Abstract
Objectives
Inherited genetic variability contributes to susceptibility to cervical cancer. We investigated the association of single nucleotide polymorphisms (SNPs) in the human epidermal growth factor receptor (ErbB) family with cervical cancer.
Methods
We used the transmission disequilibrium test (TDT) to look for excessive transmission of tag single nucleotide polymorphisms (tSNPs) in ERBB family members EGFR, ERBB2, ERBB3, and ERBB4 in a large sample of women with invasive and in situ cervical cancer and their biological parents (628 trios). The study used a discovery set of trios (244) analyzed by Ilumina GoldenGate in which SNPs reaching a P< .05 were re-tested by TaqMan in the combined set of 628. We also explored collaborative effects of different ERBB alleles.
Results
Based on single SNP TDT tests we identified 16 SNPs significant in the discover stage and six of 14 SNPs that could be assayed by TaqMan were significantly overtransmitted in women with cervical cancer in the combined replication set. Four SNPs were located in intron 1 of EGFR and two SNPs in intron 24 of ERBB4 The EGFR variants are located near multiple enhancers, silencers, and the previously identified functional common polymorphisms in intron 1.
Conclusions
Our data provide evidence for the involvement of intron 1 EGFR variants and intron 24 ERBB4 variants in modulating risk for the development of in situ and invasive cervical cancer. These variants should be examined in additional populations and functional studies would be needed to confirm this hypothesis.
Keywords: EGFR, ERBB4, cervical cancer, polymorphism, Transmission disequilibrium test
Introduction
Persistent infection by high-risk human papillomavirus (HPV) is important for the progression of HPV infection to cervical cancer [1]. However, only a small proportion of HPV-infected women develop cervical cancer [2], which suggests that variation in the host genome is also important. We and others have previously identified inherited genetic polymorphisms in host immunoregulatory and HPV-interacting genes [3–6]. Here, we present additional data on genetic variation in the ERBB family of tyrosine kinase transmembrane receptors using a family-based study. By obtaining case-control information from the genotypes of cervical cancer patients, their fathers and mothers, the study matches each proband and its controls by ethnicity. The study examines whether the frequency of transmission of parental marker alleles to affected offspring deviates from the expected Mendelian frequency of 50%.
The ErbB family of membrane-bound tyrosine kinase receptors consists of Egfr, ErbB2, ErbB3, and ErbB4, which activate potent signaling pathways. Homo-or hetero-dimerization of family members induces phosphorylation of tyrosine residues on the intracellular portion of the receptors, activating downstream signal transduction cascades. Dysregulation of ERBB genes play key roles in the development of cancer by affecting cell proliferation, apoptosis, and angiogenesis [7].
All four receptors are expressed in cervical cancer [8–11], and overexpression of ERBB family members and expression of alternative Egfr isoforms are associated with advanced stage cancers and more aggressive cancers [10, 12–14]. Moreover, the HPV oncoproteins have been linked to activation of Egfr. Because activating mutations within the catalytic kinase domain of Egfr are uncommon in cervical intraepithelial neoplasia (CIN) and cervical cancer, interaction of highrisk HPV proteins with inherited polymorphisms may be an important factor in the dysregulation of EGFR in cervical cancer [14].
Heterodimerization of ErbB family members increases the diversity of ligands recognized by individual receptors, boosting the repertoire of signaling pathways that can be activated by a given receptor. Taking into account the important differential heterodimerization among the ERBB receptors and the strong association with the development of cancer, we determined whether there was an association between SNPs in any of the four ERBB genes with susceptibility to in situ (CIN3) or invasive cervical cancer [7]. The ability and importance of the ERBB genes to form heterodimers for signal diversity lead us to explore genetic variability within the family of genes and susceptibility to cervical cancer.
Methods
Proband Characteristics
We obtained informed consent from all subjects in the 628 family trios. Each trio consisted of a proband – a woman with invasive or in situ cervical cancer – and either her biological parents or one parent and one or more siblings. Proband characteristics are provided in Table 1, which shows that the two sets were comparable in ethnicity, mean age of diagnosis, smoking status, and histology. Washington University’s Human Research Protection Office and the Medical College of Wisconsin’s Institutional Review Board approved the study.
Table 1.
