Abstract
The opportunistic pathogen Aspergillus fumigatus is ubiquitous in the environment and predominantly infects immunocompromised patients. The functions of many genes remain unknown despite sequencing of the fungal genome. A putative translation elongation factor 1Bγ (eEF1Bγ, termed elfA; 750 bp) is expressed, and exhibits glutathione S-transferase activity, in A. fumigatus. Here, we demonstrate the role of ElfA in the oxidative stress response, as well as a possible involvement in translation and actin cytoskeleton organization, respectively. Comparative proteomics, in addition to phenotypic analysis, under basal and oxidative stress conditions, demonstrated a role for A. fumigatus elfA in the oxidative stress response. An elfA-deficient strain (A. fumigatus ΔelfA) was significantly more sensitive to the oxidants H2O2, diamide, and 4,4′-dipyridyl disulfide (DPS) than the wild-type. This was further supported with the identification of differentially expressed proteins of the oxidative stress response, including; mitochondrial peroxiredoxin Prx1, molecular chaperone Hsp70 and mitochondrial glycerol-3-phosphate dehydrogenase. Phenotypic analysis also revealed that A. fumigatus ΔelfA was significantly more tolerant to voriconazole than the wild-type. The differential expression of two aminoacyl-tRNA synthetases suggests a role for A. fumigatus elfA in translation, while the identification of actin-bundling protein Sac6 and vacuolar dynamin-like GTPase VpsA link A. fumigatus elfA to the actin cytoskeleton. Overall, this work highlights the diverse roles of A. fumigatus elfA, with respect to translation, oxidative stress and actin cytoskeleton organization. In addition to this, the strategy of combining targeted gene deletion with comparative proteomics for elucidating the role of proteins of unknown function is further revealed.
Keywords: proteomics, translation, GST, redox, elongation factor, glutathione
Introduction
Translation is regulated in response to intracellular growth, extracellular signals and stress conditions.1–3 In response to stress, protein synthesis is usually reduced to prevent translation errors, and molecular chaperones are often upregulated to deal with any increase in denatured or misfolded proteins.1,4 This regulation of protein synthesis primarily occurs at the initiation stage; however, regulation has been observed to occur during elongation.1,4,5
The eukaryotic elongation factor 1 (eEF1) complex delivers all aminoacyl-tRNAs to the ribosome except the initiator tRNA and selenocysteine tRNAs.6 The eEF1 complex is composed of two subunits, eEF1A and eEF1B.7 eEF1A binds and recruits aminoacyl-tRNA to the ribosomal A site where, once a codon/anti-codon match is detected, it deposits the aminoacyl-tRNA.6,8 eEF1A is a G-protein and requires a guanine nucleotide exchange factor (GEF). eEF1B is the GEF for eEF1A and is composed of two subunits; eEF1Bα and eEF1Bγ.8 In metazoans, a third subunit of eEF1B, eEF1Bβ, is present.8,9 In Saccharomyces cerevisiae, the eEF1Bα subunit is the nucleotide exchange factor, while the function of eEF1Bγ has not been fully elucidated.7
In addition to delivering aminoacyl-tRNA to the elongating ribosome, eEF1A is also an actin binding and bundling protein, an interaction that is conserved from yeast to mammals.7,10 Indeed, it has been estimated that greater than 60% of eEF1A in the cell is associated with the actin cytoskeleton.10 Actin is essential and is involved in a variety of cellular processes ranging from growth and differentiation to stress response.11,12 The actin binding and bundling activity of eEF1A is independent of GTP and does not take place in the presence of aminoacyl-tRNA, suggesting mutual exclusivity of these two binding factors.13,14 In vitro, eEF1Bα reduces the actin bundling activity of eEF1A and may act as a regulator in directing eEF1A function to translation elongation.7
The crystal structure of the N-terminal region of S. cerevisiae eEF1Bγ contains a glutathione S-transferase (GST)-like domain, although GST activity was not observed for this domain.9 GST activity, however, has been observed in eEF1Bγ proteins from different organisms (e.g., Aspergillus fumigatus15). In addition to A. fumigatus eEF1Bγ, orthologs from rice and the silk worm Bombyx mori, expressed in Escherichia coli, were also amenable to glutathione (GSH)-affinity purification,16,17 and eEF1Bγ from both these organisms exhibited GST activity.
In A. fumigatus, eEF1Bγ is encoded by A. fumigatus elfA, first identified following GSH-Sepharose affinity chromatography and two-dimensional electrophoresis (2DE).15 Further analysis determined that ElfA was present as a monomer in the cell at 20 kDa. GST activity of native ElfA represented the first demonstration of such activity in native eEF1Bγ, as previously this activity had only been determined in recombinant eEF1Bγ.15–17 The relevance, and presence, of a GST domain in an elongation factor requires further investigation with respect to a possible role in redox control and oxidative stress. In other organisms (e.g., S. cerevisiae and mammalian epithelial cells), eEF1Bγ has been shown to interact with the membranes and the cytoskeleton;18,19 however, it is not known whether A. fumigatus elfA interacts with either the membrane network or the actin cytoskeleton.
Comparative proteomics is a powerful tool for investigating the impact a gene deletion has on the cell as a whole, that is, for studying fungal systems biology. Thus, we employed targeted gene deletion in conjunction with comparative proteomics to gain a better understanding of A. fumigatus elfA.
Results
Deletion and complementation of elfA in A. fumigatus
An A. fumigatus elfA deletion strain (ΔelfA) was generated by homologous recombination using the bi-partite method20 to facilitate functional analysis of elfA. Gene deletion, and subsequent complementation was confirmed by Southern blot analysis (Supporting Information Fig. 1). qRT-PCR confirmed the presence of elfA transcripts in A. fumigatus ATCC46645 and elfAC, and its absence in ΔelfA (Supporting Information Fig. 1). Expression of elfA was higher (1.6-fold) in elfAC than in the wild-type strain. Finally, ElfA was isolated from elfAC thereby confirming expression of the protein in the complemented strain (Supporting Information Fig. 2).
A. fumigatus elfA is involved in the oxidative stress response and redox control
qRT-PCR analysis of elfA expression in A. fumigatus subjected to H2O2-induced oxidative stress (2 mM) for 15–60 min revealed that elfA expression was significantly increased (P = 0.0009) after 15 min [Fig. 1(A)]. After 30 min, expression of elfA was still higher than basal levels, and decreased to below basal levels at 60-min post-exposure.
Figure 1.
