Abstract
To address how changes in the subclass of antibody molecules affect their thermodynamic stability, we prepared three types of four monoclonal antibody molecules (chimeric, humanized, and human) and analyzed their structural stability under thermal stress by using size-exclusion chromatography, differential scanning calorimetry (DSC), circular dichroism (CD), and differential scanning fluoroscopy (DSF) with SYPRO Orange as a dye probe. All four molecules showed the same trend in change of structural stability; the order of the total amount of aggregates was IgG1 < IgG2 < IgG4. We thus successfully cross-validated the effects of subclass change on the structural stability of antibodies under thermal stress by using four methods. The Th values obtained with DSF were well correlated with the onset temperatures obtained with DSC and CD, suggesting that structural perturbation of the CH2 region could be monitored by using DSF. Our results suggested that variable domains dominated changes in structural stability and that the physicochemical properties of the constant regions of IgG were not altered, regardless of the variable regions fused.
Keywords: antibody, circular dichroism, differential scanning calorimetry, differential scanning fluoroscopy, subclass change, thermodynamic stability
Introduction
Monoclonal antibodies (mAbs) are very versatile reagents and are under extensive development for therapeutic use. To date, more than 30 therapeutic antibodies have been approved for clinical use and more than 240 molecules are under development.1–3
Antibodies are heterodimers composed of two heavy chains and two light chains, which are linked through disulfide bonds. As classified on the basis of the constant region of the heavy chain there are five classes of antibody, namely IgA, IgD, IgE, IgG, and IgM. The constant heavy regions of IgA, IgD, and IgG have three IgG domains and a hinge region to give flexibility. Despite this range of choice of antibody classes to select from in the development of therapeutic antibodies, all recombinant antibodies under development to date have belonged to the IgG class because it has the highest serum half-life among all the classes.4,5
Four subclasses of IgG antibody have been identified, and each subclass has, in principle, highly homologous constant region sequences. However, the abilities of the IgGs in the four subclasses to trigger effector functions differ distinctly. For therapeutic applications the subclass needs to be carefully chosen; to achieve the desired therapeutic effects and avoid side effects, subclass selection is primarily dictated by the effector functions needed. IgG1 and IgG3 have greater ability to activate antibody-dependent cellular cytotoxicity and complement-dependent cellular cytotoxicity than do IgG2 and IgG4.6–8 If the effector function is required to eliminate targets (e.g., to destroy tumor cells in oncology applications), one would expect IgG1 or IgG3 to be used. However, in practice, the IgG3 subclass has not been used as a therapeutic candidate owing to its short half-life and long hinge region, which makes it susceptible to proteolysis, along with its allotypic polymorphism.9 Most of the therapeutic antibodies approved to date have belonged to the IgG1 subclass; only a few IgG2 and IgG4 antibodies have reached the market.3 Nevertheless, subclasses IgG2 and IgG4 are increasingly being developed as therapeutic antibodies because of their blocking or inhibitory functions.9 Selection of antibody subclass is therefore considered increasingly critical in the development of therapeutic antibodies.9
Products have been developed that have reduced effector functions compared with those seen with the IgG1 subclass.10–12 IgG2 and IgG4 have unique physicochemical properties. Human IgG2 antibodies can form covalent dimers and structural isoforms via disulfide shuffling in vivo.13–17 IgG4 antibodies can form half-antibodies (i.e., those with one heavy chain and one light chain) that lack inter-heavy chain disulfide bonds and form intra-chain disulfide bonds. Recent studies have shown that a half-antibody can exchange with another IgG4 half-antibody, resulting in the production of a bispecific antibody.18–20 Moreover, IgG2 and IgG4 are unstable in terms of tendency to aggregate.21,22 Among the physicochemical properties of antibodies, homogeneity and stability are particularly critical issues limiting the successful development of therapeutic antibodies.
Here, we focus on the effects of changes in IgG subclass on the conformational and physicochemical stability of antibodies. We prepared three types of four antibody molecules, namely chimeric, humanized, and human. From our results, we concluded that changes in variable domains dominate changes in thermodynamic stability and that the trends in thermodynamic stability caused by changes due to subclass switching are in principle identical among antibody molecules.
Results
Size-exclusion chromatography and SDS-PAGE
To compare differences in aggregation and degradation tendency among subclasses, we investigated aggregation and degradation behavior in the pH range of 4.0–7.0 at 25°C and 40°C. Samples taken at different incubation times were analyzed by using size-exclusion chromatography (SEC).
We tried to construct and analyze subclass-substituted molecules of mAb-A, mAb-B, mAb-C, and mAb-D. Here, we first give the results for mAb-C, followed by those of the other antibody molecules.
Supporting Information Figures 1 to 1–6 give representative SEC chromatograms of mAb-C (chromatographic UV profile) for various glutamate formulations as a function of pH. The intact non-aggregated antibody (monomer) was eluted at approximately 31.5 min. Soluble aggregate (high-molecular-weight species, HMWS) was eluted earlier, and degraded species (low-molecular-weight species, LMWS) were eluted later. The total amounts of degraded species were enhanced with increasing incubation time at 40°C.
All subclasses were most stable at pH 5.5, and the total HMWS content was enhanced with increasing pH in the range 5.5–7.0. In the case of IgG1, the lower pH samples exhibited greater physical stability, as evidenced by less aggregation. In contrast, IgG2AAAS, IgG4, and IgG4PE showed greater aggregation at lower pH (pH 4.0 and 5.5).
We examined the percentages of HMWS, as revealed by SEC analysis as a function of pH after storage at 25°C or 40°C for 1 or 3 months (Fig. 1). IgG1 showed no increase in HMWS content over time at pH 4.0 at 40°C from time zero (Initial) to 3 months. In contrast, IgG2 (IgG2AAAS) and IgG4 (IgG4PE) showed an increase in HMWS content over time at 40°C during the 3-month period at all pH values. The rate of aggregation was strongly dependent on the antibody subclass (IgG1 vs. IgG2 vs. IgG4). At 40°C, all IgG1s were resistant to low-pH aggregation, but the IgG2s and IgG4s were aggregated.
Figure 1.
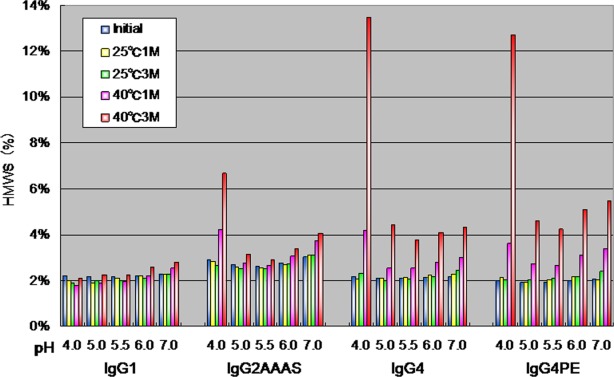
Size-exclusion high-performance liquid chromatographic analysis of percentages of HMWS as a function of pH after storage at 25 or 40°C for 1 or 3 months.