Proband Characteristics
| Discover dataset | Additional probands | Combined Validation dataset | |
|---|---|---|---|
| Number of trios | 244 | 384 | 628 |
| Genotyping platform | GoldenGate | TaqMan | |
| Cervical cancer | Invasive: 97% In situ: 3% |
Invasive: 34% In situ: 66% |
Invasive: 59% In situ: 41% |
| Caucasian | 93% | 88% | 90% |
| Age of diagnosis (mean) | 35.9 ± 7.4 | 33.2 ± 9.3 | 34.3 ± 8.7 |
| Ever smoked | 49% | 46% | 47% |
| Probands with HPV 16, 31, 52, 18 and/or 45 | 92% | 86.7% | 88% |
| Histology* | Squamous: 64% Adeno: 30% Other 6% |
Squamous: 74% Adeno: 21% Other 5% |
Squamous: 70% Adeno: 25% Other 5% |
Squamous denotes carcinoma-in-situ or invasive squamous cell carcinoma; Adeno denotes adeno-in-situ or adenocarcinoma
SNP Selection and Genotyping
SNPs in the discovery data set were chosen as tag SNPs selected by Haploview based on the CEU population of the HapMap Project database (http://www.hapmap.org) [15]. The selected SNPs were in the EGFR, ERBB2, ERBB3 and ERBB4 genes and an additional 5000 base pairs upstream and downstream of defined gene boundaries. Thus, the 5′ UTR promoter region and transcription factor-binding sites, as well as the 3′ UTR potential regulatory sequences are included in the current analyses. The haplotype blocks were defined by r2≥0.8 (for ERBB4, r2 ≥0.5; due to the large number of SNPs), and the minor allele frequencies of the tag SNPs were >0.05. As a result, 79 EGFR SNPs, 4 ERBB2 SNPs, 5 ERBB3 SNPs, and 149 ERBB4 SNPs were included in the discovery analysis.
Genomic DNA extracted from buccal cells or blood was genotyped, using two platforms. The discovery sample set (244 trios) was typed using the Illumina GoldenGate assay (Illumina, Inc., San Diego CA). Then SNPs found to be significantly (P <0.05) overtransmitted were genotyped by applying the TaqMan assay (Life Technologies Corp, Carlsbad CA) in the combined dataset of 628 trios. Genotyping results obtained by the two technologies for the 244 discovery trios were then compared to validate experimental assays. Discrepant genotypes were confirmed by bidirectional sequencing on an Applied Biosystems 3730 Genetic Analyzer as described previously [6].
Family-Based Association Analysis
We used the family-based test of association that is implemented in the program TRANSMIT version 2.5.4 because it is robust to population stratification. The program estimates the overtransmission or undertransmission of single ERBB alleles from heterozygous parents to affected women with cervical cancer in each family trio. By obtaining case-control information from the genotypes of cervical cancer patients, fathers, and mothers the study automatically matches each subject and its controls by ethnicity. The TDT evaluates whether the frequency of transmission of parental marker alleles to affected offspring deviates from the expected Mendelian frequency of 50%. The final genotypes were analyzed for transmission consistency between parents and offspring, and genotypes that showed Mendelian error were deleted and not used in analysis.
We also used that software to look for epistasis—overtransmission of combinations of different ERBB alleles [16]. A two-stage design was used to evaluate genetic variation in the ERBB genes. First, 237 SNPs were screened in a discovery set of 244 trios. SNPs significant at a nominal P = 0.05 were then typed in validation set of all 628 trios. We reported the results of discovery testing and our analysis of the combined validation samples for optimal power [17]. We analyzed the discovery and the combined replication datasets separately. Asymptotic P values are reported in the results (Table 2). We conducted follow-up tests by subdividing the sample into HPV16-and HPV18-related types, stage and race as supported by previous studies [3, 5, 6].
Table 2.