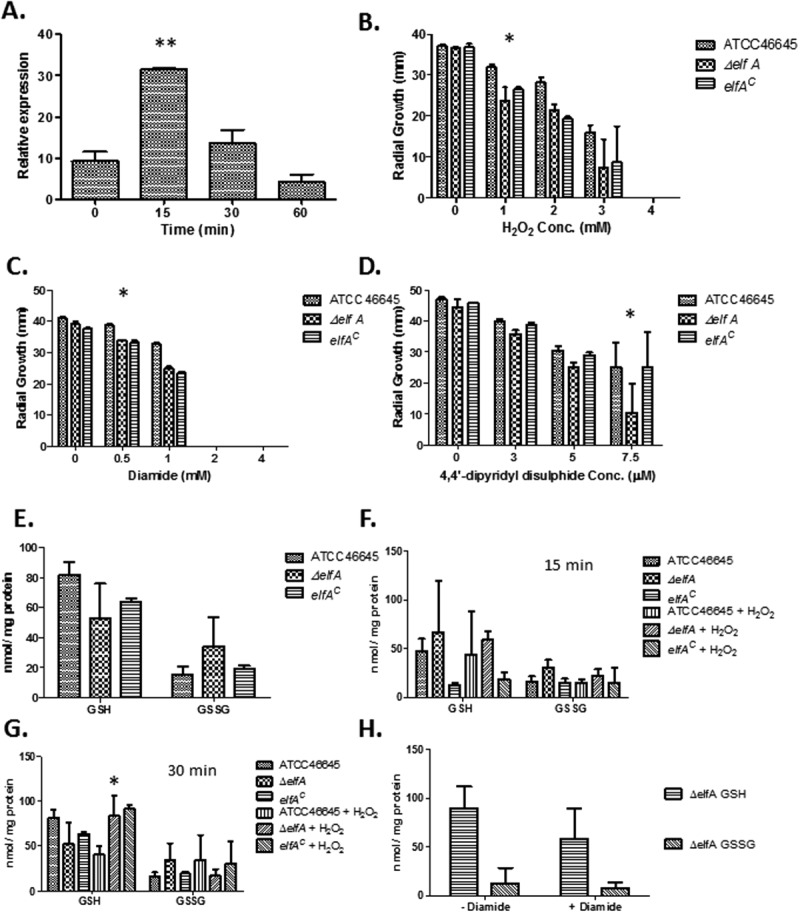
(A) H2O2-induced oxidative stress increases elfA expression, particularly following 15 min exposure (**P = 0.0009). (B) Impact of H2O2 (0–4 mM) on the growth of A. fumigatus ATCC46645, ΔelfA, and elfAC, respectively, after 72 h. (C) Growth of A. fumigatus ATCC46645, ΔelfA, and elfAC in response to diamide (0–4 mM) exposure. (D) The impact of 4,4′-dipyridyl disulfide (0–7.5 µM) on the growth of A. fumigatus ATCC46645, ΔelfA, and elfAC. (E) GSH and GSSG determination in A. fumigatus ATCC46645, ΔelfA, and elfAC under normal growth conditions. (F) Relative amounts of GSH and GSSG in A. fumigatus ATCC46645, ΔelfA, and elfAC in the absence and presence of 2 mM H2O2 for 15 min. (G) GSH and GSSG amounts in A. fumigatus ATCC46645, ΔelfA, and elfAC in the absence and presence of 2 mM H2O2 for 30 min. (H) Exposure to diamide causes a reduction in GSH levels in A. fumigatus ΔelfA.
A. fumigatus ΔelfA was investigated to ascertain any altered phenotype in response to the oxidative stress-inducing agents, menadione (0–50 µM), H2O2 (0–5 mM), diamide (0–4 mM), and 4,4′-dipyridyl disulfide (DPS; 0–7.5 µM). A. fumigatus ΔelfA was found to be significantly more sensitive to H2O2, diamide and DPS (Supporting Information Fig. 3), but not to menadione (data not shown), indicating a possible role for elfA in the oxidative stress response to specific oxidizers. After 72 h exposure, A. fumigatus ΔelfA was significantly more sensitive to 1 mM H2O2 (P = 0.0006) [Fig. 1(B)]. A. fumigatus ΔelfA was also significantly more sensitive to 0.5 mM diamide (P = 0.0001) after 72 h exposure [Fig. 1(C)]. After 96 h exposure, it was observed that A. fumigatus ΔelfA was significantly more sensitive to 7.5 µM DPS (P = 0.0007) compared to A. fumigatus ATCC46645 or elfAC [Fig. 1(D)]. A. fumigatus ΔelfA displayed an increased tolerance (P = 0.0251) to voriconazole (0.5 µg/mL) compared to A. fumigatus ATCC46645 and elfAC after 72 h exposure (Supporting Information Fig. 4).
In some cases, it was observed that complementation did not restore the wild-type phenotype while in others, i.e., in the presence of DPS and voriconazole, the wild-type phenotype was restored. As described earlier, Southern blot analysis determined a single targeted integration of the complementation construct (Supporting Information Fig. 1) and ElfA was isolated from elfAC (Supporting Information Fig. 2). In addition to this, sequencing of elfA in the complemented strain did not reveal any mutations (data not shown). Importantly, qRT-PCR for the genes flanking elfA showed the identical expression levels in wild-type and ΔelfA confirming that the observed phenotypes were not due to a disruption of the flanking genes (Supporting Information Fig. 5). Conceivably, the genomic architecture of the complemented locus may be altered whereby the hygromycin resistance cassette has a positional effect on elfA expression.
A reduced GSH/GSSG ratio was observed in ΔelfA under normal growth conditions compared to the wild-type and elfAC (wild-type; 5.6, ΔelfA; 3.3, elfAC; 6.0), indicative of cells under oxidative stress (Supporting Information Table 1). This resulted from decreased GSH levels and increased GSSG levels in ΔelfA [Fig. 1(E)]. When exposed to H2O2 for 15 and 30 min, the effect on the GSH/GSSG ratios in wild-type and ΔelfA differed. The ratios decreased in wild-type which is indicative of an oxidative stress response (0 min; 5.66, 15 min; 4.51, 30 min; 2.05) (Supporting Information Table 1). However, in ΔelfA, the GSH/GSSG ratio increased following 15 min exposure to H2O2 (from 3.33 to 4.61), and subsequently decreased (to 1.90) following exposure for 30 min. The increased GSH/GSSG ratio following 15 min exposure of A. fumigatus ΔelfA to H2O2 resulted from decreased GSH and GSSG levels [Fig. 1(F)]. While after 30 min exposure to H2O2, an increase in the amount of GSH in A. fumigatus ΔelfA and a decrease in the amount of GSSG resulted in the decreased GSH/GSSG ratio [Fig. 1(G)]. Significantly increased levels of GSH (P < 0.05) were present in ΔelfA compared to wild-type following exposure to H2O2 for 30 min, while the other trends, although not significant were reproducible (n = 3).
Measurement of GSH levels in A. fumigatus ΔelfA cultured for 24 h prior to exposure to diamide (1 mM) for 2 h, confirmed that diamide decreases cellular GSH levels [Fig. 1(H)].
