymorphism in 10% or more of their DNA sequences in cross-strain comparisons of known sequences. However, a low degree of discrimination was found in each of 11 different genes tested, and the initial objective was not satisfied. The focus of our study thus shifted to examining the population genetics of L. monocytogenes and to understanding the biological underpinnings that may explain the lack of discriminatory power of the 11-gene multilocus sequencing analysis developed and implemented as described before.
Multilocus sequence typing was invented to enhance the study of the population genetics of bacteria (15). Multilocus sequence typing data can be analyzed to show probable recombination of genes between lineages in the evaluated population (15) and to probe for indications of recombination within genes (7). By combining ribotype and limited DNA sequence information, Wiedmann et al. (32) previously defined three distinct lineages of L. monocytogenes. Further DNA sequence analyses have confirmed the three lineages (4, 25), but the basis for the distinction of the lineages has not yet been defined. The multilocus sequence analysis described here allowed us to evaluate inheritance, mutation, and horizontal gene transfer in L. monocytogenes and uncover the recombinant construction of mosaic genes in three different patterns that were unique for each of the three previously described L. monocytogenes lineages.
MATERIALS AND METHODS
Whole-genome sequence analyses.
The file (accession no. AL591824.ffn) containing the DNA sequences of the annotated open reading frames of L. monocytogenes strain EGD-e (9) was retrieved from the Genome Database at the National Center for Biotechnology Information. A file with preliminary data on the genomic DNA sequence of L. monocytogenes ATCC 19115 (serotype 4b) was obtained from the Institute of Genomic Research at www.TIGR.org. The genomic data file was converted into a BLAST database and searched with the gene file by use of BLASTn (1). The output file was scanned to determine the percent identity of matches to the genes. Only genes that aligned along 90% or more of their length were considered, and the rest were rejected; thus, those alignments made with less than 100% of their length gave a slightly exaggerated level of identity because of the portion of the gene that was not considered.
Nine genes (Table 1) with 80 to 90% identity between the two strains of L. monocytogenes and two genes (gyrB and rnhB) with approximately 97% identity were chosen for further study. Primers were designed in conserved sequence segments to amplify these genes by PCR to give amplicons from 332 bp to as much as 1,331 bp.
TABLE 1.
Primers used for PCR and sequencing
| Gene | Locationa | Primerb | Annealing temp (°C) | Sequence lengthc (bp) |
|---|---|---|---|---|
| gyrB | 6,056-6,747 | f-5′ACAAGAAAATGCTTCAGATT3′ | 49 | 627 |
| r-5′GCTCCACGTAAGAACGA3′ | ||||
| hisJ | 607,296-607,939 | f-5′AAAAATAGTTGCTTATGG3′ | 45 | 530 |
| r-5′GCTGAAACTTCTGACAC3′ | ||||
| cbiE | 1,220,918-1,221,483 | f-5′GATTGGACCAGGAGAT3′ | 49 | 537 |
| r-5′CAATCACCACTACATTCA3′ | ||||
| rnhB | 1,296,153-1,296,484 | f-5′GCTATCGGAGTAGGG3′ | 47 | 275 |
| r-5′CCAATTGTATCTAAACCA3′ | ||||
| truB | 1,356,774-1,357,734 | f-5′CGGCATTATCCCACT3′ | 47 | 800 |
| r-5′AAATTCGAATTCCTCTCA3′ | ||||
| ribC | 1,357,735-1,358,785 | f-5′AGAGGAGAAGTGGCAAAA3′ | 51 | 885 |
| r-5′GGTGAGTCGCAAAAGC3′ | ||||
| comEA | 1,518,181-1,518,790 | f-5′ACTCCCTTATGATTTGAT3′ | 47 | 482 |
| r-5′TGTGCTGGTTTAGTTTAT3′ | ||||
| purM | 1,838,742-1,839,645 | f-5′CTGCCAACACCATACCAA3′ | 54 | 720 |
| r-5′AGTAAAGCAGGCGTGGAC3′ | ||||
| aroE | 1,999,439-1,998,282 | f-5′AATAAACAAGGGCTGGTT3′ | 49 | 951 |
| r-5′CATCATGCCAATACGG3′ | ||||
| hisC | 2,000,571-2,001,623 | f-5′TTCAAAAACGCCTCCAA3′ | 48 | 809 |
| r-5′CGCGAAGAAGAAGTGATG3′ | ||||
| addB | 2,358,489-2,359,819 | f-5′TCTTTTTCCCATTTCCAT3′ | 47 | 1,048 |
| r-5′CATATGTTCGGTGGTGAG3′ |
Location of primer as annotated for the L. monocytogenes EGD genome (6).
f, forward primer; r, reverse primer.
Length of analyzed sequence; the PCR product is longer.
Bacterial isolates.
Fifteen strains of L. monocytogenes, including five strains of each of the three lineages previously defined by ribotyping (4), were obtained from the Cornell University (Ithaca, N.Y.) Listeria strain collection. In addition, another 19 strains of L. monocytogenes from a ready-to-eat poultry processing facility (3) were used for sequence generation and were serotyped by the enzyme-linked immunosorbent assay-based procedure described by Palumbo et al. (24).
Template preparation and DNA sequencing.