Significant SNPs identified from the Discovery and Combined replication datasets
| SNP rs# | chr | Location* | # of transmission |
Minor allele |
MAF** | Over transmitted allele |
P-value | # of transmission |
Minor allele |
MAF | Over transmitted allele |
P-value | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EGFR | 11770506 | 7 | 55090379 | 238 | C | 0.335 | T | 0.0014 | 541 | C | 0.334 | T | 0.0066 |
| EGFR | 763317 | 7 | 55095197 | 230 | A | 0.472 | A | 0.0349 | 547 | A | 0.306 | A | 0.016 |
| EGFR | 6956366 | 7 | 55101501 | 231 | C | 0.333 | G | 0.0110 | 579 | C | 0.329 | G | 0.0044 |
| EGFR | 12668421 | 7 | 55109177 | 235 | A | 0.197 | T | 0.0003 | 600 | A | 0.209 | T | 0.0261 |
| EGFR | 723527 | 7 | 55134872 | 237 | G | 0.406 | A | 0.0120 | 600 | G | 0.427 | A | 0.0507 |
| EGFR | 11487218 | 7 | 55141540 | 236 | C | 0.373 | C | 0.027 | 583 | C | 0.353 | C | 0.5382 |
| EGFR | 7780270 | 7 | 55151886 | 235 | T | 0.475 | G | 0.0247 | 592 | T | 0.489 | G | 0.0882 |
| ERBB4 | 1836721 *** | 2 | 212252194 | 238 | A | 0.101 | T | 0.0467 | |||||
| ERBB4 | 12052398 | 2 | 212355840 | 230 | G | 0.305 | A | 0.0280 | 577 | G | 0.293 | A | 0.0672 |
| ERBB4 | 13424871 | 2 | 212531352 | 230 | A | 0.243 | A | 0.0091 | 574 | A | 0.249 | A | 0.1802 |
| ERBB4 | 11892696 | 2 | 212710146 | 236 | C | 0.206 | T | 0.0003 | 598 | C | 0.220 | T | 0.0364 |
| ERBB4 | 16847082 | 2 | 212718670 | 235 | C | 0.197 | T | 0.0003 | 600 | C | 0.209 | T | 0.0261 |
| ERBB4 | 1978873 | 2 | 212765835 | 238 | A | 0.082 | G | 0.0186 | 606 | A | 0.080 | G | 0.1191 |
| ERBB4 | 16847416 *** | 2 | 212847605 | 238 | G | 0.026 | A | 0.0225 |
Location is based on human genome assembly GRCh37/hg19
MAF – minor allele frequency
Unable to generate ABI TaqMan probes for AT1836721 and AG16847416
Results
Single SNP TDT analyses
The Illumina GoldenGate assay of the discovery data set indicated that 16 of 237 genotyped SNPs in EGFR, ERBB2, ERBB3, and ERBB4 were significantly overtransmitted to women with cervical cancer (P<0.05; Table 2). Fourteen of the 16 were genotyped in the extended set of all the family trios, using the TaqMan assay. We were unable to generate TaqMan probes for the other two SNPs (rs1836721 and rs16847416). Reproducibility was >99% for 12/14 SNPs in the 244 trios genotyped on both platforms. The Mendelian error rate was <5%, and the questionable trios were deleted from the TDT analyses.
A comparison of the GoldenGate and TaqMan data for rs11238349 and rs1801200 gave highly inconsistent results. Therefore, we sequenced those two SNPs by first developing primers across the two regions and then sequencing the resulting PCR products from 50 samples. The results showed that neither of the SNP assays could genotype these two SNPs accurately. In both cases, previously unknown neighboring SNPs (3–4 bp from the initial target SNP) may have interfered with genotyping. Therefore, both rs11238349 and rs1801200 were excluded from further analysis and from the list of significantly overtransmitted SNPs shown in Table 2.
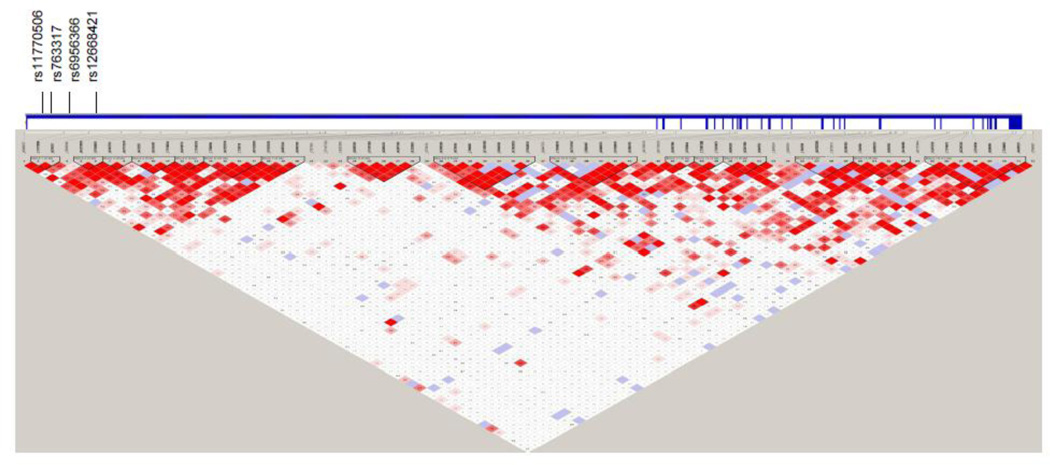
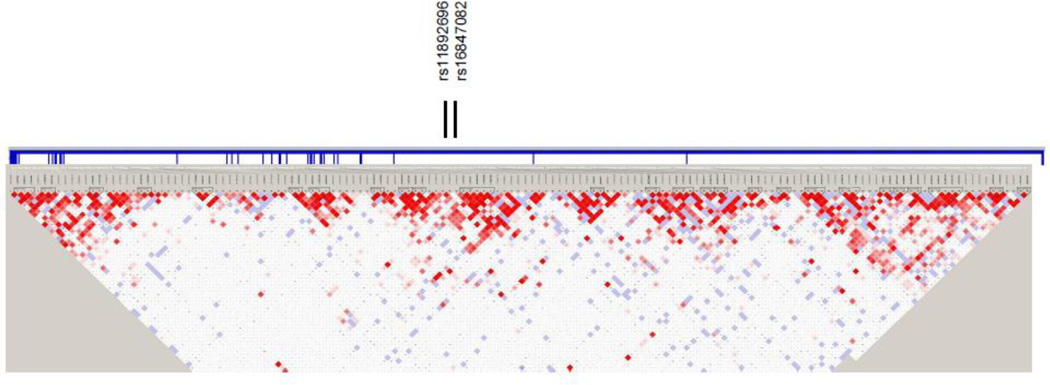
Six of the SNPs retained their significance in the combined dataset, and in all the same allele was overtransmitted: four SNPs in intron 1 of EGFR and two SNPs in intron 24 of ERBB4 (Table 2; Figures 1 and 2). The four EGFR SNPs were located in intron 1 within 18.8 kb.