Proteome remodelling occurs as a consequence of elfA deletion in A. fumigatus
Thirteen proteins were found to be differentially expressed when A. fumigatus ATCC46645 and ΔelfA were cultured for 24 h under basal conditions; eight proteins had a fold increase >1.5 (P < 0.05) in A. fumigatus ΔelfA while five proteins had a fold decrease >1.5 (P < 0.05) when compared to A. fumigatus ATCC46645 [Table I; Fig. 2(A,B)]. Specifically, mitochondrial peroxiredoxin Prx1 and molecular chaperone Hsp70 exhibited increased expression of 2.2- and 1.7-fold, respectively, while Hsc70 co-chaperone (SGT) was decreased 1.7-fold in expression. Tyrosyl-tRNA synthetase was upregulated 2.7-fold. In addition to this, actin-bundling protein Sac6 and Sec13 displayed a 1.9- and 1.7-fold increase in expression.
Table I.
Proteins (n = 13) with a Fold Change in A. fumigatus ΔelfA Compared with A. fumigatus Under Normal Growth Conditions Following Identification by 2DE and LC-MS/MS
| Spot no. | Annotation name | CADRE IDa | Fold change | tpI | tMW | % Sequence coverage |
|---|---|---|---|---|---|---|
| 16 | Tyrosyl-tRNA synthetase | AFUA_5G10640 | +2.7 | 6.18 | 43588 | 20 |
| 25 | Regulatory protein SUAPRGA1 | AFUA_3G09030 | +2.4 | 4.88 | 40070 | 32 |
| 31 | Mitochondrial peroxiredoxin Prx1 | AFUA_4G08580 | +2.2 | 5.38 | 23378 | 33 |
| 43 | Polysaccharide deacetylase family protein | AFUA_5G09130 | +2.0 | 5.39 | 35188 | 42 |
| 48 | Actin-bundling protein Sac6 | AFUA_2G07420 | +1.9 | 5.78 | 72434 | 12 |
| 56 | Nuclear pore complex subunit SEC13 | AFUA_4G06090 | +1.7 | 6.17 | 33856 | 28 |
| 57 | Molecular chaperone Hsp70 | AFUA_1G07440 | +1.7 | 5.09 | 69618 | 56 |
| 70 | MRS7 family protein | AFUA_3G08230 | +1.5 | 6.42 | 68745 | 18 |
| 5 | Ketol-acid reductoisomerase | AFUA_3G14490 | −4.9 | 9.32 | 56318 | 21 |
| 14 | Phoshoribosylaminoimidazole carboxamide formyltransferase/IMP cyclohydrolase | AFUA_4G07690 | −3.0 | 6.39 | 64987 | 33 |
| 59 | Transcription factor RfeF | AFUA_4G10200 | −1.7 | 4.91 | 71649 | 18 |
| 60 | Hsc70 cochaperone (SGT) | AFUA_1G09830 | −1.7 | 4.74 | 35721 | 38 |
| 68 | Short chain dehydrogenase | AFUA_4G08710 | −1.6 | 5.28 | 30909 | 38 |
Figure 2.
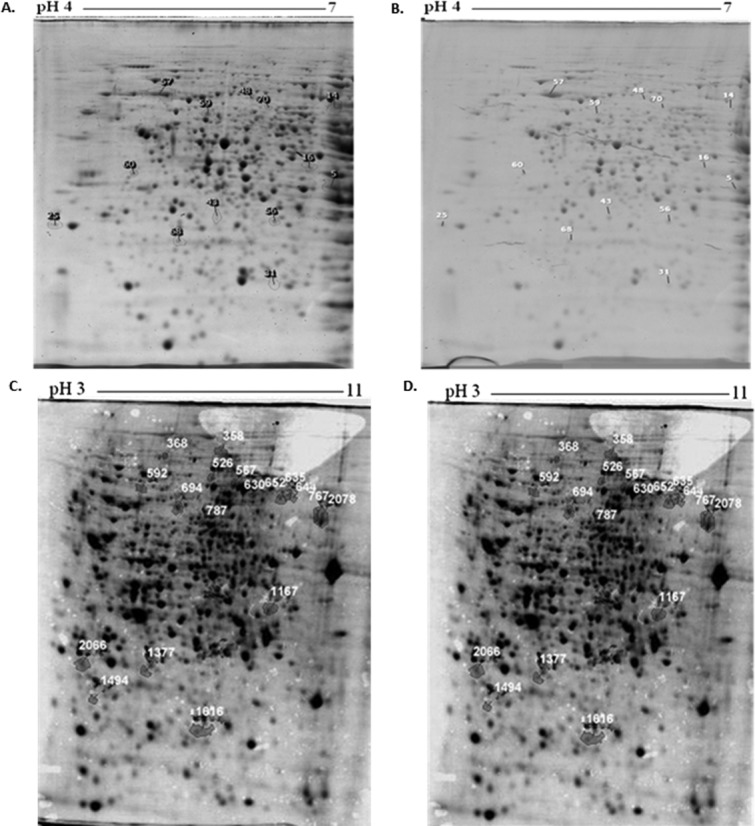
Proteomic remodeling in A. fumigatus ΔelfA in the absence and presence of oxidative stress. 2DE analysis of A. fumigatus ATCC46645 (A) and ΔelfA (B) under normal growth conditions. 2DE analysis of A. fumigatus ATCC46645 (C) and ΔelfA (D) following exposure to 2 mM H2O2 for 1 h. The proteins found to be differentially expressed after analysis using Progenesis™ SameSpot software are numbered.
Oxidative stress induces differential protein expression in A. fumigatus ΔelfA
A. fumigatus ATCC46645 and ΔelfA were cultured for 24 h in AMM before H2O2 (2 mM final concentration) was added for 1 h. Following LC-MS/MS of 18 protein spots that were found to be differentially expressed, 10 proteins were identified as having a fold increase >1.5 (P < 0.05) in A. fumigatus ΔelfA [Table II; Fig. 2(C,D)] while five proteins exhibited a fold decrease >1.5 (P < 0.05) compared to A. fumigatus ATCC46645 in response to 2 mM H2O2 exposure for 1 h. Vacuolar dynamin-like GTPase VpsA was identified from three different spots with fold increases of 7.8, 4.3, and 3.0 in A. fumigatus ΔelfA, while ATP citrate lyase, subunit 1 was identified from two spots which were upregulated 1.9- and 1.5-fold in A. fumigatus ΔelfA. The isoforms of both vacuolar dynamin-like GTPase VpsA and ATP citrate lyase, subunit 1 had the same molecular mass but displayed slight changes in pI. Cobalamin-independent methionine synthase Met H/D was upregulated 1.7-fold, while cysteinyl-tRNA synthetase and glycerol-3-phosphate dehydrogenase were upregulated 1.5-fold in A. fumigatus ΔelfA. Meanwhile, Ras small monomeric GTPase RasA and proteosome regulatory particle Subunit (RpnL) underwent decreased expression of 2.2- and 1.8-fold, respectively.
Table II.
