Abstract
Chlorinated hydroquinones of biological origin are fully dechlorinated to 1,4-dihydroquinone by anaerobic bacteria such as Desulfitobacterium spp. (C. E. Milliken, G. P. Meier, J. E. M. Watts, K. R. Sowers, and H. D. May, Appl. Environ. Microbiol. 70:385-392, 2004). In the present study, mixed microbial communities from Baltimore Harbor sediment and a pure culture of Desulfitobacterium sp. strain PCE1 were discovered to demethylate, reductively dehydroxylate, and dechlorinate chlorinated hydroquinones into chlorophenols. Mixed microbial cultures from a freshwater source and several other desulfitobacteria in pure culture did not perform these reactions. Desulfitobacterium sp. strain PCE1 degraded 2,3,5,6-tetrachloro-4-methoxyphenol, a metabolite of basidiomycete fungi, to 2,3,5,6-tetrachlorophenol and 2,3,5-trichlorophenol, recalcitrant compounds that are primarily synthesized anthropogenically.
The presence of halogenated compounds in the environment is due primarily to anthropogenic activity, but recent investigations have demonstrated that a significant number of organohalides are biologically synthesized (5, 7) and degraded within the global halogen cycle (see reviews in references 8 and 12). Among the biosynthesized organohalides are the extensively chlorinated hydroquinone metabolites (CHMs) that are produced by basidiomycete fungi (5), such as the prevalent metabolite 2,3,5,6-tetrachloro-4-methoxyphenol (TCMP) (3, 17). The primary pathway for the biodegradation of TCMP by desulfitobacteria is the removal of the two chlorines adjacent to the hydroxylated carbon, followed by demethylation and, finally, full dechlorination (11). However, an examination of mixed microbial communities from estuarine sediment and a pure culture of Desulfitobacterium sp. strain PCE1 revealed a novel route of biodegradation ending with the accumulation of chlorophenols. Chlorophenols in the environment are present primarily due to anthropogenic activity. The U.S. Environmental Protection Agency has documented chlorophenols at many National Priority Sites (Superfund sites), and the Environmental Protection Agency and the U.S. Agency for Toxic Substances and Disease Registry list chlorophenols as priority pollutants (1, 2; see also www.atsdr.cdc.gov).
Mixed cultures and several pure cultures of anaerobic bacteria were examined in this study. Inocula for the mixed cultures were prepared by transferring the supernatant from a sediment-containing culture, initiated with sediment from either Baltimore Harbor (39°16.8′N, 76°36.1′W) or a freshwater pond in the Cumberland Mountains (36°21.72′N, 84°42.07′W), as described previously (11). Seven Desulfitobacterium sp. strains (DSM11544, ATCC 51507, DSM10664, DSM13498, ATCC 700357, DSM12704, and DSM10344) and Sulfurospirillum multivorans (DSM12446; previously Dehalospirillum multivorans) were grown in DSMZ medium 720 (German Collection of Microorganisms and Cell Cultures [www.dsmz.de]), which is a basic mineral medium plus 0.1% (wt/vol) pyruvate and 0.1% (wt/vol) yeast extract, and transfers were made as described previously (11). Organochlorides were added to separate cultures to achieve a final concentration of 173 μM TCMP, a 183 μM concentration of each 2,3,5,6-tetrachlorohydroquinone (TCHQ) and 2,3,5,6-tetrachlorophenol (2,3,5,6-TCP), a 202 μM concentration of each trichlorohydroquinone (TriCHQ) and 2,3,5-trichlorophenol (2,3,5-TriCP), a 239 μM concentration of each dichlorohydroquinone (DCHQ), a 155 μM concentration of monochlorohydroquinone, and a 205 μM concentration of 1,4-dihydroquinone. The 2,3,5,6-TCP was purchased from Supelco, the 2,3,6-TriCP was purchased from Sigma-Aldrich, and the 2,3,5-TriCP was purchased from Acros. All other compounds were synthesized as described previously (11).