One loopful of cells was taken from an overnight culture of each strain grown on BHI plates at 35°C. The cells were washed once in 0.5 ml of sterile distilled water and resuspended in 0.5 ml of sterile distilled water. The cells were then placed in a 100°C dry bath for 10 min. One microliter of the lysed cells was used for the PCR template. The conditions for PCR were as follows: denaturation at 95°C for 1 min, annealing at the appropriate temperature for each primer (Table 1) for 30 s, and extension at 72°C for 1 min, for a total of 30 cycles. PCR products were purified with the Qiagen PCR purification system (Qiagen, Valencia, Calif.), and Big Dye terminator sequencing reactions were performed with the protocol recommended by the manufacturer (Perkin-Elmer, Boston, Mass.) and the same primers that generated the templates for forward and reverse sequence determination. Sequences were read with an ABI 3700 capillary automated DNA sequencer (Perkin-Elmer), and a minimum of one forward and one reverse sequence reaction was used for each sequence. Table 1 indicates the lengths of the sequences that were analyzed, ranging from 275 bp to 1,048 bp.
Alignments and sequence analyses.
DNA sequences were aligned with ClustalX (28). No gaps were introduced into any of the alignments, and no additional editing was necessary. Clustering of the sequences was performed by the neighbor-joining algorithm with Jukes-Cantor distances by using the computer program PAUP* version 4.0b10 (27). Trees were rooted with sequences from the genomic sequence of Listeria innocua (9). Nucleotide diversity scores were determined by the method of Nei (20) as implemented in DnaSP (26). Sawyer's runs test for detecting recombination intervals based on the detection of shared patterns of polymorphisms was performed with the computer program GENECONV (S. A. Sawyer, 1999, Washington University, St. Louis, Mo., available at http://www.math.wustl.edu/∼sawyer). Each allotype, the genetically determined individual type, was assigned a two-part code. The first part of the code represented the allogroup, which was determined by cluster analysis of all the allotypes as described below, and the second part represented an identifier for each unique sequence.
RESULTS AND DISCUSSION
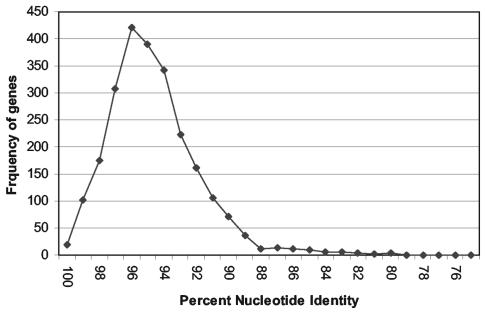
The available genomic sequences of L. monocytogenes strains EGD-e (9) and ATCC 19115 were compared with a simple BLAST search to identify hypervariable fragments. Matches were found for 2,418 genes. The distribution of gene identities as determined with BLASTn was nearly normal, with a mode at 96% identity and slight overrepresentation of identities between 80 and 90% (Fig. 1). These numbers were slightly exaggerated because for many genes, as much as 10% of the gene alignment may not have been considered by the BLASTn algorithm used. One-hundred seventy-one genes that had from 80 to 90% DNA sequence identity were identified. From this group of genes, nine genes that had been annotated as probable housekeeping genes in L. monocytogenes EGD (9) were chosen for further study (hisJ, cbiE, truB, ribC, comEA, purM, aroE, hisC, and addB). Two additional genes (gyrB and rnhB) with approximately 97% identity were chosen as more-conserved references.
FIG. 1.
Distribution of differences between paired genes of L. monocytogenes EGD and L. monocytogenes ATCC 19115 serotype 4b.
The DNA sequences of the 11 gene fragments were determined for 34 strains of L. monocytogenes, and the corresponding sequences from L. monocytogenes EGD were added to our database. Optimal alignments of all the sequences were obtained without the need for any gaps (indel sites). Ribbon diagrams of representative sequences (i.e., allogroups; see below) demonstrate the patterns of polymorphisms for each gene (Fig. 2). It was not surprising that polymorphisms were not uniformly distributed within a gene, and some regions of the genes were more conserved than others. Summary statistics of polymorphisms and diversity were determined (Table 2). These statistics may not be reflective of the global population of L. monocytogenes because the Cornell University collection overrepresents the number of lineage III organisms and the remaining isolates were all from one processing plant. Therefore, it was helpful to interpret the diversity statistics when they were broken down into the lineage categories (Table 2).
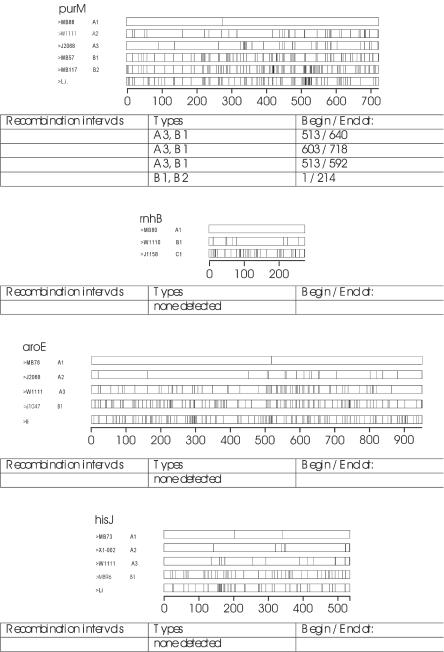
FIG. 2.