Proteins (n = 15) with a Fold Change in A. fumigatus ΔelfA Compared with A. fumigatus After Exposure to 2 mM H2O2 Following Identification by 2DE and LC-MS/MS
| Spot no. | Annotation name | CADRE IDa | Fold change | tpI | tMW | % Sequence coverage |
|---|---|---|---|---|---|---|
| 644 | Vacuolar dynamin-like GTPase VpsA | AFUA_5G02360 | +7.8 | 8.30 | 78,425 | 18 |
| 635 | Vacuolar dynamin-like GTPase VpsA | AFUA_5G02360 | +4.3 | 8.30 | 78,425 | 3 |
| 652 | Vacuolar dynamin-like GTPase VpsA | AFUA_5G02360 | + 3.0 | 8.30 | 78,425 | 19 |
| 592 | Mitochondrial outer membrane translocase receptor (TOM70) putative | AFUA_2G01660 | +1.9 | 5.40 | 70,009 | 10 |
| 767 | ATP citrate lyase, subunit 1, putative | AFUA_6G10650 | +1.9 | 8.60 | 78,828 | 33 |
| 368 | Carbamoyl-phosphate synthase, large subunit | AFUA_2G10070 | +1.7 | 6.10 | 129,214 | 25 |
| 567 | Cobalamin-independent methionine synthase Met H/D | AFUA_4G07360 | +1.7 | 6.33 | 87,072 | 31 |
| 1377 | Metallo-β-lactamase family protein | AFUA_5G12770 | +1.6 | 5.60 | 34,197 | 30 |
| 526 | Cysteinyl-tRNA synthetase | AFUA_5G09610 | +1.5 | 5.95 | 91,041 | 25 |
| 630 | Glycerol-3-phosphate dehydrogenase, mitochondrial | AFUA_1G08810 | +1.5 | 6.71 | 76,899 | 31 |
| 694 | Vacuolar ATP synthase catalytic subunit A | AFUA_5G02370 | +1.5 | 5.83 | 75,329 | 43 |
| 787 | Pyruvate decarboxylase PdcA | AFUA_3G11070 | +1.5 | 6.08 | 63,307 | 33 |
| 2078 | ATP citrate lyase, subunit 1 | AFUA_6G10650 | +1.5 | 8.66 | 79,359 | 42 |
| 1494 | RAS small monomeric GTPase RasA | AFUA_5G11230 | −2.2 | 4.97 | 24,292 | 61 |
| 1167 | Nuclear pore complex protein (SonA) | AFUA_1G09020 | −2.1 | 7.61 | 40,220 | 32 |
| 2066 | Proteasome regulatory Particle Subunit (RpnL) | AFUA_3G08940 | −1.8 | 4.9 | 31,201 | 55 |
| 358 | Pyruvate carboxylase | AFUA_4G07710 | −1.6 | 6.23 | 132,003 | 27 |
| 1616 | Haloalkanoic acid dehalogenase | AFUA_6G14460 | −1.6 | 6.19 | 26,716 | 39 |
Increased protein glutathionylation was observed in A. fumigatus ΔelfA
Due to the altered redox environment in A. fumigatus ΔelfA, it was hypothesized that global protein glutathionylation may be increased as a protective measure. Biotinylation of GSSG was confirmed by both HPLC and MALDI-ToF analysis (Supporting Information Fig. 6) and was subsequently used as a probe to detect proteins susceptible to glutathionylation as the addition of GSSG leads to thiol-disulfide exchange reactions with these proteins, while the biotin moiety allows for their detection by Western analysis.23 Increased protein glutathionylation in A. fumigatus ΔelfA compared to A. fumigatus ATCC46645 was observed [Fig. 3(A,B)], which was supported by image analysis that determined a 24% increase in chemiluminescent signal intensity in A. fumigatus ΔelfA compared to wild-type signals [Fig. 3(C)]. Addition of DTT and treatment of whole cell lysates with water acted as controls to ensure that any proteins detected were as a direct result of glutathionylation by biotin-GSSG [Fig. 3(B)], while equal protein loading [Fig. 3(A)] confirmed observed signal intensity differences between the strains.
Figure 3.
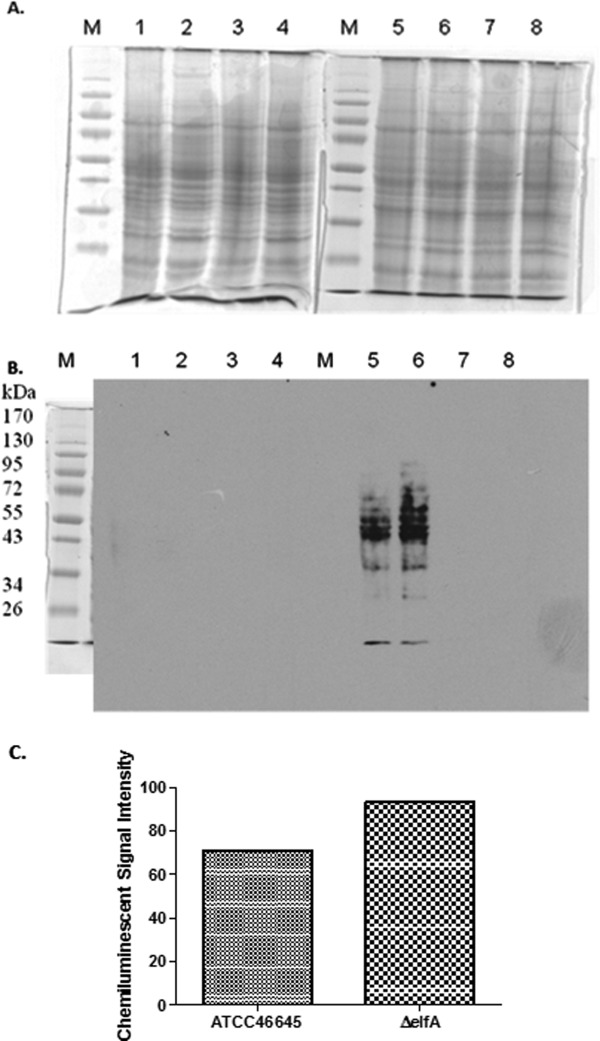
Western blot analysis of protein glutathionylation in A. fumigatus ATCC46645 and ΔelfA. (A) SDS-PAGE of A. fumigatus ATCC46645 (Lanes 1, 3, 5, 7) and ΔelfA (Lanes 2, 4, 6, 8) whole cell lysates (50 µg) under reducing (Lanes 1–4) and nonreducing conditions (Lanes 5–8). The samples in Lanes 1, 2, 5, and 6 were incubated with biotin-GSSG, while the samples in Lanes 3, 4, 7, and 8 were incubated with water as a control. (B) Western blot probed with streptavidin-HRP. Proteins glutathionylated upon addition of biotin-GSSG were detected in A. fumigatus ATCC46645 (Lane 5) and ΔelfA (Lane 6). There were more glutathionylated proteins detected in A. fumigatus ΔelfA (Lane 6) than in A. fumigatus ATCC46645 (Lane 5). No glutathionylated proteins were detected on the reduced blot (Lanes 1–4) or in the samples treated with water (Lanes 7–8). (C) Image analysis indicates a 24% increase in chemiluminescent signal intensity in A. fumigatus ΔelfA compared to A. fumigatus ATCC46645 supporting the observation that there was a higher level of protein glutathionylation in A. fumigatus ΔelfA.