Figure 1.
Haploview linkage disequilibrium (LD) structure map of SNPs genotyped within EGFR. The 4 SNPs that reached significance level of P<0.05 in the combined replication set are marked above the map. EGFR exons are denoted by thick vertical lines between the SNPs and map. Depth of shading in the map represents D´ values (dark=high inter-SNP D´; white=low inter-SNP D´). Pairwise correlation structure analyzed by Haploview software (Haploview: http://www.broadinstitute.org/haploview/haploview)
Figure 2.
Haploview linkage disequilibrium (LD) structure map of SNPs genotyped within ERBB4. The 2 SNPs that reached significance level of P<0.05 in the combined replication set are marked above the map. ERBB4 exons are denoted by thick vertical lines between the SNPs and map. Depth of shading in the map represents D´ values (dark=high inter-SNP D´ white=low inter-SNP D´). Pairwise correlation structure analyzed by Haploview software (Haploview: http://www.broadinstitute.org/haploview/haploview)
The haplotype analyses of the SNPs did not add additional information. The single SNP associations were not significantly altered by HPV type, race, or stage.
Epistasis
We also looked for possible epistatic effects of alleles from different ErbB family members that segregate together to affected women, resulting in 15,765 TDT tests.
Three combinations met the stringent Bonferroni corrected significance threshold. All three were interactions between SNPs in EGFR and ERBB4 (Table 3). One of the ERBB4 intronic SNPs, rs16847082, was identified in both single and two-SNP analyses.
Table 3.
Two SNP epistasis analyses based on TDT
| Discovery | ||||||
|---|---|---|---|---|---|---|
|
EGFR SNP rs# |
chr | Location* |
ERBB4 SNP rs# |
chr | Location* | P-value |
| rs2075112 | 7 | 55219611 | rs10497968 | 2 | 213114645 | 2.18E-07 |
| rs2075112 | 7 | 55219611 | rs16847082 | 2 | 212718670 | 1.34E-06 |
| rs845552 | 7 | 55245507 | rs1978873 | 2 | 212765835 | 4.11E-07 |
Location is based on human genome assembly GRCh37/hg19
Discussion
The current study identified polymorphisms in intron 1 of EGFR and intron 24 of ERBB4 that are overtransmitted from heterozygous parents to probands with in situ or invasive cervical cancer. We also detected haplotypes of EGFR and ERBB4 SNPs in association with cervical cancer indicating possible epistatic effects. We used a family-based study to identify genetic markers for susceptibility to cervical cancer. Unlike traditional case-control methods, family-based tests use within-family data to avoid identification of spurious associations that may result from population admixture.
We further explored SNP-SNP interactions within the ERBB family of genes. The two-SNP analyses also provide evidence for epistatic effects or statistical interaction between SNPs in EGFR (rs2075112; rs845552) and ERBB4 (rs10497968; rs1978873; rs16847082) (Table 3). EGFR SNP rs2075112 is in linkage disequilibrium (LD) with the EGFR furin-like cysteine-rich region, whose function is not clear. Rs845552 is in LD with the EGFR tyrosine kinase region. Rs10497968 is an intron SNP in ERBB4, and is not in LD with any known functional regions of ERBB4. Rs1978873 and rs16847082 are ERBB4 intron SNPs that lie between two exons coding for the ligand binding domain. Rs16847082 is the only SNP from the two-SNP analysis that also showed significant association in the single SNP analyses (Table 2).