Ribbon diagrams of polymorphism sites for representatives of allogroups for each locus that was sequenced. Vertical bars within the locus indicate sites that differ from the consensus sequence. The analyses were weighted so that allogroup A1 for each locus is closest to the consensus. Under each set of ribbons are the results of Sawyer's analysis of recombination. The numbers indicate the regions of the sequences that are possible recombination junctions. An asterisk indicates recombination junctions that were not significant (P > 0.05) after Bonferroni's correction was applied.
TABLE 2.
Summary of sequence diversity for each gene for all the data and by lineage type
| Lineage (no. of isolates) | Gene (sequence length, bp) | No. of polymorphic sitesa (% of total) | No. of synonymous/ nonsynonymous sitesb | No. of allotypes | Allotype (gene) diversityc | Nucleotide diversityd (per site) | Avg. no. of differences (% of total) |
|---|---|---|---|---|---|---|---|
| All (35) | gyrB (627) | 39 (6.3) | 38/0 | 17 | 0.938 | 0.01801 | 11.292 (1.80) |
| hisJ (530) | 74 (13.9) | 53/21 | 15 | 0.882 | 0.04615 | 24.461 (4.62) | |
| cbiE (537) | 107 (19.3) | 67/46 | 13 | 0.845 | 0.07207 | 38.703 (7.21) | |
| rnhB (275) | 40 (14.6) | 28/13 | 9 | 0.755 | 0.02446 | 6.726 (2.45) | |
| truB (800) | 163 (20.4) | 115/52 | 23 | 0.945 | 0.06516 | 52.126 (6.52) | |
| ribC (885) | 155 (17.5) | 126/33 | 19 | 0.931 | 0.06014 | 53.22 (6.01) | |
| comEA (482) | 93 (19.3) | 46/47 | 7 | 0.689 | 0.08435 | 40.655 (8.44) | |
| purM (720) | 150 (20.8) | 118/37 | 23 | 0.953 | 0.05815 | 41.871 (5.82) | |
| aroE (951) | 194 (20.4) | 171/23 | 26 | 0.958 | 0.0657 | 62.26 (6.55) | |
| hisC (809) | 141 (17.4) | 110/35 | 22 | 0.948 | 0.05943 | 48.081 (5.94) | |
| addB (1,048) | 185 (17.7) | 129/65 | 17 | 0.852 | 0.0744 | 77.97 (7.44) | |
| Lineage I (17) | gyrB | 10 | 6 | 0.816 | 0.0053 | 3.324 (0.53) | |
| hisJ | 2 | 3 | 0.640 | 0.00142 | 0.75 (0.01) | ||
| cbiE | 9 | 5 | 0.728 | 0.0049 | 2.632 (0.49) | ||
| rnhB | 3 | 4 | 0.331 | 0.00128 | 0.353 (0.01) | ||
| truB | 9 | 8 | 0.801 | 0.00274 | 2.191 (0.27) | ||
| ribC | 7 | 7 | 0.794 | 0.00311 | 2.75 (0.31) | ||
| comEA | 2 | 2 | 0.309 | 0.00128 | 0.618 (0.13) | ||
| purM | 9 | 9 | 0.831 | 0.00343 | 2.471 (0.34) | ||
| aroE | 13 | 9 | 0.824 | 0.00407 | 3.868 (0.41) | ||
| hisC | 7 | 8 | 0.816 | 0.00245 | 1.985 (0.25) | ||
| addB | 2 | 3 | 0.404 | 0.00041 | 0.426 (0.04) | ||
| Lineage II (13) | gyrB | 15 | 7 | 0.872 | 0.01014 | 6.359 (1.01) | |
| hisJ | 11 | 7 | 0.731 | 0.00421 | 2.231 (0.42) | ||
| cbiE | 72 | 3 | 0.295 | 0.0211 | 11.333 (2.11) | ||
| rnhB | 2 | 3 | 0.564 | 0.00224 | 0.615 (0.22) | ||
| truB | 119 | 10 | 0.923 | 0.06806 | 54.449 (6.81) | ||
| ribC | 103 | 7 | 0.833 | 0.02256 | 19.962 (2.26) | ||
| comEA | 2 | 3 | 0.295 | 0.00064 | 0.308 (0.06) | ||
| purM | 134 | 10 | 0.949 | 0.06537 | 47.064 (6.54) | ||
| aroE | 122 | 12 | 0.987 | 0.03594 | 34.179 (3.59) | ||
| hisC | 104 | 9 | 0.923 | 0.02956 | 23.91 (2.96) | ||
| addB | 11 | 9 | 0.91 | 0.00203 | 2.128 (0.20) | ||
| Lineage III (5) | gyrB | 20 | 5 | 1 | 0.01499 | 9.4 (1.50) | |
| hisJ | 27 | 5 | 1 | 0.02491 | 13.2 (2.49) | ||
| cbiE | 57 | 5 | 1 | 0.0473 | 25.4 (4.73) | ||
| rnhB | 36 | 4 | 0.9 | 0.05345 | 14.7 (5.35) | ||
| truB | 56 | 5 | 1 | 0.03725 | 29.8 (3.73) | ||
| ribC | 59 | 5 | 1 | 0.03412 | 30.2 (3.41) | ||
| comEA | 81 | 3 | 0.7 | 0.09398 | 45.3 (9.40) | ||
| purM | 48 | 5 | 1 | 0.03556 | 25.6 (3.56) | ||
| aroE | 66 | 5 | 1 | 0.04143 | 39.4 (4.14) | ||
| hisC | 66 | 5 | 1 | 0.04734 | 38.3 (4.73) | ||
| addB | 68 | 5 | 1 | 0.03473 | 36.4 (3.47) |
Number of polymorphic sites, total number of base positions in aligned sequences with more than one base.