Discussion
The work described here reveals the functional characterization of ElfA, an eEF1Bγ from A. fumigatus. eEF1Bγ is a member of the eEF1 complex required for translation and, along with eEF1Bα, forms the eEF1B component of the eEF1 complex which provides nucleotide exchange factor activity to the eEF1A subunit.7,8 Comparative proteomics highlighted that under normal growth conditions, proteins involved in stress responses (i.e., mitochondrial peroxiredoxin Prx1, molecular chaperone Hsp70, and Hsc70 co-chaperone) were differentially expressed, with Prx1 and Hsp70 expression increased while the expression of Hsc70 was decreased. The observation of these differentially expressed proteins indicates that in the absence of A. fumigatus elfA, a stressed intracellular environment exists. This is confirmed by altered redox homeostasis in A. fumigatus ΔelfA where, under normal growth conditions the GSH/GSSG ratio of A. fumigatus ΔelfA was decreased compared to that of A. fumigatus ATCC46645, strongly indicative of oxidative stress.24 Further evidence of an altered redox state is the increased level of protein glutathionylation in A. fumigatus ΔelfA. Combined, these data suggest that in the absence of ElfA, translation is partially disrupted (as evidenced by altered levels of certain tRNA synthetases and enzymes dealing with protein aggregation), either resulting in, or consequent to, oxidative stress, allied to dysregulation of GSH/GSSG in A. fumigatus.
The increased expression of mitochondrial peroxiredoxin Prx1 in A. fumigatus ΔelfA was especially interesting because in S. cerevisiae, mitochondrial peroxiredoxin Prx1 requires GSH-mediated reduction for antioxidant activity.25 Indeed, both GSH and a thioredoxin were required to maintain the mitochondrial peroxiredoxin Prx1 in its reduced, active state. Specifically, the catalytic cysteine of peroxiredoxin is reactivated by glutathionylation and subsequent reduction by thioredoxin reductase coupled with GSH.26 The increased expression of mitochondrial peroxiredoxin Prx1 in A. fumigatus ΔelfA may be consequent to the decreased GSH levels observed.
The significant increase in expression of A. fumigatus elfA after 15 min of exposure to H2O2 confirmed our initial hypothesis that elfA is involved in the oxidative stress response in A. fumigatus. Consolidating this hypothesis, A. fumigatus ΔelfA was found to be significantly more sensitive than wild-type to the oxidants H2O2, diamide, and DPS. Both diamide and DPS are thiol-specific oxidants which react directly with GSH.26 The reduced GSH levels in A. fumigatus ΔelfA compared to those in A. fumigatus ATCC46645 confirm that GSH levels are depleted to a greater extent in A. fumigatus ΔelfA in the presence of these oxidants resulting in the increased sensitivity of A. fumigatus ΔelfA. Conversely, in S. cerevisiae, an eEF1Bγ deletion strain was more resistant to the oxidants; H2O2, menadione and CdSO4.27 However, in S. cerevisiae, the GST domain of eEF1Bγ is not active,9 unlike in A. fumigatus.15 Thus, the data presented here lead to the conclusion that the eEF1Bγ proteins in A. fumigatus and S. cerevisiae, respectively, mediate differential responses consequent to oxidative stress which may be due to the presence of an active GST domain in ElfA.
The effects of H2O2-induced oxidative stress on the proteome of A. fumigatus ΔelfA was investigated and compared to that of A. fumigatus ATCC46645. Proteins which participate in the oxidative stress response such as cobalamin-independent methionine synthase and mitochondrial glycerol-3-phosphate dehydrogenase exhibited increased expression, while Ras small monomeric GTPase RasA and the proteasome regulatory particle underwent decreased expression.28–33
Cobalamin-independent methionine synthase, which exhibited increased expression in A. fumigatus ΔelfA, is essential for S-adenosylmethionine (SAM) cycle occurrence and homocysteine, an intermediate of the SAM cycle, can be converted to glutathione.34 This is particularly relevant since GSH/GSSG determination in A. fumigatus ΔelfA, following exposure to H2O2, revealed an increase in GSH levels. Thus, a clear systems link is established between the proteomic and phenotypic analyses. Because GSH levels in A. fumigatus ΔelfA were lower than those in the wild-type due to the stressed environment, we speculate that cobalamin-independent methionine synthase expression was upregulated to increase the GSH levels following exposure to H2O2. Interestingly, it has been observed that addition of methionine to Penicillium chrysogenum cultures also resulted in increased intracellular GSH levels,35 and if this is also the case in A. fumigatus, it may represent a universal response to GSH depletion consequent to oxidative stress in filamentous fungi.
Further analysis of differentially expressed proteins identified in A. fumigatus ΔelfA under both normal growth conditions and following exposure to H2O2, revealed a number of proteins involved in protein folding and protein degradation were differentially expressed. Under normal growth conditions, molecular chaperone Hsp70 and nuclear pore complex Sec13 exhibited increased expression in A. fumigatus ΔelfA. The Hsp protein family is required for the disassembly of protein aggregates, protein folding, and the degradation of misfolded proteins,36,37 while Sec13 functions in the coat protein complex II (COPII), vesicular trafficking, nuclear pore function, and endoplasmic reticulum-associated degradation (ERAD).38 The COPII complex also transports misfolded proteins to the ERAD,39 which in turn is involved in the retrotranslocation of misfolded proteins from the ER to the cytosol for degradation by proteasomes.40 Previously, Sec13 was found to be upregulated in S. cerevisiae by the unfolded protein response (UPR),41 which has been shown to target genes involved in the ERAD.42,43 The increased expression of these proteins under normal growth conditions is indicative of a greater requirement for protein degradation in A. fumigatus ΔelfA which is in accordance with the stressed environment evident due to aberrant translation and decreased GSH/GSSG ratio.
One of the consequences of oxidative stress is an increase in misfolded and irreversibly damaged proteins which, unless removed from the cell, are highly toxic.26,44 Following exposure to H2O2, the increased expression of vacuolar dynamin-like GTPase VpsA and the decreased expression of the proteasome regulatory particle subunit (RpnL) pointed toward an increased requirement for protein degradation in A. fumigatus ΔelfA. One of the functions of vacuolar dynamin-like GTPase VpsA is in vacuole fission.28 Vacuoles, along with proteasomes, degrade proteins exported from the ER by the ERAD.18,40,45 The proteasome regulatory particle subunit forms the lid of the 19S regulatory subunit which is required for degradation of ubiquitinylated proteins but inhibits degradation of oxidized proteins.31,33 This decreased expression of the proteasome regulatory particle subunit in A. fumigatus ΔelfA is indicative of an attenuation of proteasome regulation to facilitate degradation of the increased levels of misfolded or oxidized proteins present in the cell, consequent to loss of ElfA. Interestingly, in S. cerevisiae, increased accumulation of oxidized proteins was observed in an eEF1Bγ deletion strain which also exhibited altered vacuole morphology and altered expression of Hsp proteins.18 Thus, the identification of a number of proteins involved in protein folding and degradation of misfolded proteins in A. fumigatus ΔelfA, under both normal growth conditions and following exposure to H2O2, suggests that in the absence of ElfA, the cellular protein degradation systems are activated. We hypothesize that in the absence of ElfA, a stressed environment ensues resulting in increased levels of misfolded and irreversibly damaged proteins, which must be removed from the cell to prevent manifestation of toxic effects.