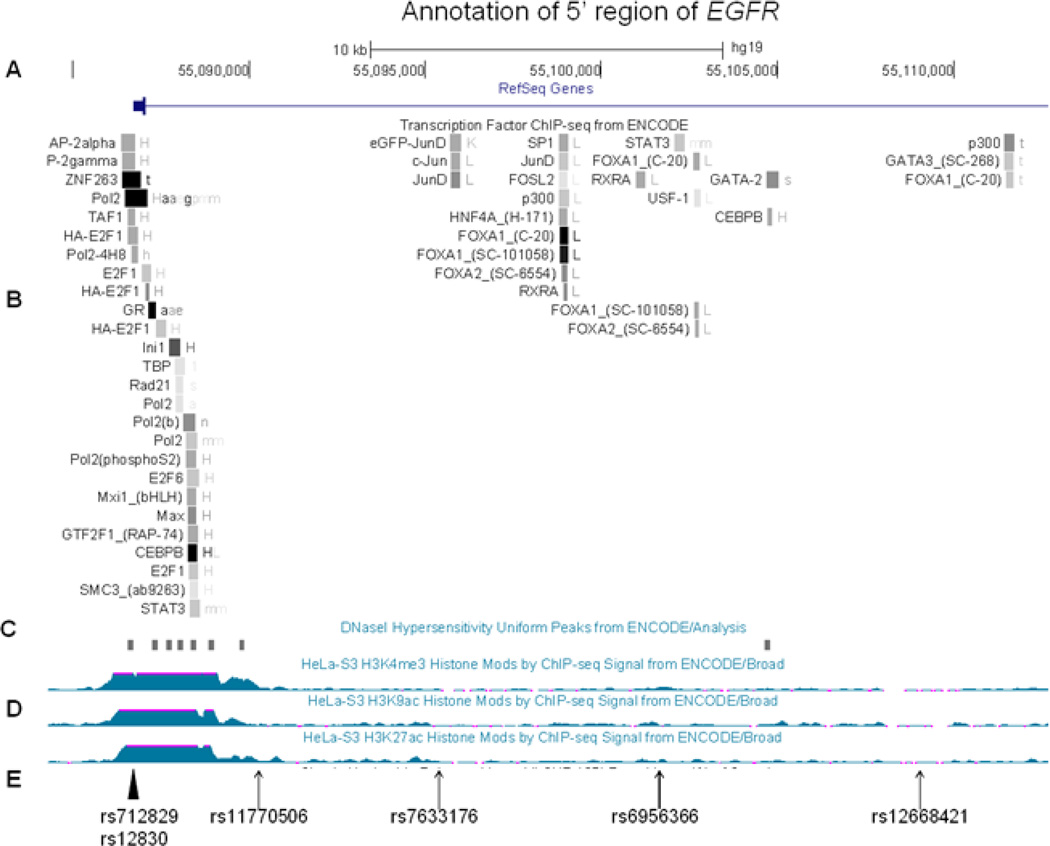
Several lines of evidence support genetic variation in EGFR and ERBB4 and susceptibility to cancer. The four intron 1 EGFR SNPs identified in this study are located near previously described sequence variants with enhancer activity that are located within DNase hypersensitivity sites and show LD with various transcription factor binding sites (Figure 3) [18–21]. One previously described functional variant is a promoter polymorphism (−216G>T; rs712829) associated with increased expression of EGFR. This polymorphism may contribute to the variability of EGFR expression and influence cellular reliance on Egfr, leading to interindividual variability in carcinogenesis [22, 23]. The second functional variant is a highly polymorphic (CA)n repeat in intron 1 (rs712830); in vitro and in vivo studies have revealed that transcriptional activity of EGFR is reduced with increasing CA repeats [24–26]. This polymorphism also has been associated with the risk of breast and lung cancer [27, 28]. Moreover, the (CA)n repeat length associates with increased occurrence of EGFR tumor mutations in lung cancer [21, 29]. The location of one of the most significant SNPs identified in our study—rs11770506—is 1500 bp downstream of the (CA)n repeat (Figure 3).
Figure 3.
Structure of the 5’ region of EGFR using the aligned annotate tracks from the Human Genome Browser (CRCh37/gh19) (http://genome.ucsc.edu/cgi-bin/hgGateway). A. Genome scale, nucleotide position and 5´EGFR. B. Transcription Factor ChIP-seq data from ENCODE-shaded squares show DNA regions where transcription factors bind as assayed by chromatin immunoprecipitation with antibodies specific to the transcription factor followed by sequencing of the precipitated DNA (ChIP-seq) C. DNaseI Hypersentivity Peaks from ENCODE-shaded squares show areas of open chromatin that defines segments of DNA that are unpacked and accessible to the regulatory factors, enzymes, and smaller molecules in the cell, D. Histone modification by ChIP-seq signal from Encode, E. The significant SNPs identified in this study are indicated by an arrow and two previously reported SNPs altering EGFR function are indicated by an arrowhead.