Some sites are counted as both, and uncertain sites are not counted. Only sites are presented and not rates because rate calculations are not valid with recombination.
Allotype diversity is the probability that two gene sequences drawn at random will have different allotypes.
Nucleotide diversity is the probability that a given base position from two gene sequences drawn at random will have different bases.
aroE had the greatest number of allotypes (26 allotypes), and purM had the greatest proportion of polymorphic sites (20.83% of total sites). truB represented the only gene analyzed in which the nucleotide diversity for lineage III was lower than that seen in lineage II. gyrB had the lowest proportion of polymorphic sites and the lowest nucleotide diversity yet still provided differentiation into 17 allotypes. comEA yielded the lowest number of allotypes (seven allotypes) but still displayed a large number of polymorphic sites (19.29% of sites). With the exception of cbiE and comEA, allotype diversity was lower among lineage I isolates than among lineage II strains. The comEA and rnhB sequences were the only ones that did not separately distinguish all five lineage III isolates, even though these two genes had the first and second highest (respectively) nucleotide diversities of any gene sequenced.
When all the data were analyzed as a combined (concatenated) sequence, 33 allotypes were distinguished among the 34 isolates characterized. The level of allotype diversity demonstrated by gyrB was surprising, given the small number of polymorphic sites in this gene and the incomplete resolving power of the other genes that were chosen based on the large differences seen in previously known sequences. In another recently completed study with the Cornell University collection of isolates, the sequences of actA demonstrated complete resolution of all 15 isolates (4), whereas in this study, the sequences of a minimum of seven genes (aroE, comEA, gyrB, hisC, purM, ribC, and rnhB) were required to achieve the same resolution.
To help understand the diversity patterns observed for the 11 genes analyzed, cluster analyses were performed for each of the genes by the neighbor-joining algorithm implemented in PAUP* with Jukes-Cantor distances, with L. innocua gene sequences used to serve as roots for the trees. The resulting trees should not be interpreted as phylogenetic reconstructions because of the evidence of recombination discussed below but do serve as a means of classifying the alleles. The trees demonstrated a spectrum of shapes that were generally characterized by the existence of two populations that were deeply removed from a common node. The spectrum of tree topologies varied from that seen for addB (Fig. 3), in which there appeared to be only very recent divergence from two clones, to that seen for truB (Fig. 3), in which there were several clusters branching closer to the root. The clonal history of the strains was greatly simplified yet still informative if the isolates were assigned to allogroups rather than individual allotypes. Allotypes were assigned the letter A if they clustered on the side of the root that was characteristic of lineage I organisms. Conversely, allotypes that were characteristic of lineage II allotypes were assigned the letter B. In two cases, aroE and rnhB, there were allotypes that branched close enough to the root to deserve a separate group designation of C.
FIG. 3.
Dendrograms for addB (left) and truB (right). The dendrograms were constructed by neighbor joining of Jukes-Cantor's distances as implemented in PAUP* version 4. The allogroups are illustrated by the ellipses.
The groups were further divided by the appending of a numeric designation to demonstrate clusters that branched off successively closer to the root. In the case of truB (Fig. 3), groups were labeled A1 for types that were characteristic of the lineage I isolates, and clusters more deeply removed from that type were successively labeled A2, A3, etc., through A7. Separate clusters on the lineage II side of the root were handled similarly to give allogroup designations of B1, B2, and so forth. The assignment of allogroups along with unique allotype identifiers allowed the tabular visualization of genes that were certain to have similar clonal histories. The allogroup designations for the 11 genes for all the isolates examined are tabulated in Table 3. Table 3 clearly demonstrated that there are several instances where allogroups that were characteristic of lineage I were found in lineage II strains. Lineage III strains were predominantly characterized by allogroups that were most similar to those of lineage I strains and a few allogroups that were more characteristic of lineage II strains. hisJ was the only locus that had no incongruent alleles (Table 3).
TABLE 3.