We examined the percentages of LMWS, as revealed by SEC analysis as a function of pH after storage at 25°C or 40°C for 1 or 3 months (Fig. 2). All subclasses were most stable at pH 5.5–6.0 and showed decreasing LMWS content with increasing pH in the range 4.0–6.0. IgG1 samples exhibited more LMWS than did IgG2 and IgG4 samples, revealing the lower chemical stability of IgG1. The amount of LMWS was highest at pH 4.0 for each subclass, although to differing extents. Reducing and non-reducing SDS-PAGE showed a high-molecular-weight ladder (data not shown); this was consistent with the heterogeneity observed within the aggregates upon SEC.
Figure 2.
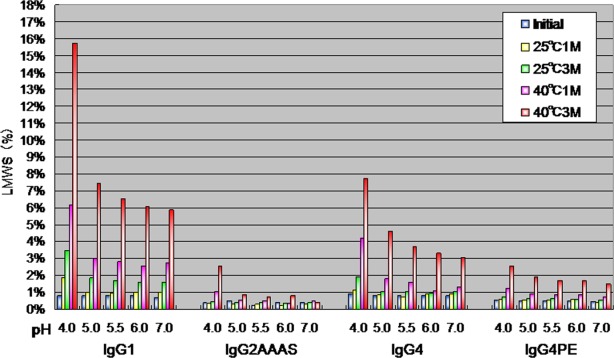
Size-exclusion high performance liquid chromatographic analysis of percentages of LMWS as a function of pH after storage at 25 or and 40°C for 1 or 3 months.
The same trend was observed for other antibodies: examples for mAb-D are given in Supporting Information Figure 1 (1-7 to 1–11).
Differential scanning calorimetry
To determine whether antibodies of different subclasses exhibited different thermodynamic stabilities, we obtained differential scanning calorimetry (DSC) thermograms for antibodies of three different subclasses, including mutations that had the same variable region. We constructed and analyzed subclass-substituted molecules of mAb-A, mAb-B, mAb-C, and mAb-D. Here, we first describe the results for mAb-C, followed by a discussion of the other mutants.
We examined the profiles of temperature-induced unfolding of all subclasses of mAb-C (IgG1, IgG2AAAS, IgG4, IgG4PE) under the same solvent conditions (Fig. 3). Table 1 lists the observed thermal unfolding midpoint (Tm) values for mAb-C. In the case of mAb-C, we prepared IgG1, IgG2AAAS, IgG4, and IgG4PE. Thermal unfolding of mAb-C G1 resulted in three partially overlapping transitions with Tm values of about 73, 78, and 84°C, respectively, at pH 5.5 under optimized formulation conditions. The results showed good agreement with previous results from an IgG1 Mab, in which the three transitions were assigned to unfolding of the CH2 domain, the antigen-binding fragment (Fab), and the CH3 domain.23,24 For mAb-C G2AAAS, G4, and G4PE, the first transition shifted to the low temperature side compared with that for IgG1. The change in the number of transitions in members of the IgG subclass with the same variable regions of heavy and light chains (VH/VL region) suggests that independent unfolding of the CH2, CH3, and Fab domains is a general property of IgGs.
Figure 3.
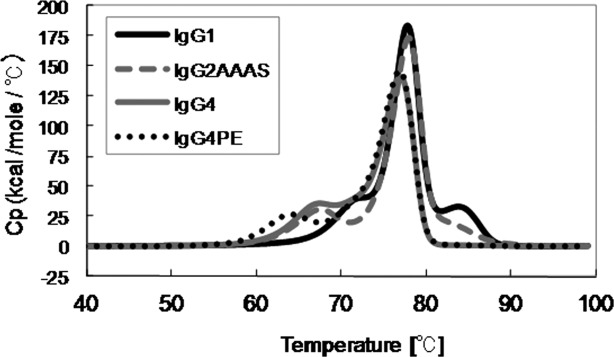
Temperatures inducing melting (unfolding) of all subclasses of mAb-C at pH 5.5, as measured by using DSC.
Table 1.
Differential scanning calorimetry measurements of melting transition of mAb-C in all subclasses as a function of pH.
| Tm (°C) | |||||||
|---|---|---|---|---|---|---|---|
| Subclass | domain | pH | 4.0 | 5.0 | 5.5 | 6.0 | 7.0 |
| G1 | |||||||
| CH2 | 60.10 | 69.41 | 72.55 | 73.23 | 73.72 | ||
| Fab | 73.91 | 77.23 | 77.87 | 78.05 | 78.06 | ||
| CH3 | 79.07 | 83.55 | 83.88 | 84.05 | 84.02 | ||
| G2AAAS | |||||||
| CH2 | 52.78 | 64.55 | 67.73 | 69.24 | 70.50 | ||
| Fab | 72.58 | 77.19 | 78.04 | 77.58 | 77.99 | ||
| CH3 | NA | NA | NA | NA | NA | ||
| G4 | |||||||
| CH2 | 54.76 | 64.07 | 67.40 | 69.04 | 69.90 | ||
| Fab | 71.07 | 76.55 | 77.05 | 76.36 | 76.06 | ||
| CH3 | NA | NA | NA | NA | NA | ||
| G4PE | |||||||
| CH2 | 53.77 | 61.42 | 64.08 | 65.79 | 67.74 | ||
| Fab | 71.25 | 76.55 | 76.90 | 76.27 | 75.56 | ||
| CH3 | NA | NA | NA | NA | NA | ||
| NA, not available; melting transition was not detected or well defined. | |||||||
The apparent Fab Tm values of mAb-C in the IgG1 and IgG2AAAS formats were similar to each other (ΔTm < 0.3°C), whereas the apparent Fab Tm values in the IgG4 and IgG4PE formats were about 1°C different from the Fab Tm values of mAb-C in the IgG1 and IgG2AAAS formats. Among the four antibodies with the same variable domains and different subclasses, the Tm values of the Fab fragments differed in the order of IgG1 ≥ IgG2 ≥ IgG4 and IgG4PE.
The same trend was observed for the other antibodies, namely mAb-A, mAb-B, and mAb-D: a list of observed Tm values is given in Supporting Information Tables 1–III and Supporting Information Figure 2. Average values of Tm for the four antibodies were calculated and plotted against pH (Fig. 4). The CH1 and Fc sequences of the IgG subclasses have some amino acid differences and different disulfide-bonding patterns (data not shown); this might suggest why the effects on the profiles of antibodies with different variable domains were identical.