All experiments were performed with duplicate cultures incubated anaerobically in the dark at 30°C. Duplicate cultures (10 ml) were extracted and analyzed by gas chromatography (GC)-mass spectrometry as described previously (11). All chlorinated organic compounds were derivatized with bis(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich) and were identified as the trimethylsilyl ether derivatives. Identification of all compounds was determined by matching GC retention times, molecular weights, relative natural isotopic abundances of 35Cl to 37Cl (19), and mass spectrometry fragmentation patterns with those of authentic standards (11). Each compound gave baseline separation by GC.
Four serially transferred, anaerobic mixed cultures, inoculated with estuarine sediment from Baltimore Harbor, produced 2,3,5-TriCP from TCMP. This required the sequential demethylation, dehydroxylation, and dechlorination of the TCMP. At 4 weeks, 10% of the TCMP had been transformed into TCHQ, 5% had been transformed into 2,3,5,6-TCP, and 5% had been transformed into 2,3,5-TriCP. The GC retention times for these chlorophenols were clearly distinguishable from those of all other chlorophenols; e.g., the absolute retention times were 8.45 min for 2,3,5-TriCP and 8.84 min for 2,3,6-TriCP. No further degradation was observed at 10 weeks. A separate experiment demonstrated that TCHQ, an apparent metabolite, was also dehydroxylated to 2,3,5,6-TCP (80% of TCHQ added) and then dechlorinated to 2,3,5-TriCP (5% of TCHQ added), with 15% of the TCHQ remaining after 4 weeks. In contrast, a series of anaerobic mixed cultures prepared with sediment from a freshwater pond did not transform TCMP or TCHQ to 2,3,5,6-TCP after 10 weeks of metabolism; instead, these microorganisms demethylated TCMP to TCHQ and then dechlorinated TCHQ to 1,4-dihydroquinone, as reported previously (11). Recovery of the total organochlorides was approximately 85% throughout these experiments.
Of the eight axenic cultures examined, only Desulfitobacterium sp. strain PCE1 produced 2,3,5,6-TCP and 2,3,5-TriCP from TCMP or TCHQ. All other bacterial strains either demethylated and dechlorinated the CHMs to 1,4-dihydroquinone or did not degrade the compounds at all, just as described previously (11). After 10 weeks of incubation, strain PCE1 had demethylated, reductively dehydroxylated, and dechlorinated a significant amount of TCMP to TCHQ, 2,3,5,6-TCP, and 2,3,5-TriCP, with 2,3,5,6-TCP as the major product (Table 1). Additionally, Desulfitobacterium sp. strain PCE1 dehydroxylated and dechlorinated TCHQ to 2,3,5,6-TCP and 2,3,5-TriCP. This path of biodegradation was nearly identical to that observed with the mixed cultures from Baltimore Harbor. To further understand this degradation pathway, strain PCE1 was grown with lesser-chlorinated hydroquinones, including 1,4-dihydroquinone, and all of the compounds were identified as degradation products of TCMP (Table 1). The growth of Desulfitobacterium sp. strain PCE1 was inhibited (i.e., turbidity was unchanged) by >200 μM 2,3,5-TriCP or 2,3,5-TriCHQ, which suggests that these compounds are toxic to the microorganism even though a small amount (5%) of the 2,3,5-TriCHQ was dechlorinated to 2,5-DCHQ. Inhibition of growth was not observed when 40 μM TriCP was added in a separate experiment, indicating that the amount of 2,3,5-TriCP produced by Desulfitobacterium sp. strain PCE1 would not prevent the growth of this organism during the transformation of TCMP in the experiments described. None of the other hydroquinone or phenolic compounds tested were degraded or inhibited microbial growth.
TABLE 1.