Allelic assignments for each strain testeda
| Strain | Lineage | Serotype | Allelic assignmenta
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gyrB | hisJ | cbiE | rnhB | truB | ribC | comEA | purM | aroE | hisC | addB | |||
| MB79 | ?b | 1/2b | A1-001 | A1-002 | A1-001 | A1-001 | A1-001 | A1-007 | A1-001 | A1-001 | A1-001 | A1-001 | A1-001 |
| FSL J1-038 | I | 1/2b | A1-005 | A1-002 | A1-002 | A1-001 | A1-006 | A1-004 | A1-001 | A1-007 | A1-002 | A1-006 | A1-003 |
| FSL J2-064 | I | 1/2b | A1-001 | A1-002 | A1-001 | A1-001 | A1-001 | A1-007 | A1-001 | A1-001 | A1-001 | A1-002 | A1-001 |
| FSL J1-051 | I | 4b | A1-003 | A1-003 | A1-003 | A1-001 | A1-004 | A1-004 | A1-001 | A1-006 | A1-004 | A1-005 | A1-001 |
| FSL J2-039 | I | 4b | A1-003 | A1-002 | A1-005 | A1-002 | A1-003 | A1-006 | A1-001 | A1-009 | A1-006 | A1-008 | A1-003 |
| FSL J2-045 | I | 4b | A1-003 | A1-003 | A1-003 | A1-004 | A1-008 | A1-005 | A1-001 | A1-005 | A1-009 | A1-007 | A1-002 |
| MB62 | I | 1/2b | A1-006 | A1-001 | A1-002 | A1-001 | A1-006 | A1-001 | A1-002 | A1-003 | A1-005 | A1-003 | A1-001 |
| MB67 | I | 4e | A1-003 | A1-003 | A1-004 | A1-001 | A1-006 | A1-003 | A1-001 | A1-008 | A1-003 | A1-004 | A1-003 |
| MB73 | I | 1/2b | A1-002 | A1-002 | A1-001 | A1-001 | A1-002 | A1-007 | A1-001 | A1-001 | A1-001 | A1-001 | A1-001 |
| MB76 | I | 1/2b | A1-006 | A1-001 | A1-002 | A1-005 | A1-002 | A1-002 | A1-003 | A1-005 | A1-003 | A1-001 | |
| MB80 | I | 1/2b | A1-004 | A1-003 | A1-002 | A1-001 | A1-001 | A1-004 | A1-001 | A1-008 | A1-008 | A1-003 | A1-001 |
| MB81 | I | 3b | A1-001 | A1-002 | A1-001 | A1-001 | A1-001 | A1-007 | A1-001 | A1-001 | A1-001 | A1-001 | A1-001 |
| MB82 | I | 1/2b | A1-004 | A1-003 | A1-002 | A1-001 | A1-007 | A1-004 | A1-001 | A1-002 | A1-009 | A1-003 | A1-001 |
| MB88 | I | 1/2b | A1-001 | A1-002 | A1-001 | A1-001 | A1-001 | A1-007 | A1-001 | A1-001 | A1-001 | A1-001 | A1-001 |
| MB113 | I | 1/2b | A1-006 | A1-001 | A1-002 | A1-001 | A1-006 | A1-001 | A1-002 | A1-004 | A1-005 | A1-003 | A1-001 |
| MB121 | I | 3b | A1-001 | A1-002 | A1-001 | A1-001 | A1-001 | A1-007 | A1-001 | A1-001 | A1-001 | A1-001 | A1-001 |
| MB125 | I | 3b | A1-001 | A1-002 | A1-001 | A1-003 | A1-001 | A1-007 | A1-001 | A1-001 | A1-001 | A1-001 | A1-001 |
| FSL C1-117 | II | 1/2a | B2-016 | B1-009 | B1-012 | B1-005 | A4-015 | B1-018 | B-005 | B1-019 | B1-020 | B1-011 | B1-010 |
| FSL J2-003 | II | 1/2a | B2-017 | B1-011 | A3-008 | B1-007 | A6-017 | B1-015 | B1-005 | A3-014 | B1-018 | B1-014 | B1-009 |
| FSL J2-017 | II | 1/2a | B2-017 | B1-010 | B1-012 | B1-005 | B2-022 | B1-014 | B1-005 | B2-024 | B1-022 | B1-011 | B1-011 |
| FSL J1-022 | II | 1/2c | B1-011 | B1-009 | B1-012 | B1-006 | B1-018 | B1-014 | B1-005 | A1-009 | B1-016 | B1-009 | B1-017 |
| FSL J1-047 | II | 1/2c | B1-011 | B1-009 | B1-012 | B1-006 | B1-018 | B1-014 | B1-005 | B1-018 | B1-017 | B1-009 | B1-017 |
| MB17 | II | 1/2a | B1-015 | B1-009 | B1-012 | B1-005 | B1-019 | B1-017 | B1-005 | B1-022 | B1-015 | B1-013 | B1-011 |
| MB57 | II | 1/2a | B1-011 | B1-009 | B1-012 | B1-006 | B1-018 | B1-014 | B1-005 | B1-017 | B1-019 | B1-010 | B1-017 |
| MB58 | II | 1/2a | B1-013 | B1-015 | B1-012 | B1-005 | A3-013 | A2-008 | B1-005 | B1-021 | B1-021 | B1-012 | B1-016 |
| MB96 | II | 1/2a | B2-016 | B1-009 | B1-012 | B1-005 | B1-020 | B1-019 | B1-005 | A3-016 | B1-024 | A3-020 | B1-012 |
| MB98 | II | 1/2a | B1-014 | B1-012 | B1-013 | B1-005 | B1-021 | B1-017 | B1-005 | B1-022 | C-025 | B1-015 | B1-014 |
| MB99 | II | 1/2a | B1-012 | B1-014 | B1-012 | B1-005 | A7-023 | B1-017 | B1-005 | B1-020 | C-025 | A2-018 | B1-013 |
| MB117 | II | 3a | B2-017 | B1-013 | B1-012 | B1-005 | A5-016 | B1-016 | B1-005 | B2-023 | B1-023 | B1-011 | B1-015 |
| FSL X1-002 | III | 4a | A2-009 | A2-004 | A3-007 | B1-005 | A3-011 | A2-009 | A3-004 | A3-015 | A2-010 | A2-017 | B2-008 |
| FSL J1158 | III | 4b | A2-007 | A3-008 | A4-010 | C-009 | A2-010 | A2-013 | A2-003 | A2-011 | A3-014 | A4-022 | B3-004 |
| FSL J2068 | III | 4c | A2-008 | A2-005 | A2-006 | B1-006 | A3-012 | A2-010 | B1-005 | A3-013 | A2-011 | A2-016 | B2-006 |
| FSL W1110 | III | 4c | A2-010 | A2-006 | A3-009 | B1-008 | A3-014 | A2-011 | B1-005 | A3-012 | A2-012 | A2-019 | B2-007 |
| FSL W1111 | III | 4c | B2-016 | A3-007 | A5-011 | B1-005 | A2-009 | A2-012 | B1-005 | A2-010 | A3-013 | A4-021 | B3-005 |
| EGDc | 1/2a | B1-011 | B1-009 | B1-012 | B1-006 | B1-018 | B1-014 | B1-006 | B1-017 | B1-019 | B1-009 | B1-017 | |
Allelic designations are formatted so that the alphanumeric to the left of the hyphen indicates an allogroup assignment as explained in the text, and the number to the right of the hyphen is an arbitrary unique allelic designation. Alleles in bold are incongruent with lineage and therefore are indications of probable recombination.