Figure 4.
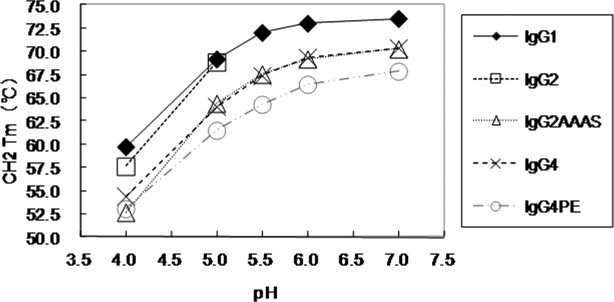
DSC measurements of CH2-domain melting transition of antibodies as a function of pH. IgG1, 4 (mAb-A, B, C, D); IgG2, 2 (mAb-A, -B); IgG2AAAS, 2 (mAb-C, -D); IgG4, 2 (mAb-C, -D); IgG4PE, 4 (mAb-A, -B, -C, -D).
Changing pH had marked effects on the DSC profiles of each antibody molecule reported here (Table 1, Supporting Information Tables 1–III). A decrease in pH resulted in broader endotherms that occurred at lower temperatures. Notably, the CH2 domain displayed greater pH dependence of thermal stability than did the CH3 domain; ΔTm was larger for IgG4 than for IgG1 and IgG2.
Previous characterization of the Fc fragment has identified an early transition due to unfolding of the CH2 domain followed by thermal unfolding of the CH3 domain.25 Previous studies indicate that the CH3 region of IgG1 has an extremely high Tm compared with other domains.25 Of the three human IgG subclasses (IgG1, IgG2, and IgG4), IgG1 has the most stable Fc, based on the Tm of its CH2 and CH3 domain.24 This subclass dependence is correlated with reduced thermodynamic stability of IgG2 and IgG4 CH2 relative to IgG1 CH2. Regardless of subclass, we found that thermodynamic stability of CH2 was an important determinant of the stability and aggregation of Fc and intact antibody molecules under acidic conditions. Some of these findings have already been reported.26,27 Notably, also, the unfolding transitions of the CH2 and CH3 domains for all four IgG1 constructs were identical as described in the results. This trend was also observed for the other subclasses, namely IgG2, IgG2AAAS, IgG4, and IgG4PE; in contrast, the Fab unfolding transitions were highly variable.
Circular dichroism
We used far-UV circular dichroism (CD) to compare the secondary structures of the subclasses (Supporting Information Figs. 3-1 to 3-3). First, we give the results for mAb-C, followed by a discussion of those for the other antibody molecules.
The CD spectra of all subclasses of mAb-C before and after heating are shown in Supporting Information Figure 3-1. The subclasses showed little difference in terms of far-UV CD before heating (Supporting Information Fig. 3-1a), regardless of pH (data not shown). The spectra were characterized by a single negative peak with a minimum at a wavelength of 217 nm, suggestive of the expected β-sheet structure of the immunoglobulin fold.28 These observations suggested that subclass changes did not alter the overall secondary structure.
To probe the thermal stability, we monitored the signal at 217 nm from 25 to 100°C and compared the thermal stabilities of the subclasses (Fig. 5). In all of the ellipticity–temperature profiles there were two steps at which the intensity decreased, suggesting that there were two distinct temperatures at which changes in secondary structure occurred. At pH 4.0, the antibody had a slightly lower transition-onset temperature than at other pH values. The transition-onset temperatures of the antibody at pH 5.0 and 6.0 were comparable, and the ellipticity did not change until the temperature reached approximately 60°C, whereas a much lower onset temperature was observed at pH 4.0. At pH 4.0, the major changes occurred at 55°C, followed by a second step at about 70°C, in all subclasses. The transition-onset temperatures were in the order of IgG4 < IgG2 < IgG1. Furthermore, in the case of the IgG4 formats, precipitation was observed after heating at pH 6.0. The temperature values corresponded well to the Tm values observed for the structural transitions in the DSC experiments. IgG4 has the lowest thermal stability among the subclasses reported here.
Figure 5.
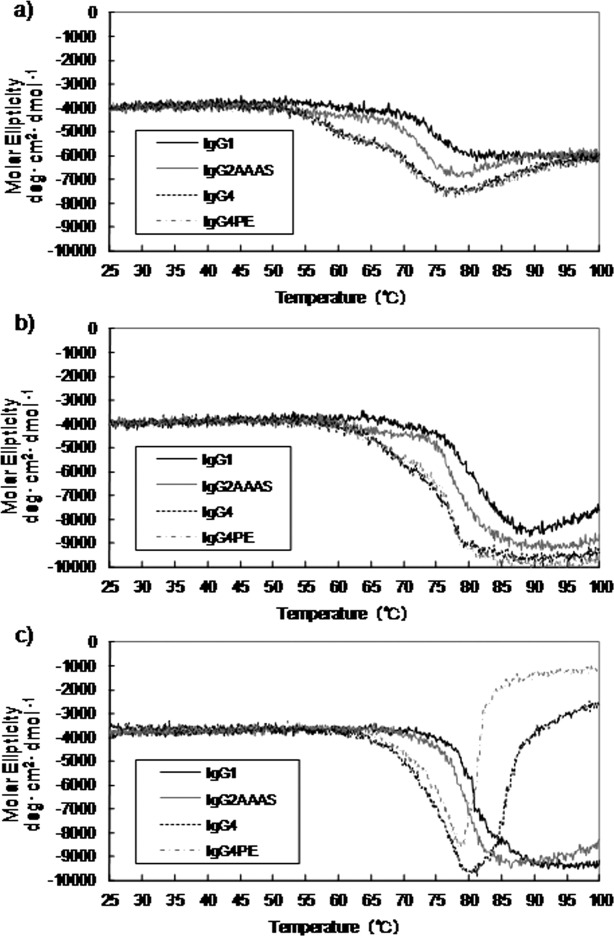
Effect of temperature and pH on molar ellipticity at a wavelength of 217 nm (a, pH 4.0; b, pH 5.5; c, pH 6.0) in the case of mAb-C.
Next, we obtained CD spectra of different subclasses of the other three antibodies, mAb-A, mAb-B, and mAb-D; the results for mAb-A and mAb-D are shown in Supporting Information Figures 3-2, 3-3 and 4-1, 4-2, respectively. The same trends as with mAb-C were observed for these antibodies.