Tyrosyl-tRNA synthetase exhibited increased expression in A. fumigatus ΔelfA under normal growth conditions, while cysteinyl-tRNA synthetase exhibited increased expression upon exposure to H2O2. We postulate that the increased expression of these aminoacyl-tRNA synthetases is indicative of altered protein synthesis in A. fumigatus ΔelfA. In particular, the increased expression of cysteinyl-tRNA suggests an increased requirement for cysteine-containing proteins, such as peroxiredoxins, which are essential for attenuation of oxidative stress within the cell.46,47
The unexpected increased expression of additional proteins (i.e., actin-bundling protein Sac6 and vacuolar dynamin-like GTPase VpsA), in A. fumigatus ΔelfA under both normal growth conditions and following oxidative stress, involved in cytoskeletal transformation, suggests the involvement of ElfA with the cytoskeleton. Under normal growth conditions, actin-bundling protein Sac6 exhibited increased expression in A. fumigatus ΔelfA. Moreover, following exposure to H2O2, increased expression of vacuolar dynamin-like GTPase VpsA, which in addition to its function in vacuole fission is also required for normal actin cytoskeleton organization in S. cerevisiae,48 was observed. The cytoskeleton plays a role in the oxidative stress response whereby it confers protection by collapsing into actin bundles that sequester actin and its associated proteins into immobile structures.11 In S. cerevisiae, Sac6 was observed to co-localize with these actin bundles during oxidative stress induced by H2O2, menadione, and diamide.11 Translation elongation and actin cytoskeleton modulation are linked, because eEF1A in addition to its canonical role of delivering aminoacyl-tRNA to the elongating ribosome, is also an actin binding and bundling protein.10 In S. cerevisiae, eEF1Bα regulates the actin binding and bundling activities of eEF1A and has been proposed to direct eEF1A toward binding aminoacyl-tRNA.7 In mammalian epithelial cells, eEF1Bγ increased the formation of keratin intermediate filament bundles in vivo.19 Moreover, overexpression of eEF1Bγ in epithelial cells resulted in a disrupted interaction between eEF1Bγ and keratin, and also reduced protein synthesis suggesting a functional link between the cytoskeletal structure and translation in epithelial cells.19,49 The observation of increased expression of Sac6 in A. fumigatus ΔelfA under normal growth conditions suggests that ElfA (an EF1Bγ) also links the actin cytoskeleton with the translational apparatus.
We have demonstrated a diverse role for ElfA with respect to the oxidative stress response, GSH/GSSG redox homeostasis, regulation of protein degradation and antifungal resistance in A. fumigatus. We hypothesize that the ensuing alteration in protein synthesis, after loss of ElfA, results in increased levels of misfolded proteins which may in turn generate a stressed environment which activates the oxidative stress response. Alternatively, loss of ElfA and the subsequent deficit in translation results in a stressed environment, which not only activates the oxidative stress response, but also results in increased levels of misfolded and irreversibly damaged proteins. The result of either outcome is an increase in protein degradation and oxidative stress response pathways in A. fumigatus.
Materials and Methods
Strains and growth conditions
A. fumigatus strains were grown at 37°C in Aspergillus minimal media (AMM), and fungal culturing was carried out as described previously.50 The bacterial strain E. coli TOP10 (Invitrogen, The Netherlands) which was cultivated in LB (1% (w/v) Bacto-tryptone, 0.5% (w/v) yeast extract, 1% (w/v) NaCl, pH 7.5) medium was used for general cloning procedures.
Deletion and complementation of A. fumigatus elfA
An A. fumigatus ΔelfA strain was generated using the bipartite marker technique51 using overlapping fragments of a pyrithiamine resistance gene (ptrA),20,52 fused to 1.2 kb of the 5′ and 3′ flanking regions of the elfA coding sequence. The primers required for generating the constructs are listed in Supporting Information Table 2. A. fumigatus ATCC46645 was transformed as described previously50 and colonies picked in order to obtain homokaryotic transformants. Single genomic integration was confirmed by Southern blot analysis following DNA purification using a ZR Fungal/Bacterial DNA Kit (Zymoresearch).
A. fumigatus ΔelfA was complemented with a construct that contained elfA and the 5′ and 3′ flanking regions, and also a hygromycin resistance gene (hph) for selection.53 Briefly, elfA and the 5′ and 3′ flanking regions were amplified using the primers oelfA5 and oelfA6, the resulting 3.1 kb product was cloned into TOPO® vector. The resulting plasmid was linearized and ligated to hph, which was released from pAN7-153 by restriction digestion, and cloned into TOPO® vector to give the complementation construct. The plasmid was linearized before transforming A. fumigatus ΔelfA protoplasts.
RNA isolation and quantitative RT-PCR
Fungal RNA isolation, DNase treatment, cDNA synthesis and qRT-PCR was performed as described previously.54 The primers used in the qRT-PCR reactions are listed in Supporting Information Table 2.
Plate assays
Conidia (106), harvested aseptically from 1-week-old AMM plates, were subject to a variety of phenotypic assays by point inoculating on AMM plates containing a stressor, as shown in Supporting Information Table 3. Plates were incubated at 37°C. Colony diameter was measured periodically and statistical analysis was carried out using one-way ANOVA.