The lineage type based on the ribotype could not be determined for strain 79.
Sequences for L. monocytogenes EGD were obtained from the GenBank file (accession no. AL591824.ffn); the lineage type for this strain was not previously determined, but the data are consistent with lineage II.
No single gene could be used to reliably differentiate all three lineages, but analysis of both addB and hisJ sequences allowed differentiation of the three lineages. All of the addB sequences for lineage I clustered in a single allogroup, and addB sequences for lineage II clustered in another allogroup, but the addB sequences for lineage III clustered close to the lineage II strains. The clustering of the hisJ sequences placed all of the lineage II strains in a single allogroup, and the hisJ sequences of the lineage III strains clustered close to those of the lineage I strains. It is probable that sequences of a larger population will make lineage discrimination more difficult unless there are specific barriers to sharing specific genes. All of the lineage I organisms had allotypes that were tightly clustered into a single allogroup for each gene analyzed. This allotype distribution was indicative of clonal development of the lineage I isolates. The lineage II and lineage III organisms differed by the side of the root on which most of the genes fell; lineage II organisms were mostly very different from lineage I organisms, and lineage III organisms had allogroups that were mostly more similar to lineage I types.
The differences in the tree topologies could be explained with respect to the effects of bottlenecks and recombination on the origins of the allogroups. The tree seen for addB (Fig. 3) would be expected for a recent severe bottleneck that only allowed two clones to survive. Since housekeeping genes are very unlikely to be subject to loss and recovery, a purifying selection that would put only one housekeeping gene through a bottleneck cannot occur; so all the genes (i.e., all the housekeeping genes in the genome) were put through a bottleneck at the same time. Assuming that such a bottleneck did occur, the allogroups that clustered apart from the characteristic types may have arisen by one of two possible mechanisms: disparate allogroups may have been pulled through the bottlenecks by virtue of linkage to the trait needed for success in the bottleneck, or the noncharacteristic allogroups may represent recombination junctions within the genes that were sequenced, resulting in mosaic genes, and thus average the distance between the extremely different genes.
Recombination analyses were performed with the data from all the sequenced genes to test the latter possibility. Sawyer's test for recombination (http://www.math.wustl.edu/∼sawyer), implemented in the program GENECONV, was performed with each of the sequence alignments. The results are illustrated in Fig. 2. Sawyer's test is not the most sensitive method of detecting recombination, but it is one of the few tests that annotate the breakpoints in individual sequences, also illustrated in Fig. 2. Many of the indicated recombination intervals were not significant (P > 0.05) after correction when Bonferroni's correction for multiple pairwise comparisons (http://www.math.wustl.edu/∼sawyer) was applied (denoted by *), but inspection of the ribbon diagrams demonstrates that some of the putative recombination junctions may not have been statistically significant due to the short length of the possible recombinant fragment (such as seen with addB, comEA, and ribC) or the paucity of polymorphisms in the possible recombinant region (e.g., in cbiE and gyrB).
The only test for recent bottlenecks relies on the overrepresentation of heterozygotes and hence is limited to diploid organisms. Therefore, we could only rely on the structure of cluster analysis dendrograms (especially Fig. 3, addB) to conclude that there was a bottleneck. Recombination will result in the reconstruction of trees that are not accurate phylograms. The effect of recombination is seen as a homogenization of the populations, meaning that the average difference between the populations will be reduced. Therefore, recombination will obscure signs of bottlenecks in phylogenetic trees, and to still have a clear signal of bottlenecks in such a phylogram may indicate a bottleneck of great magnitude or one that occurred fairly recently.