Degradation of fungal metabolites, chlorinated hydroquinones, and chlorophenols by Desulfitobacterium sp. strain PCE1 at 10 weeks
| Substrate | Growtha | Product(s) (% substrate converted) |
|---|---|---|
| TCMP | + | 2,3,5,6-TCHQ (10), 2,3,5,6-TCP (70), 2,3,5-TriCP (5) |
| TCHQ | + | 2,3,5,6-TCP (80), 2,3,5-TriCP (5) |
| 2,3,5,6-TCP | + | 2,3,5-TriCP (5) |
| 2,3,5-TriCP | − | None |
| 2,3,5-Trichlorohydroquinone | − | 2,5-DCHQ (5%) |
| 2,5-DCHQ | + | None |
| 2,6-Dichlorohydroquinone | + | None |
| 2-Chloro-1,4-hydroquinone | + | None |
| 1,4-Hydroquinone | + | None |
Growth of strain PCE1 was present (+) or absent (−) on indicated substrate.
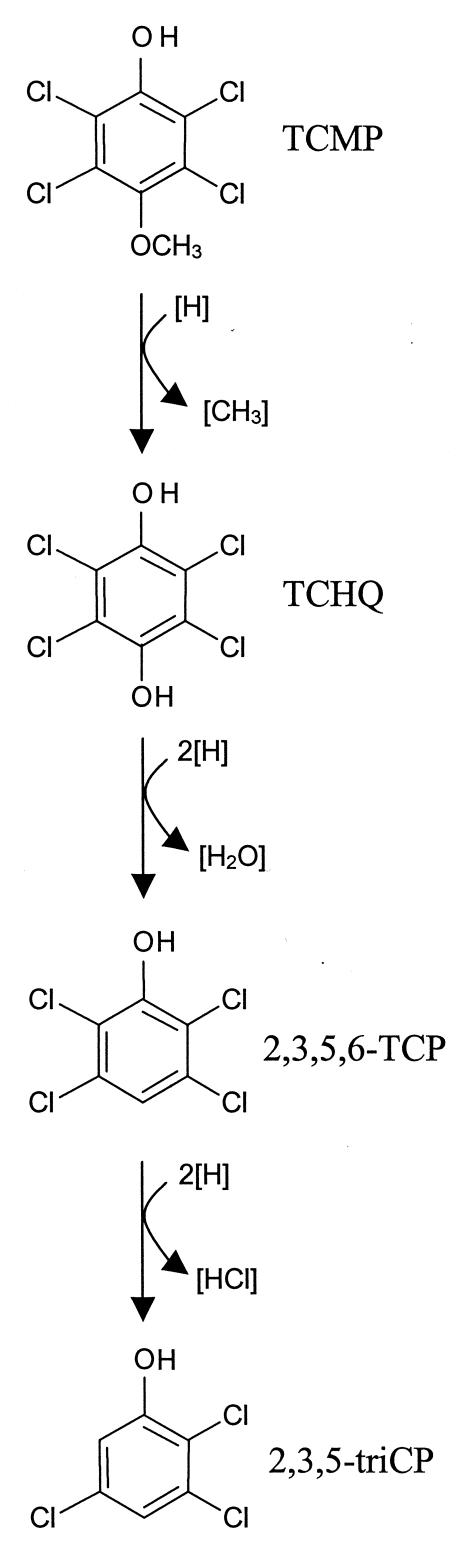
Figure 1 summarizes the pathway of chlorophenol production by Desulfitobacterium sp. strain PCE1 and the mixed cultures obtained from Baltimore Harbor sediment. The results of this study indicate that dehydroxylation by Desulfitobacterium sp. strain PCE1 occurs only when all ortho and meta positions of hydroquinone are chlorinated, since TCP and 1,4-dihydroquinone were not dehydroxylated by this organism (Table 1). It is unusual to find this degree of specificity for reductive dehydroxylation by a microorganism. The reductive dehydroxylation of aromatic compounds has been reported for phenols (13), hydroquinones (13, 16), hydroxybenzoates (10, 14, 15), and 3-chlorobenzoate (4), and it has been proposed that this metabolic activity can be coupled to anaerobic bacterial growth (16). This is the first report on the reductive dehydroxylation of a chlorinated hydroquinone by a microorganism, and it remains to be determined whether Desulfitobacterium sp. strain PCE1 or any other microorganism can respire and grow by the dehydroxylation of a hydroxylated organochloride.