The interaction of Egfr and ErbB4 with HPV oncogenes has been examined in several studies. Signaling through Egfr is important for the growth of HPV-infected cells and progression to cancer. Egfr is overexpressed in the epithelium soon after HPV infection, and it increases with advancing stage [11, 30, 31]. Moreover, several HPV oncogenes rely on Egfr signaling for their tumorigenic properties. Woodworth et al. reported that Egfr signaling is important for immortalization induced by HPV 16 E6 and E7 and for progression to papillomas and carcinomas in mouse epithelial cells [32]. Egfr is also required for the induction of epithelial hyperplasia by HPV 16 E5 in a transgenic mouse model [33]. In addition, E5 affects Egfr level by reducing degradation of the receptor and increasing its recycling to the cell surface [34]. HPV 16 E5 protein can form a complex with ErbB4 by binding to the extracellular and transmembrane domains. E5 inhibits ErbB4-induced c-Jun expression and phosphorylation, increasing cell proliferation [35].
Studies examining the distribution of the four ERBB receptors in cervical tissue showed significant co-expression of Egfr and ErbB4 receptors in squamous cell carcinoma awhich is not seen with ErbB2 or ErB3 [8, 36]. There is also a high correlation of the HPV16 E5 oncoprotein with Egfr and ErbB4 levels in CIN and invasive cancers[37]. The authors suggest that E5 may associate with activation of the signaling pathway through homo-or hetero-dimerization of Egfr and ErbB4.
Our study also highlights the importance of accurate SNP genotyping calls in association studies and the need to validate across different experimental platforms. We used both Illumina GoldenGate and TaqMan assays to genotype all subjects for the 16 significant SNPs. We did not obtain identical genotyping data of subjects between the Illumina and TaqMan assays for two SNPs. The genotyping errors were probably due to interference by previously unknown neighboring SNPs, which we identified by further sequencing the region.
Limitations of this study include the disproportionate number of in situ and ICC cases in the discovery and combined replication datasets. The trays were prepared as families were assembled and invasive cancer was represented at a higher proportion on the discovery trays. This may be important if genetic susceptibility is different for in situ and ICC. However, many association studies use > CIN2 as the definition for cervical cancer. We have carefully validated the pathology on all probands and confirmed in situ or invasive cervical cancer. Because ERBB4 is very large, we were able to genotype only 149 of its tagSNPs, obtaining an r2 ≥0.5. Further genotyping of ERBB4 might identify additional genetic associations. Moreover, future work needs to validate the results in additional populations and functional studies to confirm hypothesis. With better understanding of the LD structures and genetic variations within the LD blocks and with regional sequencing, it should be possible to test the validity of the currently identified variants and to identify functional variations in the ERBB gene family.
Our study is unique because it associates susceptibility to cervical cancer with single SNPs and suggests epistasis between EGFR and ERBB4. Though our significant findings are limited to tag SNPs, the next generation of sequencing technology will enable us to deep-sequence the promising regions to distinguish the functional variation as well as to further address the underlying mechanisms and pathways.
Highlights.
We examine ERBB family of polymorphisms using a family-based association study
We identify variants in EGFR and ERBB4 in susceptibility to cervical cancer
We provide genotyping validation across different experimental platforms
Acknowledgements
This work was supported by National Cancer Institute grant R01CA095713.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict(s) of Interest/Disclosure(s) Statement
None
References
- 1.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 2.Hariri S, Unger ER, Sternberg M, Dunne EF, Swan D, Patel S, Markowitz LE. Prevalence of genital human papillomavirus among females in the United States, the National Health And Nutrition Examination Survey 2003–2006. J Infect Dis. 2011;204:566–573. doi: 10.1093/infdis/jir341. [DOI] [PubMed] [Google Scholar]