It can also be concluded that the lineage I L. monocytogenes strains are clonal and the lineage II organisms are active in recombination, with recombination intervals that transfer entire genes (evident from Table 3) and in events that create mosaic genes (as seen in Fig. 2). The lineage III L. monocytogenes were more similar to the lineage I organisms than the lineage II organisms, but they also demonstrated evidence of recombination with lineage II organisms at the whole-gene level (Table 3). The only significant (P < 0.05 after Bonferroni's correction) recombination interval within the sequences found for lineage III organisms was found in truB. Since lineage III L. monocytogenes strains are rare, it is possible that some allotypes were pulled through the bottlenecks, but we believe that they probably represent mosaic genes constructed in recombination-active organisms that otherwise would have been lineage I. Perhaps sequence analysis of a larger population will clarify this incomplete analysis.
Given a clonally developing lineage, the number of base changes between isolates can be used to estimate the time since the last common ancestor. Since the lineage I organisms appeared to be clonal, we applied evolutionary clock analysis to estimate when the founder to this lineage existed. With a divergence rate for synonymous nucleotide changes of 0.90% per million years (22, 23) and Li's estimate of the rate of synonymous base changes (26), the time since the founder of lineage I existed was estimated to be about one-half million years. If Nei and Gojobori's estimate of the rate of synonymous base changes (21) is used instead, the estimated time since the founder existed was extended to about one million years ago. This observation may indicate that lineage I L. monocytogenes arose before any influence from high-density human populations could be expected. Similar analyses to find the most recent common ancestor for lineage I organisms and organisms that have all B1 allotypes (thus, lineage II organisms with the least amount of evident recombination with lineage I) gave estimates of 30 to 67 million years by the two methods of estimation of the synonymous substitution rate. However, the times to the most recent common ancestor could be influenced by the selection of the genes that were sequenced.
The sequences used in the analyses presented here (i.e., sequences that were chosen as being hypervariable) may be diverging at a rate approximating two to threefold that seen for most L. monocytogenes genes. Also, the clock that was applied was developed for Salmonella enterica and Escherichia coli, and the accuracy of this clock would be affected by the population size and the number of generations the organism has per year. Since L. monocytogenes can replicate at lower temperatures than Salmonella spp. or E. coli, it is possible that there are more generations per year for Listeria in environmental niches, and thus the estimates of the time to the founder member of the clones are overly long. Our estimate for the time to the most recent common ancestor of lineage I and lineage II L. monocytogenes was about half as long as that since the last common ancestor of S. enterica serovar Typhimurium and E. coli (23).
It is also noteworthy that we were able to isolate 17 different strains of L. monocytogenes representing both lineage I and lineage II organisms from one food-processing plant. While we are unable to describe any environmental factors that may segregate these two types, it is possible that strains representing lineages occupy microniches within a food-processing plant that are not easily defined. Given the wide variety of environments in which L. monocytogenes is found and the variety of conditions in which the organism can grow (6), we would expect that bottlenecks due to environmental constraints would affect only a limited portion of the clones existing at the time, so an environmental bottleneck seems unlikely. An alternative explanation is that a clone arose that had a greatly increased selective advantage and replaced all the noncompetitive clones. If this successful clone was a lineage I organism, it is possible that lineage II and III organisms, because of their recombination ability, were able to obtain the gene(s) required for the selective advantage and thus were able to avoid extinction.
The population structure that we observed for L. monocytogenes is similar to the one described for species in the genus Mycobacterium. The commonly pathogenic mycobacteria (M. tuberculosis, M. leprae, and M. avium subsp. paratuberculosis) appeared to have suffered a bottleneck as recently as 10 to 15 thousand years ago, and the species of mycobacteria that are commonly environmental were not clonal (8, 14). L. monocytogenes lineage I strains are significantly more common among human clinical cases and have been responsible for more than 80% of human listeriosis outbreaks (12). Furthermore, the vast majority of serotype 1/2b and 4b strains are lineage I (19), and these two serotypes are the most common ones responsible for human listeriosis outbreaks (32). All epidemic-associated serotype 4b and 1/2b strains characterized to date were classified as lineage I (12; M. Wiedmann, B. Saunders, E. Fortes, and K. Windham, unpublished data). While some human listeriosis outbreaks have been associated with lineage II strains (i.e., serotype 1/2a strains), no human outbreaks have been linked to lineage III strains (12, 31). Lineage III strains are occasionally involved in human cases (12) but represented about 10% of the animal clinical isolates characterized by Jeffers et al. (12).
Interestingly, our study shows that for L. monocytogenes, as was seen for Mycobacterium spp., the more virulent strains are clonal. According to Muller's ratchet hypothesis, asexual populations will accumulate deleterious mutations over time through random genetic drift (2). It can therefore be concluded that the clonal lineages have become inextricably linked in their association with a host because of losses of capabilities for more general environmental proliferation. This is not to say that lineage I strains are dependent on humans for survival, but that there may be a human-related bottleneck that results in the cohesion of the observed lineages. An assumption of Muller's ratchet hypothesis is that all mutations are neutral or deleterious, i.e., advantageous mutations are rare enough to be ignored entirely (2). However, advantageous mutations do occur, albeit rarely. It is possible that rare advantageous mutations gave a single clone a selective advantage that allowed the clone to sweep the pathogenic niche (16), but the apparent simultaneous bottleneck in less virulent L. monocytogenes strains suggests that replacement of clones with a fitter clone was not likely due to a new advantageous mutation.
