Abstract
In this report we highlight the latest trends in phasing methods used to solve alpha helical membrane protein structures and analyze the use of heavy atom metals for the purpose of experimental phasing. Our results reveal that molecular replacement is emerging as the most successful method for phasing alpha helical membrane proteins, with the notable exception of the transporter family, where experimentally derived phase information still remains the most effective method. To facilitate selection of heavy atoms salts for experimental phasing an analysis of these was undertaken and indicates that organic mercury salts are still the most successful heavy atoms reagents. Interestingly the use of seleno-l-methionine incorporated protein has increased since earlier studies into membrane protein phasing, so too the use of SAD and MAD as techniques for phase determination. Taken together this study provides a brief snapshot of phasing methods for alpha helical membrane proteins and suggests possible routes for heavy atom selection and phasing methods based on currently available data.
Keywords: alpha helical membrane proteins, protein crystallography, phasing
Introduction
Membrane protein structural biology is currently undergoing rapid expansion with many developments in new technologies and beam line instrumentation that are making a real, significant impact on the number of new structures reported.1 The bottlenecks in membrane protein structure determination are well known2 and for obvious reasons substantial resource has gone into understanding recombinant expression,3,4 protein stabilization, crystallization,5,6 and data collection. Given the often poorly diffracting, anisotropic nature of crystals grown from detergent solubilized samples, further developments in phasing practices and methodologies are perhaps the next area where more insight could make a difference. Using our previous database compiled to analyze crystallization conditions for alpha helical membrane proteins (MPs),7 we present here a complimentary analysis of the current trends in phasing practice and heavy atom use for these proteins. The information updates previous studies conducted before the recent increase in the number of alpha helical MP structures and provides indications as to which heavy atom salts and phasing methods might be tried following successful crystallization and native data collection.
Results
Current trends in phasing methods
Our crystallization database contains 360 entries for alpha helical membrane protein structures. Of these, 229 had initial phase information determined using Molecular Replacement (63%) whilst the remaining 131 (37%) required the use of experimental phase determination methods (Fig. 1 inset). This analysis indicates that almost all types of alpha helices MP are amenable to molecular replacement, with more than 60% of structures for the Respiratory Complexes (61%), Channels (69%), Photosynthetic and Light Harvesting Complexes (60%), G-Protein Coupled Receptors (GPCRs) (93%), ATPases (63%), and Bacterial Rhodopsins (94%) having their initial structures phased using this method. The remarkable success in the application of protein fusions and lipid cubic phase crystallization methodology in the GPCR family in particular has resulted in all of the recent structures being phased using MR rather than through the use of experimental methods. Of note however is that for the transporters the application of MR has proved far less successful. Although accounting for a quarter of all structures in the database, the same as for the Channel proteins, only 40% of these structures were determined using MR. Perhaps this is due to the irregular helical structures observed in this family, which make construction of an accurate search model(s) difficult from distantly related proteins. It is also becoming apparent that within the same structural family of transporters, such as the Major Facilitator Superfamily (MFS), different sub-families have different localized regions of helical flexibility, making their use as MR search models dubious. Our results indicate that where a representative structure from the same sub-family is available as an MR search model, this is likely to be far more effective than using a distantly related structure, even if the distantly related structure is likely to share the same overall fold.
Figure 1.
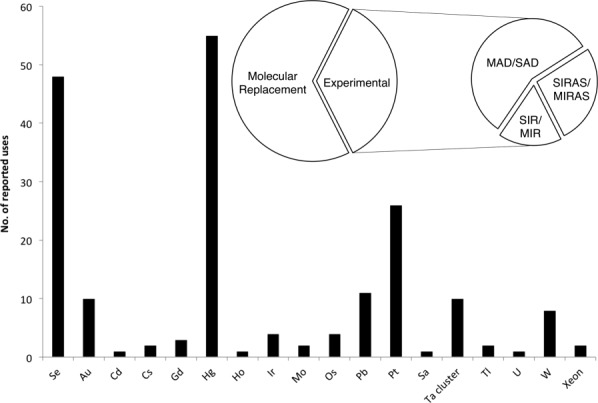
Histogram showing the uses of different elements in heavy atom derivatives of alpha helical membrane proteins. Inset, pie of pie showing the proportion of alpha helical membrane protein structures phased using molecular replacement versus experimental methods. Experimental methods have been further subdivided by technique.