GSH/GSSG determination
A. fumigatus ATCC46645 and ΔelfA (1 × 105 cfu/mL) were cultured for 24 h in AMM before addition of H2O2 (2 mM final) for 15 and 30 min, respectively. A. fumigatus ATCC46645 and ΔelfA not exposed to H2O2 were used as controls. Mycelia were harvested through miracloth and dried before glutathione reduced/glutathione disulfide (GSH/GSSG) determination.55
Protein extraction
A. fumigatus ATCC46645 and ΔelfA (1 × 105 cfu/mL) were grown for 24 h in AMM (100 mL cultures), at 37°C with shaking at 200 rpm. After 24 h, H2O2 (2 mM final) was added. Following 1 h incubation, mycelia were harvested, washed with cold deionized water, dried in tissue and snap frozen in liquid N2. The mycelia were ground into a fine powder under liquid N2. Ground mycelia (250 mg) were added to 10% (w/v) TCA (1.5 mL), incubated on ice for 30 min and sonicated with a sonication probe (Bandelin Sonopuls, Bandelin electronic, Berlin) with cooling. Lysed mycelia were incubated on ice for a further 30 min before centrifugation at 12,000g for 10 min at 4°C. Supernatants were discarded and 60 µL H2O was added to protein and vortexed. Following acetone washes, protein was resuspended in IEF Buffer (500 µL), incubated at room temperature for 1 h before centrifugation at 13,000g for 3 min. The supernatants were removed, protein concentration determined and analyzed by 2DE. Protein was quantified using Bradford reagent (BioRad Laboratories). ElfA isolation were carried out as described previously.15
Two-dimensional electrophoresis and LC-MS
2DE was carried out as described previously.15,54 Gels were stained with Colloidal Coomassie® Blue G-250 (Serva Electrophoresis, Germany) and scanned using a Typhoon Trio Variable Mode Imager or ImageScanner III (GE Healthcare, Freiburg, Germany). Five replicate gels of each, wild-type or ΔelfA were used for analysis using the Progenesis™ SameSpot Software (Nonlinear Dynamics, UK) to identify differentially regulated spots. Proteins were excised from 2DE gels and digested with trypsin and analyzed as previously described.54,56 LC-MS Analysis was carried out on a 6340 Ion-trap LC Mass Spectrometer using electrospray ionization (Agilent Technologies). MSn analysis was carried out on the 3 most abundant peptide precursor ions in each sample, by automatic selection. Peptides from the MSn spectra were compared to the NCBI nr database using MASCOT (http://www.matrixscience.com) for protein identification.54
Biotinylation of glutathione disulfide (Biotin-GSSG)
Glutathione disulfide was glutathionylated following a published protocol.23 To confirm the successful biotinylation of GSSG, the Biotin-GSSG was analysed by RP–HPLC with UV detection (Agilent 1200 system), using a C18 RP–HPLC column (Agilent Zorbax Eclipse XDB-C18; 5 mm particle size; 4.6 × 15 mm) and MALDI-ToF analysis (Ettan MALDI-ToF Pro mass spectrometer (Amersham Biosciences)).
Analysis of protein glutathionylation
Biotin-GSSG was incubated with whole protein lysates, extracted under nonreducing conditions in a 1/10 ratio for 10 min at room temperature followed by SDS-PAGE under nonreducing conditions and Western Blot was carried out using Streptavidin-HRP and ECL for detection. The analysis was also carried out under reducing conditions and whole cell lysates were also treated with water as controls to ensure that any proteins detected were as a direct result of glutathionylation by biotin-GSSG.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Deplazes A, Mockli N, Luke B, Auerbach D, Peter M. Yeast Uri1p promotes translation initiation and may provide a link to cotranslational quality control. EMBO J. 2009;28:1429–1441. doi: 10.1038/emboj.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodnina MV, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Curr Opin Cell Biol. 2009;21:435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Preiss T, Hentze MW. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays. 2003;25:1201–1211. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- 4.Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- 5.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mateyak MK, Kinzy TG. eEF1A: thinking outside the ribosome. J Biol Chem. 2010;285:21209–21213. doi: 10.1074/jbc.R110.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pittman YR, Kandl K, Lewis M, Valente L, Kinzy TG. Coordination of eukaryotic translation elongation factor 1A (eEF1A) function in actin organization and translation elongation by the guanine nucleotide exchange factor eEF1Balpha. J Biol Chem. 2009;284:4739–47. doi: 10.1074/jbc.M807945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozturk SB, Kinzy TG. Guanine nucleotide exchange factor independence of the G-protein eEF1A through novel mutant forms and biochemical properties. J Biol Chem. 2008;283:23244–53. doi: 10.1074/jbc.M801095200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeppesen MG, Ortiz P, Shepard W, Kinzy TG, Nyborg J, Andersen GR. The Crystal structure of the glutathione S-transferase-like domain of elongation factor 1Bγ from Saccharomyces cerevisiae. J Biol Chem. 2003;278:47190–47198. doi: 10.1074/jbc.M306630200. [DOI] [PubMed] [Google Scholar]
- 10.Gross SR, Kinzy TG. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol. 2005;12:772–8. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- 11.Farah ME, Sirotkin V, Haarer B, Kakhniashvili D, Amberg DC. Diverse protective roles of the actin cytoskeleton during oxidative stress. Cytoskeleton. 2011;68:340–354. doi: 10.1002/cm.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kummasook A, Tzarphmaag A, Thirach S, Pongpom M, Cooper CR, Jr, Vanittanakom N. Penicillium marneffei actin expression during phase transition, oxidative stress, and macrophage infection. Mol Biol Rep. 2011;38:2813–2819. doi: 10.1007/s11033-010-0427-1. [DOI] [PubMed] [Google Scholar]
- 13.Edmonds BT, Bell A, Wyckoff J, Condeelis J, Leyh TS. The effect of F-actin on the binding and hydrolysis of guanine nucleotide by Dictyostelium elongation factor 1A. J Biol Chem. 1998;273:10288–10295. doi: 10.1074/jbc.273.17.10288. [DOI] [PubMed] [Google Scholar]
- 14.Liu G, Tang J, Edmonds BT, Murray J, Levin S, Condeelis J. F-actin sequesters elongation factor 1 alpha from interaction with aminoacyl-tRNA in a pH-dependent reaction. J Cell Biol. 1996;135:953–963. doi: 10.1083/jcb.135.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carberry S, Neville C, Kavanagh K, Doyle S. Analysis of major intracellular proteins of Aspergillus fumigatus by MALDI mass spectrometry: identification and characterisation of an elongation factor 1B protein with glutathione transferase activity. Biochem Biophys Res Commun. 2006;341:1096–1104. doi: 10.1016/j.bbrc.2006.01.078. [DOI] [PubMed] [Google Scholar]
- 16.Kamiie K, Nomura Y, Kobayashi S, Taira H, Kobayashi K, Yamashita T, Kidou S-i, Ejiri S-i. Cloning and expression of Bombyx mori silk gland elongation factor 1gamma; in Escherichia coli. Biosci Biotechnol Biochem. 2002;66:558–565. doi: 10.1271/bbb.66.558. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi S, Kidou S, Ejiri S-i. Detection and characterisation of glutathione S-transferase in rice EF-1betabeta'gamma and EF-1gamma expressed in Escherichia coli. Biochem Biophys Res Commun. 2001;288:509–514. doi: 10.1006/bbrc.2001.5799. [DOI] [PubMed] [Google Scholar]
- 18.Esposito AM, Kinzy TG. The eukaryotic translation elongation factor 1Bgamma has a non-guanine nucleotide exchange factor role in protein metabolism. J Biol Chem. 2010;285:37995–38004. doi: 10.1074/jbc.M110.160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Kellner J, Lee C-H, Coulombe PA. Interaction between the keratin cytoskeleton and eEF1B[gamma] affects protein synthesis in epithelial cells. Nat Struct Mol Biol. 2007;14:982–983. doi: 10.1038/nsmb1301. [DOI] [PubMed] [Google Scholar]