Differential scanning fluoroscopy
The typical differential scanning fluoroscopy (DSF) profile of an IgG molecule consists of a sharp sigmoid-like increase to the maximum level, followed by a decrease in fluorescence intensity. This profile represents changes in the environment of the dye. The initial increase in fluorescence most likely results from exposure of the dye to the hydrophobic area of the protein during thermal unfolding; the degree of exposure depends on the rate of transition from the folded to the unfolded state. Fluorescence quenching at increased solution temperature—a nonspecific transition—also occurs and contributes to the overall sigmoidal shape.29–31 Here, we used SYPRO Orange as a probe in the DSF system.
We investigated four mAb molecules: mAb-A and mAb-B (IgG1, IgG2, IgG4PE) and mAb-C and mAb-D (IgG1, IgG2AAAS, IgG4, IgG4PE). We used a variety of formulation pH values to generate DSF profiles for each mAb's subclasses.
Here, we give the results for mAb-C as an example. The fluorescence profiles and temperatures of hydrophobic exposure (Th) of each subclass-changed IgG are shown in Supporting Information Figures 5-1 to 5-5 and Table II, respectively. Comparison of fluorescence profiles at pH 5.5 revealed that the fluorescence intensity increased at about 55°C; after two-step transitions it then decreased at about 80°C. These findings were similar to the results reported previously.29,30 The changes in intensity started in the order of IgG4PE, IgG4, IgG2AAAS, and IgG1. IgG1 had the highest Th1 value, followed by IgG2AAAS, IgG4, and IgG4PE. The order was almost the same in the case of the Th2 values.
Table II.
Temperature of Hydrophobic Exposure (Th) Values of Antibodies Obtained From Differential Scanning Fluoroscopy Measurements, as a Function of pH
| Subclass | Tm (°C) | ||||||
|---|---|---|---|---|---|---|---|
| pH | 4.0 | 5.0 | 5.5 | 6.0 | 7.0 | ||
| Mab-A | |||||||
| G1 | |||||||
| Th1 | 63 | 68 | 69.5 | 70 | 70.5 | ||
| Th2 | 73 | 74.5 | 74.5 | NA | NA | ||
| G2 | |||||||
| Th1 | 62.5 | 68 | 69.5 | 70 | 70.5 | ||
| Th2 | 71.5 | 73 | NA | NA | NA | ||
| G4PE | |||||||
| Th1 | 56.5 | 60 | 61 | 61.5 | 62 | ||
| Th2 | 67.5 | 69.5 | 70 | 70 | 70 | ||
| Mab-B | |||||||
| G1 | |||||||
| Mab-C | |||||||
| G1 | |||||||
| Th1 | 62.5 | 67.5 | 69 | 69.5 | 70 | ||
| Th2 | 74 | 75 | 75 | 74.5 | 74 | ||
| G2 | |||||||
| Th1 | 61.5 | 67 | 69 | 70 | 70.5 | ||
| Th2 | 73.5 | 74.5 | 74.5 | 74.5 | NA | ||
| G4PE | |||||||
| Th1 | 56.5 | 60 | 60.5 | 61 | 62 | ||
| Th2 | 73 | 74 | 74 | 73.5 | 73.5 | ||
| Mab-C | |||||||
| G1 | |||||||
| Th1 | 63.5 | 68 | 69.5 | 70 | 70.5 | ||
| Th2 | 76.5 | 78 | 78 | 78 | 78 | ||
| G2AAAS | |||||||
| Th1 | 58.5 | 63.5 | 65 | 66 | 66 | ||
| Th2 | 74.5 | 76.5 | 77 | 77.5 | 77.5 | ||
| G4 | |||||||
| Th1 | 59 | 62.5 | 64 | 65 | 65.5 | ||
| Th2 | 74 | 75.5 | 76 | 76 | 75.5 | ||
| G4PE | |||||||
| Th1 | 56.5 | 60.5 | 61.5 | 62 | 62.5 | ||
| Th2 | 74 | 75.5 | 75.5 | 75.5 | 75.5 | ||
| Mab-D | |||||||
| G1 | |||||||
| Th1 | 63 | 68 | 69 | 70 | 70 | ||
| Th2 | 79 | 81 | 81.5 | 82 | 82.5 | ||
| G2AAAS | |||||||
| Th1 | 58 | 63 | 65 | 65.5 | 66 | ||
| Th2 | 75 | 82.5 | 82.5 | 83 | 83 | ||
| G4 | |||||||
| Th1 | 58 | 62.5 | 64.5 | 65.5 | 65.5 | ||
| Th2 | 78 | 81 | 81 | 81.5 | 81.5 | ||
| G4PE | |||||||
| Th1 | 56.5 | 60.5 | 61.5 | 62.5 | 63 | ||
| Th2 | 78.5 | 81 | 81.5 | 81.5 | 81.5 | ||
| NA,not available; melting transition was not detected or well defined. | |||||||
Tm, thermal unfolding midpoint.
Next, we examined the effects of pH on changes in Th1 and Th2 values. In all subclass-changed IgGs the Th1 and Th2 values were highest at neutral pH; a decrease in pH led to a decrease in values. Notably, our DSF analyses showed that mutations into IgG2 and IgG4 led to decreases in Th values, and that the Th1 value of IgG2AAAS at pH 4.0 was lower than that of IgG4. These results are in good accordance with those obtained from DSC. The trends were similar for the other mAbs.
Discussion
Here, we reported the effects of changes in antibody subclass on stability in terms of molecule conservation and on structural stability. We focused on four antibody molecules (chimeric, humanized, and human), and three subtypes were constructed for each of the four antibodies. We substituted another subclass for the constant region of each antibody. These subclass-changed molecules had the same variable domains but different constant regions; we could therefore evaluate how subclass affected the physicochemical properties of IgG.
The results of SEC and SDS-PAGE revealed clear differences in the physical and chemical degradation of various IgGs. IgG1s were more susceptible to fragmentation, whereas IgG2s and IgG4s were more susceptible than IgG1 to aggregation at low pH. Degradation of human IgG1 at low pH originates from non-enzymatic digestion of the upper hinge region (i.e., EPKSCDKTHT; digested site is underlined)32–35; these earlier findings are in good agreement with our results. It is plausible to assume that human IgG2 is more resistant than IgG1 to non-enzymatic proteolysis for the following reasons: (1) it is shorter at the hinge region; (2) the hinge region has a more rigid structure owing to the presence of four S-S bonds; and (3) the amino acid sequence of human IgG2 is distinctly different from that of human IgG1. Aggregate formation increased with decreasing pH, and the order of the total amounts of aggregates within the pH range 4.0 to 7.0 was IgG1 < IgG2 < IgG4. IgG4 was unstable in the acidic pH range. Our results are consistent with those described previously.21,26,27,36,37 Moreover, in these previous studies the pH-dependent effects of subclass changes on the physicochemical stability of antibodies were almost identical to the changes produced by each antibody variable domain reported here.