However there is some redundancy within our database, with multiple entries for the same protein in some cases that may artificially bias the data with regard to MR use. On closer examination of the phasing method for novel structures, where novel is determined as being the first structure for a given protein, in a given conformation from a particular organism, the percentage of structures determined using MR drops to under 40%. Looking closer at these structures 66 of them used phase information from a similar protein (either a prokaryotic counterpart or the same protein from a different organism) or indeed a portion (for example a previously solved soluble domain). This leaves only 15 novel entries that were solved by molecular replacement, these are all structures that either used a FAB fragment or were structures that employed an engineered protein where T4 lysozyme has been inserted within the protein. Our data therefore suggest that MR is gaining in popularity for determining initial phase information for membrane protein structures. However, were new structures are being sought experimental phasing is very likely to be the main route to phase determination.
With regards to experimental phasing then, which methods are the most successful? Of the 131 structures that were determined using experimental phase information by far the most successful methods were either single wavelength anomalous dispersion (SAD) (17%) or multi wavelength anomalous dispersion (MAD) (13%). These were followed by multiple isomorphous replacement (MIR) (8%), single isomorphous replacement with anomalous scattering (SIRAS) (7%), multiple isomorphous replacement with anomalous scattering (MIRAS) (8%) and someway behind was single isomorphous replacement (SIR) (2%). From the database entries there was no obvious correlation of the choice of phasing method with resolution. The average resolution for structures solved using MAD and SAD was 3.0 Å; whereas, that for MIRAS and SIRAS was 2.8 Å. MIR and SIR had the lowest average resolution at 3.3 Å.
Heavy atom choice
In the past few years the number of membrane protein structures has increased substantially. Our analysis shows that 37% of these required experimental phasing. A previous study into the preparation of heavy atom derivatives for membrane proteins was reported in 2006 for 38 alpha helical MP structures.8 Here we report an updated analysis from 131 structures and focus solely on the alpha helical class of membrane proteins. Our major findings are summarized in Figure 1. Table I shows the breakdown of the successfully reported heavy atom derivatives. Consistent with the findings of the 2006 study, we also observe mercury compounds clearly dominating the database, having been successfully used in 29% of 191 cases where heavy atoms were used to determine the phase information for structure determination. This number is higher than the total number of structures (131) as a large proportion of the structures used two or more heavy atoms or a heavy atom combined with seleno-l-methionine, for example. Following mercury the next most successful heavy atom was platinum 14% (26 cases), with lead 6% (11 cases), and gold 5% (10 cases) accounting for a small but significant subset. The most successful mercury compound is methyl mercury chloride (MMC), accounting for 23% (13 cases) of all 55 cases were mercury salts were used. This is closely followed by the ethylmercurials [acetate (4 cases), chloride (2 cases), thiosalicylate (11 cases) and phosphate salts (7 cases), which collectively account for 43% (24 cases)]. The success of the organomercurial salts is very likely due to the ability of these compounds to partition efficiently into both the lipophilic and hydrophilic environments found in alpha helical MPs to react with the cysteine side chains. More reactive mercury salts are also present in significant numbers, mercury chloride (9%), potassium mercury iodide, K2HgI4 (5%), and chloro-(4-sulfophenyl) mercury (PCMBS) (9%), indicating that these compounds are worth pursuing for dramatization purposes.
Table I.