- 3.Hu X, Zhang Z, Ma D, Huettner PC, Massad LS, Nguyen L, Borecki I, Rader JS. TP53, MDM2, NQO1, and susceptibility to cervical cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:755–761. doi: 10.1158/1055-9965.EPI-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang SS, Bratti MC, Rodriguez AC, Herrero R, Burk RD, Porras C, Gonzalez P, Sherman ME, Wacholder S, Lan ZE, Schiffman M, Chanock SJ, Hildesheim A. Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. J Infect Dis. 2009;199:20–30. doi: 10.1086/595563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu KJ, Rader JS, Borecki I, Zhang Z, Hildesheim A. CD83 polymorphisms and cervical cancer risk. Gynecol Oncol. 2009;114:319–322. doi: 10.1016/j.ygyno.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Borecki I, Nguyen L, Ma D, Smith K, Huettner PC, Mutch DG, Herzog TJ, Gibb RK, Powell MA, Grigsby PW, Massad LS, Hernandez E, Judson PL, Swisher EM, Crowder S, Li J, Gerhard DS, Rader JS. CD83 gene polymorphisms increase susceptibility to human invasive cervical cancer. Cancer Res. 2007;67:11202–11208. doi: 10.1158/0008-5472.CAN-07-2677. [DOI] [PubMed] [Google Scholar]
- 7.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs I, Vorsteher N, Buhler H, Evers K, Sehouli J, Schaller G, Kummel S. The prognostic significance of human epidermal growth factor receptor correlations in squamous cell cervical carcinoma. Anticancer Res. 2007;27:959–963. [PubMed] [Google Scholar]
- 9.Kersemaekers AM, Fleuren GJ, Kenter GG, Van den Broek LJ, Uljee SM, Hermans J, Van de Vijver MJ. Oncogene alterations in carcinomas of the uterine cervix: overexpression of the epidermal growth factor receptor is associated with poor prognosis. Clin Cancer Res. 1999;5:577–586. [PubMed] [Google Scholar]
- 10.Lee CM, Shrieve DC, Zempolich KA, Lee RJ, Hammond E, Handrahan DL, Gaffney DK. Correlation between human epidermal growth factor receptor family (EGFR, HER2, HER3, HER4), phosphorylated Akt (P-Akt), and clinical outcomes after radiation therapy in carcinoma of the cervix. Gynecol Oncol. 2005;99:415–421. doi: 10.1016/j.ygyno.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 11.Maruo T, Yamasaki M, Ladines-Llave CA, Mochizuki M. Immunohistochemical demonstration of elevated expression of epidermal growth factor receptor in the neoplastic changes of cervical squamous epithelium. Cancer. 1992;69:1182–1187. doi: 10.1002/cncr.2820690519. [DOI] [PubMed] [Google Scholar]
- 12.Halle C, Lando M, Svendsrud DH, Clancy T, Holden M, Sundfor K, Kristensen GB, Holm R, Lyng H. Membranous expression of ectodomain isoforms of the epidermal growth factor receptor predicts outcome after chemoradiotherapy of lymph node-negative cervical cancer. Clin Cancer Res. 2011;17:5501–5512. doi: 10.1158/1078-0432.CCR-11-0297. [DOI] [PubMed] [Google Scholar]
- 13.Soonthornthum T, Arias-Pulido H, Joste N, Lomo L, Muller C, Rutledge T, Verschraegen C. Epidermal growth factor receptor as a biomarker for cervical cancer. Ann Oncol. 2011;22:2166–2178. doi: 10.1093/annonc/mdq723. [DOI] [PubMed] [Google Scholar]
- 14.Arias-Pulido H, Joste N, Chavez A, Muller CY, Dai D, Smith HO, Verschraegen CF. Absence of epidermal growth factor receptor mutations in cervical cancer. Int J Gynecol Cancer. 2008;18:749–754. doi: 10.1111/j.1525-1438.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.Clayton D, Jones HB. Transmission/disequilibrium tests for extended marker haplotypes. Am J Hum Genet. 1999;65:1161–1169. doi: 10.1086/302566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 18.McInerney JM, Wilson MA, Strand KJ, Chrysogelos SA. A strong intronic enhancer element of the EGFR gene is preferentially active in high EGFR expressing breast cancer cells. J Cell Biochem. 2001;80:538–549. doi: 10.1002/1097-4644(20010315)80:4<538::aid-jcb1008>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Brandt B, Meyer-Staeckling S, Schmidt H, Agelopoulos K, Buerger H. Mechanisms of egfr gene transcription modulation: relationship to cancer risk and therapy response. Clin Cancer Res. 2006;12:7252–7260. doi: 10.1158/1078-0432.CCR-06-0626. [DOI] [PubMed] [Google Scholar]