Development of antibody formulations with increased concentrations is desirable in biotherapeutics38; however, aggregation capacity should be carefully evaluated in the case of high-concentration formulations. It has been suggested that aggregates of antibodies might be correlated with immunogenicity.39,40 The choice of subclass might determine the aggregation tendency of the antibodies to be constructed.
We compared the thermal unfolding of each subclass-changed mAb-C, as monitored by CD, DSC, and DSF (Fig. 6). The DSC profile for subclass-changed mAb-C showed two well-separated transitions. For multi-domain proteins such as antibodies, multiple DSC transitions usually suggest reduced inter-domain interactions. Similar to DSC, DSF was able to distinguish two separate transitions in the thermal unfolding of mAb-C. At several test pHs, the inflection point of the DSF curve corresponded to the peak of the DSC curve and the CD thermogram, with only a small onset. The small onset was expected, because DSF monitors the exposure of hydrophobic residues, whereas DSC measures the change in excess heat capacity changes (ΔCp) and CD measures the change in secondary structure. The Th value derived from DSF was, in principle, well correlated with the onset temperature value obtained from DSC or CD. For the other three mAbs, we observed profiles identical to those of mAb-C. We therefore proposed the Th value obtained from DSF as a parameter for evaluating the thermodynamic stability of antibodies.
Figure 6.
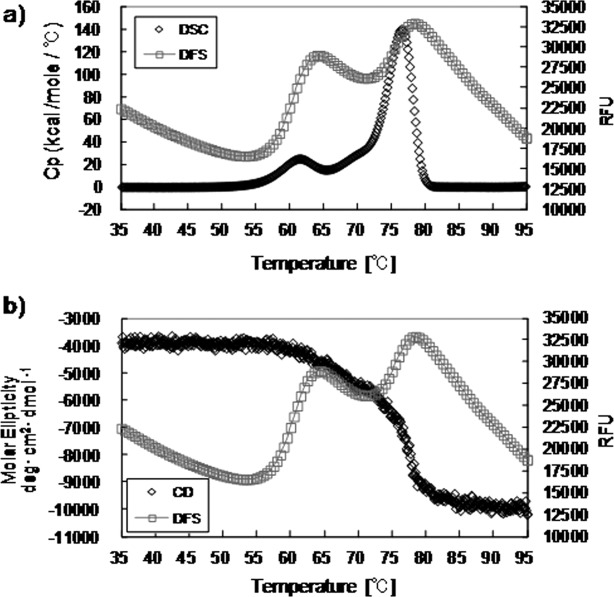
Comparison of thermal unfoldings of mAb-C IgG4PE at pH 5.0, as monitored by (a) DSC and DSF, and (b) CD and DSF.
DSC is one of the methods most commonly used to characterize conformational stability on the basis of differences in thermal stability.31,41 We compared the normalized DSC thermograms of IgG molecules and DSF data (Supporting Information Fig. 6-1 to 6-3). The Th obtained with DSF was slightly lower than the first Tm determined by DSC under all experimental conditions. At pH 4.0, the DSF profile (Supporting Information Fig. 6) seemed similar to the transition corresponding to that of the CH2 domain, which had the lowest melting temperature for these mAb molecules, as determined by DSC. The CH2 domain was more sensitive to pH changes than the other IgG domains; this was consistent with results reported previously.42 We plotted the correlation between Th and the CH2 unfolding temperature Tm for different molecules (Fig. 7). A linear relationship between Th and CH2 unfolding temperature Tm was observed. The transition of the Fab domain did not vary at this pH, as analyzed by both DSC and DSF (data not shown). At higher pH values the transition of the CH2 domain overlapped with that of the Fab domain; on the other hand, Fab domain transition did not change under the pH range tested.
Figure 7.
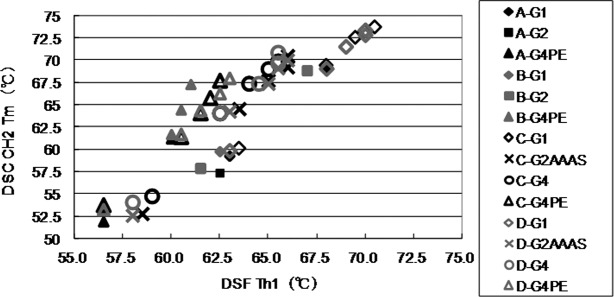
Correlation between DSF Th1 (temperature of hydrophobic exposure 1) and differential scanning calorimetry (DSC) CH2 Tm (thermal unfolding midpoint) values. DSF Th1 values were determined as the midpoint of the first fluorescence transition in DSF profiles by using the first derivative curves. DSC CH2 Tm values were determined as the lowest melting temperature in DSC thermograms. Linear regression yielded a linearity correlation coefficient of r2 = 0.9822–0.9975.
Direct comparison of DSF and DSC demonstrated that both methods detected the same trends in thermostability at different pH values and the same relative stabilities of the domain. Th was consistently lower than Tm determined by DSC. We successfully cross-validated the effects of subclass changes on the structural stability of antibodies under thermal stress by using four methods. The Th values for the CH2 domain obtained by DSF were well correlated with the onset temperatures obtained by DSC and CD, suggesting that structural perturbation of the CH2 region can be monitored by using DSF. Our evaluation of the structural stability of subclass-changed antibodies suggested that the Fc region of each subclass dominates the physicochemical characteristics of the molecules. All four molecules showed the same trend in change in structural stability, in the order of IgG1 > IgG2 > IgG4. Taken together, our findings suggest that the state of the CH2 region—the most unstable one in the domains of antibodies—is strongly correlated with the structural stability of antibodies and that aggregation tendency under acidic conditions originates from the structural stability of the CH2 domain of each subclass. Our DSF analyses suggest that exposure of the hydrophobic region upon thermal stress is correlated with the tendency of antibodies to aggregate.
Our results suggest that a tendency toward poorer physicochemical quality of antibodies is associated with the subclass type and not with the variable domain. The physicochemical properties of antibodies may therefore be conserved even if the variable domains have been exchanged with those of other IgGs. This concept might help us to select appropriate subclasses. Notably, we can exchange IgG subclasses by appropriately predicting changes in the physicochemical properties of the antibodies. Our results should provide valuable insights into antibody design using appropriate subclasses and into the process of antibody production in accordance with the physicochemical properties of different subclasses.