The most Successful Heavy Atom Compounds used to Phase Alpha Helical Membrane Protein Structures have been Listed, Grouped into Element Type
| Heavy atom usage | No. of uses in database | |
|---|---|---|
| Mercury | MMC | 13 |
| EMP | 7 | |
| EMTS | 6 | |
| HgCl2 | 5 | |
| MMA | 4 | |
| Platinum | K2PtCl4 | 10 |
| K2Pt(NO2)4 | 4 | |
| K2PtCl6 | 3 | |
| Lead | TMLA | 7 |
| Gold | KAu(CN)2 | 6 |
| KAuCl4 | 2 | |
| HA cluster | Ta6Br12 | 10 |
By far the most common platinum salt was potassium tetrachloropantinate (K2PtCl4), accounting for 38% (10 cases) of all platinum salts reported. This was followed some way behind by potassium platinum nitrate [K2Pt(NO2)4] 15% (4 cases) and potassium hexachloroplatinate (K2PtCl6) 10% (3 cases). Lead compounds favored trimethyl lead acetate, which accounted for 63% of all cases were lead was used. Potassium gold cyanide [K2Au(CN)2] was used in six cases, and accounting for 60% of all gold cases in the database. Interestingly the number of reported uses of gold compounds was equivalent to that of the tantalum bromide (Ta6Br12) cluster, which has proved successful for the phasing of large macromolecules at low resolution, including the respiratory Complex I and Na, K-ATPase.9,10
A noticeable difference from a previous analysis of heavy atom use in 20068 has been the substantial increase in the use of seleno-l-methionine incorporated protein structures in phase determination. Se incorporation now accounts for 25% of heavy atom uses reported to date (48 cases). However this figure rises to 38% if all the structures are taken into account that has used Se in combination with another HA. This is a substantial increase from the 16% of structures reported in 2006. A possible reason for this increase may be a better understanding of recombinant membrane protein expression in E. coli, that now allows for high levels of seleno-l-methionine incorporation.3,11 Advancements in beamline technology, such as highly focused X-ray beams12 and the installation of fast readout, pixel based detectors,13 have enabled crystallographers to accurately measure the weak anomalous signal from Se, whilst simultaneously reducing radiation damage.14
Discussion
MR has been steadily rising as the principle means of phasing membrane protein structures over the last few years.2 For the GPCR family in particular molecular replacement phasing has proved remarkably successful. Advances in developing and deploying the technique of lipid cubic phase crystallization to the GPCR family has also enabled high quality data to be collected from these crystals.6,15 The combination of the high quality diffraction data with the remarkable success of fusing the T4 lysozyme protein to either wild type or thermostabilized receptor molecules has resulted,16 from a phasing perspective, an ideal system for molecular replacement phasing, thus negating the need for experimental phasing procedures in this class of membrane protein.17 The use of antibodies as crystallization scaffolds has also been successful for calculating initial phases for the GPCR family18 and has proved useful for other families, as demonstrated originally for the cytochrome oxidase structure from Paracoccus denitrificans19 but also recently for AdiC transporter from Escherichia coli.20 MR has been far less successful in the transporter group of membrane proteins, most likely due to the highly dynamic nature of these proteins, although no extensive study has been carried out to date. However, it is becoming more apparent, as the number of different transporter structures increases, that the individual sub-families within the larger structural super families, such as the MFS or “LeuT-like fold”, display significantly different structural dynamics. This may be one reason why distantly related transporters, which ultimately have the same overall structure, prove ineffectual as generic MR search models. Recent examples include the eukaryotic proton coupled phosphate transporter, PipT and the nitrate-nitrite exchanger NarU, which although sharing the same overall fold as other members of the MFS were not solved using MR, but instead required experimental phasing methods.21,22
As we present in this report however, experimental phasing still accounts for over half the structures of transporter proteins, suggesting this method is likely to remain for the foreseeable future. In addition, the incorporation of seleno-l-methionine into recombinant protein using eukaryotic expression systems is far from trivial indicating an increased likelihood of the need to pursue heavy atom derivatization. In making informed decisions as to which heavy atoms are appropriate to test, Table I should provide a useful guide.