- 20.Kubodera T, Yanashita N, Nishimura A. Transformation of Aspergillus sp. and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Biosci Biotechnol Biochem. 2002;66:404–406. doi: 10.1271/bbb.66.404. [DOI] [PubMed] [Google Scholar]
- 21.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafton A, Latge JP, Li WX, Lord A, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O'Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, de Cordoba SR, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, de Aldana CRV, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 22.Mabey JE, Anderson MJ, Giles PF, Miller CJ, Attwood TK, Paton NW, Bornberg-Bauer E, Robson GD, Oliver SG, Denning DW. CADRE: the central Aspergillus data repository. Nucleic Acids Res. 2004;32:401–405. doi: 10.1093/nar/gkh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennan JP, Miller JIA, Fuller W, Wait R, Begum S, Dunn MJ, Eaton P. The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol Cell Proteomics. 2005;5.2:215–225. doi: 10.1074/mcp.M500212-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Rahman I, Kode A, Biswas SK. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc. 2006;1:3159–3165. doi: 10.1038/nprot.2006.378. [DOI] [PubMed] [Google Scholar]
- 25.Greetham D, Grant C. Antioxidant activity of the yeast mitochondrial 1-Cys peroxiredoxin is dependent on thioredoxin reductase and glutathione in vivo. Mol Cell Biol. 2009;29:3229–3240. doi: 10.1128/MCB.01918-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-Mirabal HR, Winther JR. Redox characteristics of the eukaryotic cytosol. Biochimi Biophys Acta. 2008;1783:629–640. doi: 10.1016/j.bbamcr.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Olarewaju O, Ortiz PA, Chowdhury WQ, Chatterjee I, Kinzy TG. The translation elongation factor eEF1B plays a role in the oxidative stress response pathway. RNA Biol. 2004;1:89–94. doi: 10.4161/rna.1.2.1033. [DOI] [PubMed] [Google Scholar]
- 28.Baars TL, Petri S, Peters C, Mayer A. Role of the V-ATPase in regulation of the vacuolar fission-fusion equilibrium. Mol Biol Cell. 2007;18:3873–3882. doi: 10.1091/mbc.E07-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeng S, Ko Y-J, Kim G-B, Jung K-W, Floyd A, Heitman J, Bahn Y-S. Comparative transcriptome analysis reveals novel roles of the Ras and cyclic AMP signaling pathways in environmental stress response and antifungal drug sensitivity in Cryptococcus neoformans. Eukaryot Cell. 2010;9:360–378. doi: 10.1128/EC.00309-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lessing F, Kniemeyer O, Wozniok I, Loeffler J, Kurzai O, Haertl A, Brakhage AA. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot Cell. 2007;6:2290–302. doi: 10.1128/EC.00267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pahlman I-L, Gustafsson L, Rigoulet M, Larsson C. Cytosolic redox metabolism in aerobic chemostat cultures of Saccharomyces cerevisiae. Yeast. 2001;18:611–620. doi: 10.1002/yea.709. [DOI] [PubMed] [Google Scholar]
- 32.Tone Y, Tanahashi N, Tanaka K, Fujimuro M, Yokosawa H, Toh-e A. Nob1p, a new essential protein, associates with the 26S proteasome of growing Saccharomyces cerevisiae cells. Gene. 2000;243:37–45. doi: 10.1016/s0378-1119(99)00566-1. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci Signal. 2010;3:ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fontecave M, Atta M, Mulliez E. S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Emri Ts, Pocsi In, Szentirmai A. Changes in the glutathione (GSH) metabolism of Penicillium chrysogenum grown on different nitrogen, sulphur and carbon sources. J Basic Microbiol. 1998;38:3–8. [Google Scholar]
- 36.Bursac D, Lithgow T. Jid1 is a J-protein functioning in the mitochondrial matrix, unable to directly participate in endoplasmic reticulum associated protein degradation. FEBS Lett. 2009;583:2954–2958. doi: 10.1016/j.febslet.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen AL. The coat protein complex II, COPII, protein Sec13 directly interacts with presenilin-1. Biochem Biophys Res Comm. 2009;388:571–575. doi: 10.1016/j.bbrc.2009.08.051. [DOI] [PubMed] [Google Scholar]
- 39.Fu L, Sztul E. Traffic-independent function of the Sar1p/COPII machinery in proteasomal sorting of the cystic fibrosis transmembrane conductance regulator. J Cell Biol. 2003;160:157–163. doi: 10.1083/jcb.200210086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goeckeler JL, Brodsky JL. Molecular chaperones and substrate ubiquitination control the efficiency of endoplasmic reticulum-associated degradation. Diabetes Obes Metab. 2010;12:32–38. doi: 10.1111/j.1463-1326.2010.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 42.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 44.Hayes JD, McLellan LI. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radic Res. 1999;31:273–300. doi: 10.1080/10715769900300851. [DOI] [PubMed] [Google Scholar]
- 45.Li SC, Kane PM. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta. 2009;1793:650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones DP, Go Y-M. Mapping the cysteine proteome: analysis of redox-sensing thiols. Curr Opin Chem Biol. 2011;15:103–112. doi: 10.1016/j.cbpa.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 47.Meyer A, Hell R. Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res. 2005;86:435–457. doi: 10.1007/s11120-005-8425-1. [DOI] [PubMed] [Google Scholar]
- 48.Yu X, Cai M. The yeast dynamin-related GTPase Vps1p functions in the organization of the actin cytoskeleton via interaction with Sla1p. J Cell Sci. 2004;117:3839–3853. doi: 10.1242/jcs.01239. [DOI] [PubMed] [Google Scholar]
- 49.Kim S, Coulombe PA. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat Rev Mol Cell Biol. 2010;11:75–81. doi: 10.1038/nrm2818. [DOI] [PubMed] [Google Scholar]
- 50.Schrettl M, Carberry S, Kavanagh K, Haas H, Jones GW, O'Brien J, Nolan A, Stephens J, Fenelon O, Doyle S. Self-protection against gliotoxin-A component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 2010;6:e1000952. doi: 10.1371/journal.ppat.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen ML, Albertsen L, Lettier G, Nielsen JB, Mortensen UH. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet Biol. 2006;43:54–64. doi: 10.1016/j.fgb.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 52.Kubodera T, Yamashita N, Nishimura A. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci Biotechnol Biochem. 2000;64:1416–1421. doi: 10.1271/bbb.64.1416. [DOI] [PubMed] [Google Scholar]
- 53.Woods JP, Heinecke EL, Goldman WE. Electrotransformation and expression of bacterial genes encoding hygromycin phosphotransferase and beta-galactosidase in the pathogenic fungus Histoplasma capsulatum. Infect Immun. 1998;66:1697–1707. doi: 10.1128/iai.66.4.1697-1707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Hanlon KA, Cairns T, Stack D, Schrettl M, Bignell EM, Kavanagh K, Miggin SM, O'Keeffe G, Larsen TO, Doyle S. Targeted disruption of non-ribosomal peptide synthetase pes3 augments the virulence of Aspergillus fumigatus. Infect Immun. 2011;79:3978–3992. doi: 10.1128/IAI.00192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carberry S, Molloy E, Hammel S, O'Keeffe G, Jones GW, Kavanagh K, Doyle S. Gliotoxin effects on fungal growth: mechanisms and exploitation. Fungal Genet Biol. 2012;49:302–312. doi: 10.1016/j.fgb.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 56.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2007;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