Materials and Methods
Materials
All IgG antibodies used were manufactured by Kyowa Hakko Kirin (Tokyo, Japan). All antibodies were expressed in Chinese Hamster Ovary cells under essentially identical conditions. Secreted antibodies were recovered from the culture medium and purified by using a series of chromatographic and filtration steps. Carbohydrate structures bound to all antibodies were confirmed by mass-spectroscopy, demonstrating that no major differences have been observed (data not shown).
Four IgG antibodies with different variable regions, mAb-A (human), mAb-B (human), mAb-C (chimeric), and mAb-D (humanized) have been used in this study. IgG4PE contained an amino acid point mutation of Ser228Pro and Leu235Glu in the heavy chain of IgG4 (EU-index numbering scheme used) to prevent half-antibody formation43 and reduce antibody-dependent cytotoxicity.44 IgG2AAAS contained an amino acid point mutation of Val234Ala, Gly237Ala, and Pro331Ser in the heavy chain of IgG2 to reduce effector functions.45–47 Thus, the amino acid sequences of the light chains and VH regions in each set were identical, but their subclasses were different (IgG1, IgG2 or IgG2AAAS, IgG4, or IgG4PE). Their isoelectric points were in the range of 8–9. The antibody solution contained 10 mM sodium glutamate, 262 mM D-sorbitol, and 0.05 mg/mL polysorbate 80. Glutamate was selected from the buffer compounds commonly used in clinical antibody formulation. D-sorbitol was selected from the tonicity agents commonly used in antibody formulation. Polysorbate 80 was added to prevent protein particle formation. All excipients met the criteria of the monographs in the United States Pharmacopeia and National Formulary.
All evaluated antibodies were buffer-exchanged into formulations of the desired pH values by using a desalting column (NAP25 column, GE Healthcare U.K., Buckinghamshire, England), and their concentrations were adjusted to 5.0 mg/mL. The formulated antibody solutions were sterilized with a 0.22-μm filter, and 1 mL of each solution was placed into a sterilized USP-type 5-mL glass vial, which was sealed with autoclaved rubber stopper. The prepared samples were stored in a temperature-controlled incubator at 25 or 40°C for 1 or 3 months before SEC analysis and SDS-PAGE.
Size-exclusion high-performance liquid chromatography
To detect soluble aggregates and fragments, size-exclusion high-performance liquid chromatography was performed on an Alliance 2795 device equipped with a 2487 UV detector (Waters Corporation, Mildford, MA) and a TSK G3000SWXL 7.8 × 300 mm2 column (Tosoh Biosep., Tokyo, Japan). Separation was performed with a mobile phase of 20 mM K2HPO4/KH2PO4 and 500 mM NaCl (pH 7.0) at a flow rate of 0.5 mL/min at a constant 25°C. A diluted sample was injected to obtain a total loading amount of 200 μg, and detection was performed at a wavelength of 215 nm. The resulting chromatograms were analyzed by integrating the area under each eluting peak by using Empower 2 Chromatography Data System software (Waters Corporation) and recorded as percentages of HMWS and LMWS.
SDS-PAGE
IgG samples were run on Novex 8% to 16% Tris-glycine 1.0 × 15-well precast SDS-PAGE gels (Invitrogen, Carlsbad, CA). All samples were diluted to 1 mg/mL or 0.1 mg/mL with formulation buffer and diluted further to 0.1 mg/mL or 0.01 mg/mL with a 4× solution consisting of 69 mM Tris–HCl (pH 7.0), 2.2% SDS, 0.04% bromophenol blue, and 22.2% glycerol buffer; under reducing conditions 111.1 mM dithiothreitol (DTT) was added to the 4× solution. The sample load was 1 μg or 0.1 μg. The molecular weight maker (Mark12: 200- to 2.5-kDa range) was purchased from Invitrogen.
Far-UV CD
The IgG formulations were diluted to 0.25 mg/mL with formulation buffer and quantified with a Jasco J-820 CD spectrometer in combination with a Jasco PTC-423S temperature controller (Jasco International, Tokyo, Japan) in quartz cuvettes with a path length of 1 mm at 25°C. Far-UV spectra were collected by continuous scanning from 200 to 260 nm at a scanning speed of 10 nm/min, a response time of 1 s, a bandwidth of 1 nm, a sensitivity of 100 m, steps of 0.5 nm, and an accumulation of three scans. Spectra Analysis Software (Version1.53.04, Jasco) was used to background-correct the spectra for the spectrum of the respective buffer. Data were calculated as mean residue ellipticity based on mean amino acid residue weight. The mean residue ellipticity was determined as [θ]mrw, λ = (MRW × θλ)/(10 × c × d), where MRW is the mean residue weight, θλ is the observed ellipticity (in millidegrees) at wavelength λ, c is the protein concentration in mg/mL, and d is the path length in cm. Thermal studies were conducted by raising the temperature in 0.5°C intervals from 25 to 100°C at a rate of 60°C/h. Far-UV CD spectrum after heating have been measured by incubation of the sample at 25°C for 15 min. The molar ellipticity at 217 nm was monitored for changes in the relative content of the β-sheet structure of the immunoglobulins.
DSC
The thermal stability of individual domains was evaluated by using DSC. Measurements were performed on a 1.0 mg/mL IgG solution using a capillary VP-DSC system (MicroCal LLC, Northampton, MA) with a cell volume of 0.135 mL. Temperature scans were performed from 25 to 100°C at a scan rate of 1°C/min. A buffer–buffer reference scan was subtracted from each sample scan before concentration normalization. Baselines were created in Origin 7.0 (OriginLab, Northampton, MA) by cubic interpolation of the pre- and post-transition baselines.
DSF
DSF was used to monitor IgG unfolding during temperature melting. A 96-well microplate was used during DSF, with each well containing 19.5 μL of IgG sample and 0.5 μL of SYPRO Orange (Invitrogen Inc.) that had been diluted from the purchased stock to 1:125 in water. The final dye concentration was 1/5000 of that of the initial product. A CFX96 Real-Time PCR instrument (Bio-Rad Laboratories, Hercules, CA) was used, and the “FRET” channel setting was used to record fluorescence changes during DSF measurement. Samples were incubated at 20°C for 3 min before the melting, during which time the temperature was increased from 35 to 95°C in 0.5°C increments, with an equilibration time of 10 s at each temperature. The hydrophobic exposure temperature, Tm1 or Tm2, was reported as an indication of the transition midpoint of protein unfolding. The first-order derivative curves and Tm values were determined by using CFX Manager software (Bio-Rad Laboratories).
Acknowledgments
The authors thank Dr. Masayoshi Tsukahara and Dr. Tomoyoshi Ishikawa for fruitful discussions and Ms. Rie Sato and Ms. Kazue Sasaki for help with experimental work.