- 20.Chrysogelos SA. Chromatin structure of the EGFR gene suggests a role for intron 1 sequences in its regulation in breast cancer cells. Nucleic Acids Res. 1993;21:5736–5741. doi: 10.1093/nar/21.24.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W, He L, Ramirez J, Krishnaswamy S, Kanteti R, Wang YC, Salgia R, Ratain MJ. Functional EGFR germline polymorphisms may confer risk for EGFR somatic mutations in non-small cell lung cancer, with a predominant effect on exon 19 microdeletions. Cancer Res. 2011;71:2423–2427. doi: 10.1158/0008-5472.CAN-10-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, Innocenti F, Wu MH, Desai AA, Dolan ME, Cook EH, Jr, Ratain MJ. A functional common polymorphism in a Sp1 recognition site of the epidermal growth factor receptor gene promoter. Cancer Res. 2005;65:46–53. [PubMed] [Google Scholar]
- 23.Araujo A, Ribeiro R, Azevedo I, Coelho A, Soares M, Sousa B, Pinto D, Lopes C, Medeiros R, Scagliotti GV. Genetic polymorphisms of the epidermal growth factor and related receptor in non-small cell lung cancer--a review of the literature. Oncologist. 2007;12:201–210. doi: 10.1634/theoncologist.12-2-201. [DOI] [PubMed] [Google Scholar]
- 24.Buerger H, Gebhardt F, Schmidt H, Beckmann A, Hutmacher K, Simon R, Lelle R, Boecker W, Brandt B. Length and loss of heterozygosity of an intron 1 polymorphic sequence of egfr is related to cytogenetic alterations and epithelial growth factor receptor expression. Cancer Res. 2000;60:854–857. [PubMed] [Google Scholar]
- 25.Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 26.Tidow N, Boecker A, Schmidt H, Agelopoulos K, Boecker W, Buerger H, Brandt B. Distinct amplification of an untranslated regulatory sequence in the egfr gene contributes to early steps in breast cancer development. Cancer Res. 2003;63:1172–1178. [PubMed] [Google Scholar]
- 27.Brandt B, Hermann S, Straif K, Tidow N, Buerger H, Chang-Claude J. Modification of breast cancer risk in young women by a polymorphic sequence in the egfr gene. Cancer Res. 2004;64:7–12. doi: 10.1158/0008-5472.can-03-2623. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Weissfeld JL, Romkes M, Land SR, Grandis JR, Siegfried JM. Association of the EGFR intron 1 CA repeat length with lung cancer risk. Mol Carcinog. 2007;46:372–380. doi: 10.1002/mc.20285. [DOI] [PubMed] [Google Scholar]
- 29.Sueoka-Aragane N, Imai K, Komiya K, Sato A, Tomimasu R, Hisatomi T, Sakuragi T, Mitsuoka M, Hayashi S, Nakachi K, Sueoka E. Exon 19 of EGFR mutation in relation to the CA-repeat polymorphism in intron 1. Cancer Sci. 2008;99:1180–1187. doi: 10.1111/j.1349-7006.2008.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brustmann H, Hinterholzer S, Brunner A. Expression of phosphorylated histone H2AX (gamma-H2AX) in normal and neoplastic squamous epithelia of the uterine cervix: an immunohistochemical study with epidermal growth factor receptor. Int J Gynecol Pathol. 2011;30:76–83. doi: 10.1097/PGP.0b013e3181eb2fcb. [DOI] [PubMed] [Google Scholar]
- 31.Kim SC, Park HM, Lee SN, Han WS. Expression of epidermal growth factor receptor in cervical tissue and serum in patients with cervical neoplasia. J Low Genit Tract Dis. 2004;8:292–297. doi: 10.1097/00128360-200410000-00006. [DOI] [PubMed] [Google Scholar]
- 32.Woodworth CD, Gaiotti D, Michael E, Hansen L, Nees M. Targeted disruption of the epidermal growth factor receptor inhibits development of papillomas and carcinomas from human papillomavirus-immortalized keratinocytes. Cancer Res. 2000;60:4397–4402. [PubMed] [Google Scholar]
- 33.Genther Williams SM, Disbrow GL, Schlegel R, Lee D, Threadgill DW, Lambert PF. Requirement of epidermal growth factor receptor for hyperplasia induced by E5, a high-risk human papillomavirus oncogene. Cancer Res. 2005;65:6534–6542. doi: 10.1158/0008-5472.CAN-05-0083. [DOI] [PubMed] [Google Scholar]
- 34.Venuti A, Paolini F, Nasir L, Corteggio A, Roperto S, Campo MS, Borzacchiello G. Papillomavirus E5: the smallest oncoprotein with many functions. Mol Cancer. 2011;10:140. doi: 10.1186/1476-4598-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen SL, Lin ST, Tsai TC, Hsiao WC, Tsao YP. ErbB4 (JM-b/CYT 1)-induced expression and phosphorylation of c-Jun is abrogated by human papillomavirus type 16 E5 protein. Oncogene. 2007;26:42–53. doi: 10.1038/sj.onc.1209768. [DOI] [PubMed] [Google Scholar]
- 36.Chang JL, Tsao YP, Liu DW, Han CP, Lee WH, Chen SL. The expression of type I growth factor receptors in the squamous neoplastic changes of uterine cervix. Gynecol Oncol. 1999;73:62–71. doi: 10.1006/gyno.1998.5301. [DOI] [PubMed] [Google Scholar]
- 37.Chang JL, Tsao YP, Liu DW, Huang SJ, Lee WH, Chen SL. The expression of HPV-16 E5 protein in squamous neoplastic changes in the uterine cervix. J Biomed Sci. 2001;8:206–213. doi: 10.1007/BF02256414. [DOI] [PubMed] [Google Scholar]