Variation in the mutation rate over different fragments of a genome has been observed (13), and the distance of our “hypervariable” genes between lineage I and lineage II may be due to localized increased mutation rate. As noted above, the frequency of genes with a 10 to 20% difference between the two types showed a slight overrepresentation that may indicate a distinct population of genes, possibly a set of genes that were involved in cross-species recombination. However, recombination clearly occurred within sequenced conserved genes (gyrB and rnhB) (Table 3) as well, and recombination appears evident only in strains representing lineages II and III. More data from related species will be beneficial in demonstrating the role of cross-species recombination. While a study with a larger population would be helpful to further probe the population structure and recombination history of L. monocytogenes lineages, a widely and randomly collected L. monocytogenes isolate set is not currently available. Further studies on the different L. monocytogenes lineages may be helpful to improve our understanding of host-parasite adaptation and the food-borne transmission characteristics of the three L. monocytogenes lineages.
Acknowledgments
This work was supported in part by National Institutes of Health award R01GM63259 (to M.W.).
REFERENCES
- 1.Altschul, S. E., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, D. T., and D. Hughes. 1996. Muller's ratchet decreases fitness of a DNA-based microbe. Proc. Natl. Acad. Sci. USA 93:906-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berrang, M. E., R. J. Meinersmann, J. K. Northcutt, and D. P. Smith. 2002. Molecular characterization of Listeria monocytogenes isolated form a poultry further processing facility and fully cooked product. J. Food Prot. 65:1574-1579. [DOI] [PubMed] [Google Scholar]
- 4.Cai, S., D. Y. Kabuki, Y. Kuaye, T. G. Cargioli, M. S. Chung, R. Nielsen, and M. Wiedmann. 2002. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes. J. Clin. Microbiol. 40:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Cesare, A., J. L. Bruce, T. R. Dambaugh, M. E. Guerzoni, and M. Wiedmann. 2001. Automated ribotyping using different enzymes to improve discrimination of Listeria monocytogenes isolates, with a particular focus on serotype 4b strains. J. Clin. Microbiol. 39:3002-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 8.Frothingham, R. 1999. Evolutionary bottlenecks in the agents of tuberculosis, leprosy, and paratuberculosis. Med. Hypoth. 52:95-99. [DOI] [PubMed] [Google Scholar]
- 9.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 10.Graves, L. M., B. Swaminathan, M. W. Reeves, S. B. Hunter, R. E. Weaver, B. D. Plikaytis, and A. Schuchat. 1994. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J. Clin. Microbiol. 32:2936-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 12.Jeffers, G. T., J. L. Bruce, P. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 13.Jordon, I. K., I. B. Rogozin, Y. I. Wolf, and E. V. Koonin. 2002. Microevolutionary genomics of bacteria. Theor. Pop. Biol. 61:435-447. [DOI] [PubMed] [Google Scholar]
- 14.Kapur, V., T. S. Whittam, and J. M. Musser. 1994. Is Mycobacterium tuberculosis 15,000 years old? J. Infect. Dis. 17:1348-1349. [DOI] [PubMed] [Google Scholar]
- 15.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maynard Smith, J., and J. Haigh. 1974. The hitch-hiking effect of a favorable gene. Genet. Res. 23:23-35. [PubMed] [Google Scholar]
- 17.McLauchlin, J., M. D. Hampton, S. Shah, E. J. Threlfall, A. A. Wieneke, and G. D. Curtis. 1997. Subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibility. J. Appl. Microbiol. 83:381-388. [DOI] [PubMed] [Google Scholar]
- 18.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, N.Y.
- 21.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 22.Ochman, H., S. Elwyn, and N. A. Moran. 1999. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. USA 96:12638-12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochman, H., and A. C. Wilson. 1987. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J. Mol. Evol. 26:74-86. [DOI] [PubMed] [Google Scholar]
- 24.Palumbo, J. D., M. K. Borucki, R. E. Mandrell, and L. Gorski. 2003. Serotyping of Listeria monocytogenes by enzyme-linked immunosorbent assay and identification of mixed-serotype cultures by colony immunoblotting. J. Clin. Microbiol. 41:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 26.Rozas, J., and R. Rozas. 1995. DnaSP, DNA sequence polymorphism: an interactive program for estimating Population Genetics parameters from DNA sequence data. Comput. Appl. Biosci. 11:621-625. [DOI] [PubMed] [Google Scholar]
- 27.Swofford, D. L. 1998. Paup* 4.0b4: phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Mee-Marquet, N., M. Loessner, and A. Audurier. 1997. Evaluation of seven experimental phages for inclusion in the international phage set for the epidemiological typing of Listeria monocytogenes. Appl. Environ. Microbiol. 63:3374-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wernars, K., P. Boerlin, A. Audurier, E. G. Russell, G. D. Curtis, L. Herman, and N. van der Mee-Marquet. 1996. The WHO multicenter study on Listeria monocytogenes subtyping: random amplification of polymorphic DNA (RAPD). Int. J. Food Microbiol. 32:325-341. [DOI] [PubMed] [Google Scholar]
- 31.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524-531. [PubMed] [Google Scholar]
- 32.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng, W., and S. Kathariou. 1995. Differentiation of epidemic-associated strains of Listeria monocytogenes by restriction fragment length polymorphism in a gene region essential for growth at low temperatures (4oC). Appl. Environ. Microbiol. 61:4310-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]