FIG. 1.
Proposed pathway of chlorophenol production from fungal CHMs by Desulfitobacterium sp. strain PCE1. The fate of the methyl moiety indicated in the first reaction was not determined.
Hoekstra et al. observed the production of chlorophenols following the incubation of forest soil with Na37Cl for 1 year (9). The authors speculated that fungi in the soil could have been responsible for the synthesis of the chlorophenols. Fungi are not known to synthesize chlorophenols but do produce chlorinated anisyl metabolites (CAMs) and CHMs (5). Anaerobic microbial communities have been shown to convert a CAM (3,5-dichloro-p-anisyl alcohol) into a dichlorophenol (18). This activity includes the oxidation of the CAM to a benzyl alcohol, which is then oxidized to a 3,5-dichloro-4-hydroxybenzoate. The chlorinated hydroxybenzoate is then reductively decarboxylated to 3,5-dichlorophenol. Reductive dehydroxylation, as observed with CHMs in this study, has not been reported for the anaerobic degradation of CAMs. Therefore, the previous studies and the results presented herein suggest that the anaerobic degradation of a chlorinated fungal metabolite is a plausible route for the natural formation of chlorophenols. Desulfitobacterium sp. strain PCE1 is the first anaerobic microorganism in pure culture that has been demonstrated to perform such a series of reactions.
In addition to its dehydroxylation activity, Desulfitobacterium sp. strain PCE1 has a limited ability to dechlorinate chlorophenols (it will dechlorinate 2,4,6-TCP and 2-chlorophenol) (6), which suggests that the organism can contribute to the decrease in levels of chlorophenols in the environment. However, since this microbe is capable of demethylating and dehydroxylating biological CHMs, it may also add to the environmental burden of chlorophenols. The biological production of chlorophenols has environmental implications, as this class of organochloride compound is frequently used to detect and identify pollution sources based on the premise that chlorophenols are ordinarily synthesized industrially. Furthermore, the combination of demethylation and reductive dehydroxylation of fungal CHMs provides dechlorinating anaerobes with a natural source of chlorophenols, and this biogenic source of organohalides may have promoted the evolution of microbial dehalogenation of chlorinated phenolic compounds well before humans introduced other organochlorides. The observations described here provide a new perspective on the biosynthesis and biodegradation of toxic chlorophenols and the genesis of dehalogenases and also suggest that the unique reactions exhibited by these microbes may make them useful in other applications of biocatalysis.
Acknowledgments
This study was supported by the National Science Foundation (MCB-0078133).
REFERENCES
- 1.Agency for Toxic Substances and Disease Registry. 2001. Toxicological profile for pentachlorophenol. U.S. Department of Health and Human Services, Atlanta, Ga. [PubMed]
- 2.Agency for Toxic Substances and Disease Registry. 1999. Toxicological profile for chlorophenols. U.S. Department of Health and Human Services, Atlanta, Ga. [PubMed]
- 3.Anchel, M. 1952. Identification of drosophilin A as p-methoxytetrachlorophenol. J. Am. Chem. Soc. 74:2493. [Google Scholar]
- 4.Becker, J. G., D. A. Stahl, and B. E. Rittman. 1999. Reductive dehalogenation and conversion of 2-chlorophenol to 3-chlorobenzoate in a methanogenic sediment community: implications for predicting the environmental fate of chlorinated pollutants. Appl. Environ. Microbiol. 65:5169-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Jong, E., and J. A. Field. 1997. Sulfur tuft and turkey tail: biosynthesis and biodegradation of organohalogens by basidiomycetes. Annu. Rev. Microbiol. 51:375-414. [DOI] [PubMed] [Google Scholar]
- 6.Gerritse, J., V. Renard, T. M. Pedro Gomes, P. A. Lawson, M. D. Collins, and J. C. Gottschal. 1996. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch. Microbiol. 165:132-140. [DOI] [PubMed] [Google Scholar]
- 7.Gribble, G. W. 1998. Naturally occurring organohalogen compounds. Acc. Chem. Res. 31:141-152. [Google Scholar]
- 8.Häggblom, M. M., and I. D. Bossert. 2003. Halogenated organic compounds—a global perspective, p. 3-29. In M. M. Häggblom and I. D. Bossert (ed.), Dehalogenation: microbial processes and environmental applications. Kluwer Academic Publishers, Boston, Mass.