Glossary
- CD
circular dichroism
- DSC
differential scanning calorimetry
- DSF
differential scanning fluoroscopy
- DTT
dithiothreitol
- HMWS
high-molecular-weight species
- LMWS
low-molecular-weight species
- MRW
mean residue weight
- SEC
size-exclusion chromatography
- VH
variable regions of heavy chains
- VL
variable regions of light chains.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat Rev Immunol. 2010;10:301–316. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- 2.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol. 2010;10:345–352. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- 3.Reichert JM. Marketed therapeutic antibodies compendium. mAbs. 2012;4:413–415. doi: 10.4161/mabs.19931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies DR, Metzger H. Structural basis of antibody function. Annu Rev Immunol. 1983;1:87–117. doi: 10.1146/annurev.iy.01.040183.000511. [DOI] [PubMed] [Google Scholar]
- 5.Cambier JC, Bedzyk W, Campbell K, Chien N, Friedrich J, Harwood A, Jensen W, Pleiman C, Clark MR. The B-Cell antigen receptor: structure and function of primary, secondary, tertiary and quaternary components. Immunol Rev. 1993;132:85–106. doi: 10.1111/j.1600-065x.1993.tb00838.x. [DOI] [PubMed] [Google Scholar]
- 6.Jefferis R. Antibody therapeutics: isotype and glycoform selection. Exp Opin Biol Ther. 2007;7:1401–1413. doi: 10.1517/14712598.7.9.1401. [DOI] [PubMed] [Google Scholar]
- 7.Clark MR. IgG effector mechanism. Chem Immunol. 1997;7:715–725. [PubMed] [Google Scholar]
- 8.Bell E. Antibodies: IgG effector function: a question of balance. Nat Rev Immunol. 2006;6:4–5. [Google Scholar]
- 9.Salfeld JG. Isotype selection in antibody engineering. Nat Biotechnol. 2007;25:1369–1372. doi: 10.1038/nbt1207-1369. [DOI] [PubMed] [Google Scholar]
- 10.Labrijn AF, Aalberse RC, Schuurman J. When binding is enough: nonactivating antibody formats. Curr Opin Immunol. 2008;20:479–485. doi: 10.1016/j.coi.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Jiang XR, Song A, Bergelson S, Arroll T, Parekh B, May K, Chung S, Strouse R, Mire-Sluis A, Schenerman M. Advances in the assessment and control of the effector functions of therapeutic antibodies. Nat Rev Drug Discov. 2011;10:101–109. doi: 10.1038/nrd3365. [DOI] [PubMed] [Google Scholar]
- 12.Dall'Acqua WF, Cook KE, Damschroder MM, Woods RM, Wu H. Modulation of the effector functions of a human IgG1 through engineering of its hinge region. J Immunol. 2006;177:1129–1138. doi: 10.4049/jimmunol.177.2.1129. [DOI] [PubMed] [Google Scholar]
- 13.Yoo EM, Wims LA, Chan LA, Morrison SL. Human IgG2 can form covalent dimers. J Immunol. 2003;170:3134–3138. doi: 10.4049/jimmunol.170.6.3134. [DOI] [PubMed] [Google Scholar]
- 14.Wypych J, Li M, Guo A, Zhang Z, Martinez T, Allen MJ, Fodor S, Kelner DN, Flynn GC, Liu YD, Bondarenko PV, Ricci MS, Dillon TM, Balland A. Human IgG2 antibodies display disulfide-mediated structural isoforms. J Biol Chem. 2008;283:16194–16205. doi: 10.1074/jbc.M709987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dillon TM, Ricci MS, Vezina C, Flynn GC, Liu YD, Rehder DS, Plant M, Henkle B, Li Y, Deechongkit S, Varnum B, Wypych J, Balland A, Bondarenko PV. Structural and functional characterization of disulfide isoforms of the human IgG2 subclass. J Biol Chem. 2008;283:16206–16215. doi: 10.1074/jbc.M709988200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu YD, Chen X, Enk JZ, Plant M, Dillon TM, Flynn GC. Human IgG2 antibody disulfide rearrangement in vivo. J Biol Chem. 2008;283:29266–29272. doi: 10.1074/jbc.M804787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez T, Guo A, Allen MJ, Han M, Pace D, Jones J, Gillespie R, Ketchem RR, Zhang Y, Balland A. Disulfide connectivity of human immunoglobulin G2 structural isoforms. Biochemistry. 2008;47:7496–7508. doi: 10.1021/bi800576c. [DOI] [PubMed] [Google Scholar]
- 18.Van der Neut Kolfxchoten M, Schuurman J, Losen M, Bleeker WK, Martinez-Martinez P, Vermeulen E, den Bleker TH, Wiegman L, Vink T, Aarden LA, De Baets MH, van de Winkel JG, Aalberse RC, Parren PW. Anti-inflammatory activity of human IgG4 antibodies by dynamic Fab arm exchange. Science. 2007;317:1554–1557. doi: 10.1126/science.1144603. [DOI] [PubMed] [Google Scholar]
- 19.Van der Zee JS, van Swieten P, Aalberse RC. Serologic aspects of IgG4 antibodies. IgG4 antibodies form small, nonprecipitating immune complexes due to functional monovalency. J Immunol. 1986;137:3566–3571. [PubMed] [Google Scholar]
- 20.Schuurman J, Van Ree R, Perdok GJ, Van Doorn HR, Tan KY, Aalberse RC. Normal human immunogloblin G4 is bispecific: it has two different antigen-combining sites. Immunology. 1997;97:693–698. doi: 10.1046/j.1365-2567.1999.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ejima D, Tsumoto K, Fukuda H, Yumioka R, Nagase K, Arakawa T, Philo JS. Effects of acid exposure on the conformation, stability, and aggregation of monoclonal antibodies. Proteins. 2007;66:954–962. doi: 10.1002/prot.21243. [DOI] [PubMed] [Google Scholar]
- 22.Franey H, Brych SR, Kolvenbach CG, Rajan RS. Increased aggregation propensity of IgG2 subclass over IgG1: role of conformational changes and covalent character in isolated aggregates. Protein Sci. 2010;19:1601–1615. doi: 10.1002/pro.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vermeer AW, Norder W. The thermal stability of immunoglobulin: unfolding and aggregation of a multi-domain protein. Biophys J. 2000;78:394–404. doi: 10.1016/S0006-3495(00)76602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garber E, Demarest JD. A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem Biophys Res Commun. 2007;355:751–757. doi: 10.1016/j.bbrc.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 25.Demarest SJ, Rogers J, Hansen G. Optimization of the antibody CH3 domain by residue frequency analysis of IgG sequences. J Mol Biol. 2004;335:41–48. doi: 10.1016/j.jmb.2003.10.040. [DOI] [PubMed] [Google Scholar]
- 26.Hari SB, Lau H, Razinkov VI, Chen S, Latypov RF. Acid-induced aggregation of human monoclonal IgG1 and IgG2: molecular mechanism and the effect of solution composition. Biochemistry. 2010;49:9328–9338. doi: 10.1021/bi100841u. [DOI] [PubMed] [Google Scholar]
- 27.Latypov RF, Hogan S, Lau H, Gadgil H, Liu D. Elucidation of acid-induced unfolding and aggregation of human immunoglobulin IgG1 and IgG2 Fc. J Biol Chem. 2012;287:1381–1396. doi: 10.1074/jbc.M111.297697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen CB, Manno M, Rischel C, Thórólfsson M, Martorana V. Aggregation of multidomain protein: a coagulation mechanism governs aggregation of a model IgG1 antibody under weak thermal stress. Protein Sci. 2010;19:279–290. doi: 10.1002/pro.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He F, Hogan S, Latypov RF, Narhi LO, Razinkov VI. High throughput thermostability screening of monoclonal antibody formulations. J Pharm Sci. 2010;99:1707–1720. doi: 10.1002/jps.21955. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg DS, Bishop SM, Shah AU, Sathish HA. Formulation development of therapeutic monoclonal antibodies using high-throughput fluorescence and static light scattering techniques: role of conformational and colloidal stability. J Pharm Sci. 2010;100:1306–1315. doi: 10.1002/jps.22371. [DOI] [PubMed] [Google Scholar]
- 31.King AC, Woods M, Liu W, Lu Z, Gill D, Krebs MR. High-throughput measurement, correlation analysis, and machine-learning predictions for pH and thermal stabilities of Pfizer-generated antibodies. Protein Sci. 2011;20:1546–1557. doi: 10.1002/pro.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen SL, Price C, Vlasak J. Beta-elimination and peptide bond hydrolysis: two distinct mechanisms of human IgG1 hinge fragmentation upon storage. J Am Chem Soc. 2007;129:6976–6977. doi: 10.1021/ja0705994. [DOI] [PubMed] [Google Scholar]
- 33.Xiang T, Lundell E, Sun Z, Liu H. Structural effect of a recombinant monoclonal antibody on hinge region peptide bond hydrolysis. J Chromatogr. 2007;858:254–262. doi: 10.1016/j.jchromb.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 34.Cordoba AJ, Shyong BJ, Breen D, Harris RJ. Non-Enzymatic hinge region fragmentation of antibodies in solution. J Chromatogr. 2005;818:115–121. doi: 10.1016/j.jchromb.2004.12.033. [DOI] [PubMed] [Google Scholar]
- 35.Gaza-Bulseco G, Liu H. Fragmentation of a recombinant monoclonal antibody at various pH. Pharm Res. 2008;25:1881–1890. doi: 10.1007/s11095-008-9606-3. [DOI] [PubMed] [Google Scholar]
- 36.Ishikawa T, Ito T, Endo R, Nakagawa K, Sawa E, Wakamatsu K. Influence of pH on heat-induced aggregation and degradation of therapeutic monoclonal antibodies. Biol Pharm Bull. 2010;33:1413–1417. doi: 10.1248/bpb.33.1413. [DOI] [PubMed] [Google Scholar]
- 37.Franey H, Brych SR, Kolvenbach CG, Rajan RS. Increased aggregation propensity of IgG2 subclass over IgG1: role of conformational change and covalent character in isolated aggregates. Protein Sci. 2010;19:1601–1615. doi: 10.1002/pro.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daugherty AL, Mrsny RJ. Formulation and delivery issue for monoclonal antibody therapeutics. Adv Drug Deliv Rev. 2006;58:686–706. doi: 10.1016/j.addr.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Fradkin AH, Carpenter JF, Randolph TW. Immunogenicity of aggregates of recombinant human growth hormone in mouse models. J Pharm Sci. 2009;98:3247–3264. doi: 10.1002/jps.21834. [DOI] [PubMed] [Google Scholar]
- 40.Rosenberg AS. Effects of protein aggregates: an immunologic perspective. AAPS J. 2006;8:E501–E507. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ionescu RM, Vlasak J, Price C, Kirchmeier M. Contribution of variable domain to the stability of humanized IgG1 monoclonal antibodies. J Pharm Sci. 2008;97:1414–1426. doi: 10.1002/jps.21104. [DOI] [PubMed] [Google Scholar]
- 42.Welfle K, Misselwitz R, Hausdorf G, Höhne W, Welfle H. Conformation, pH-induced conformational changes, and thermal unfolding of anti p24 (HIV-1) monoclonal antibody CB 4–1 and its Fab and Fc fragments. Biochim Biophys Acta. 1999;1431:120–131. doi: 10.1016/s0167-4838(99)00046-1. [DOI] [PubMed] [Google Scholar]
- 43.Angal S, King DJ, Bodmer MW, Turner A, Lawson AD, Roberts G, Pedley B, Adair JR. A single amino acid substitution abolishes the heterogeneity of chimeric mouse/human (IgG4) antibody. Mol Immunol. 1993;30:105–108. doi: 10.1016/0161-5890(93)90432-b. [DOI] [PubMed] [Google Scholar]
- 44.Reddy MP, Kinney CA, Chaikin MA, Payne A, Fishman-Lobell J, Tsui P, Dal Monte PR, Doyle ML, Brigham-Burke MR, Anderson D, Reff M, Newman R, Hanna N, Sweet RW, Truneh A. Elimination of Fc receptor-dependent effector functions of a modified IgG4 monoclonal antibody to human CD4. J Immunol. 2000;164:1925–1933. doi: 10.4049/jimmunol.164.4.1925. [DOI] [PubMed] [Google Scholar]
- 45.Cole MS, Anasetti C, Tso JY. Human IgG2 variants of chimeric anti-CD3 are nonmitogenic to T cells. J Immunol. 1997;159:3613–3621. [PubMed] [Google Scholar]
- 46.Oganesyan V, Gao C, Shirinian L, Wu H, Dall'Acqua WF. Structural characterization of a human Fc fragment engineered for lack of effector functions. Acta Cryst. 2008;D64:700–704. doi: 10.1107/S0907444908007877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An Z, Forrest G, Moore R, Cukan M, Haytko P, Huang L, Vitelli S, Zhao JZ, Lu P, Hua J, Gibson CR, Harvey BR, Montgomery D, Zaller D, Wang F, Strohl W. IgG2m4, an engineered antibody isotype with reduced Fc function. Mabs. 2009;1:572–579. doi: 10.4161/mabs.1.6.10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