- 9.Hoekstra, E. J., H. De Weerd, E. W. B. De Leer, and U. A. T. Brinkman. 1999. Natural formation of chlorinated phenols, dibenzo-p-dioxins, and dibenzofurans in soil of a Douglas fir forest. Environ. Sci. Technol. 33:2543-2549. [Google Scholar]
- 10.Londry, K. L., and P. M. Fedorak. 1993. Use of fluorinated compounds to detect aromatic metabolites from m-cresol in a methanogenic consortium: evidence for a demethylation reaction. Appl. Environ. Microbiol. 59:2229-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milliken, C. E., G. P. Meier, J. E. M. Watts, K. R. Sowers, and H. D. May. 2004. Microbial anaerobic demethylation and dechlorination of chlorinated hydroquinone metabolites synthesized by basidiomycete fungi. Appl. Environ. Microbiol. 70:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Öberg, G. 2002. The natural chlorine cycle—fitting the scattered pieces. Appl. Microbiol. Biotechnol. 58:565-581. [DOI] [PubMed] [Google Scholar]
- 13.O'Connor, O. A., and L. Y. Young. 1996. Effects of six different functional groups and their position on the bacterial metabolism of monosubstituted phenols under anaerobic conditions. Environ. Sci. Technol. 30:1419-1428. [Google Scholar]
- 14.Sharak Genthner, B. R., G. T. Townsend, and P. J. Chapman. 1989. Anaerobic transformation of phenol to benzoate via para-carboxylation: use of fluorinated analogues to elucidate the mechanism of transformation. Biochem. Biophys. Res. Commun. 162:945-951. [DOI] [PubMed] [Google Scholar]
- 15.Sharak Genthner, B. R., G. T. Townsend, and P. J. Chapman. 1991. para-Hydroxybenzoate as an intermediate in the anaerobic transformation of phenol to benzoate. FEMS Microbiol. Lett. 78:265-270. [DOI] [PubMed] [Google Scholar]
- 16.Szewzyk, U., R. Szewzyk, and B. Schink. 1985. Methanogenic degradation of hydroquinone and catechol via reductive dehydroxylation to phenol. FEMS Microbiol. Ecol. 31:79-87. [Google Scholar]
- 17.Teunissen, P. J. M., H. J. Swarts, and J. A. Field. 1997. The de novo production of drosophilin A (tetrachloro-4-methoxyphenol) and drosophilin A methyl ether (tetrachloro-1,4-dimethoxybenzene) by ligninolytic basidiomycetes. Appl. Microbiol. Biotechnol. 47:695-700. [DOI] [PubMed] [Google Scholar]
- 18.Verhagen, F. J. M., H. J. Swarts, J. B. P. A. Wijnberg, and J. A. Field. 1998. Biotransformation of the major fungal metabolite 3,5-dichloro-p-anisyl alcohol under anaerobic conditions and its role in formation of bis(3,5-dichloro-4-hydroxyphenyl)methane. Appl. Environ. Microbiol. 64:3225-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson, J. T. 1985. Introduction to mass spectrometry, p. 153-172. Raven Press, New York, N.Y.