Although individual proteins will have unique phasing solutions, our analysis identifies some general trends and routes to try when designing experimental set-ups for phasing. Derivatization of native cysteine residues using an organic mercury salt has proved very successful, as has the introduction of cysteine residues for this purpose, possibly as a substitution of serine. However, our own experience has indicated that the presence of >5 cysteine residues tends to result in heavy protein precipitation following this procedure. Our data also suggest that for membrane proteins containing large extra membrane domains, such as the ATPase family, ABC transporters and respiratory complexes, using more “exotic” phasing compounds, such as the “Tantalum Bromide cluster” have proved successful, although often as part of a wide range of different phasing data sets.9,23,24 Indeed, the requirement of using multiple sources of phase information is a recurrent theme in the database (Supporting Information database: MP_phasing.xls), suggesting that a combination of phasing methods should always be considered and pursued.25
As more data become available we will continue to update the current analyses on heavy atom selection and hope this will lead to a more rational approach to one of the most challenging hurdles in membrane protein structural biology.
Methods
An analysis of the deposited structures for alpha helical MPs was carried out as described previously7 to include phasing method and heavy atom information. A copy of the database incorporating the phasing information used in the present analysis is available as Supporting Information MP_phasing.xls.
Acknowledgments
This research was funded through the Medical Research Council (MRC) Career Development Award grant G0900399 to SN. The authors declare they have no conflict of interest with regard to the information and analyses presented herein.
Glossary
- GPCRs
G-Protein Coupled Receptors
- K2Au(CN)2
potassium gold cyanide
- K2PtCl6
potassium hexachloroplatinate
- K2Pt(NO2)4
potassium platinum nitrate
- K2PtCl4
potassium tetrachloropantinate
- MFS
Major Facilitator Superfamily
- MIR
multiple isomorphous replacement
- MIRAS
multiple isomorphous replacement with anomalous scattering
- MMC
methyl mercury chloride
- MRC
Medical Research Council
- MPs
membrane proteins
- SIR
single isomorphous replacement
- SIRAS
single isomorphous replacement with anomalous scattering
- Ta6Br12
tantalum bromide.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.White S. Membrane proteins of known 3D structure. Protein Sci. 2007;13:1948–1949. doi: 10.1110/ps.04712004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bill RM, Henderson PJF, Iwata S, Kunji ERS, Michel H, Neutze R, Newstead S, Poolman B, Tate CG, Vogel H. Overcoming barriers to membrane protein structure determination. Nat Biotechnol. 2011;29:335–340. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- 3.Wagner S, Bader ML, Drew D, De Gier J-W. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006;24:364–371. doi: 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Sonoda Y, Cameron A, Newstead S, Omote H, Moriyama Y, Kasahara M, Iwata S, Drew D. Tricks of the trade used to accelerate high-resolution structure determination of membrane proteins. FEBS Lett. 2010;584:2539–2547. doi: 10.1016/j.febslet.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Newstead S, Ferrandon S, Iwata S. Rationalizing alpha-helical membrane protein crystallization. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherezov V. Lipidic cubic phase technologies for membrane protein structural studies. Curr Opin Struct Biol. 2011;21:559–566. doi: 10.1016/j.sbi.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker JL, Newstead S. Current trends in α-helical membrane protein crystallization: an update. Protein Sci. 2012;21:1358–1365. doi: 10.1002/pro.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morth JP, Sørensen TL-M, Nissen P. Membrane's eleven: heavy-atom derivatives of membrane-protein crystals. Acta Cryst. 2006;D62:877–882. doi: 10.1107/S0907444906023547. [DOI] [PubMed] [Google Scholar]
- 9.Morth JP, Pedersen BP, Toustrup-Jensen MS, Sørensen TL-M, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 10.Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 11.Schlegel S, Löfblom J, Lee C, Hjelm A, Klepsch M, Strous M, Drew D, Slotboom D-J, De Gier J-W. Optimizing membrane protein overexpression in the Escherichia coli strain Lemo21(DE3) J Mol Biol. 2012;423:648–659. doi: 10.1016/j.jmb.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 12.Bowler MW, Guijarro M, Petitdemange S, Baker I, Svensson O, Burghammer M, Mueller-Dieckmann C, Gordon EJ, Flot D, McSweeney SM, Leonard GA. Diffraction cartography: applying microbeams to macromolecular crystallography sample evaluation and data collection. Acta Cryst. 2010;D66:855–864. doi: 10.1107/S0907444910019591. [DOI] [PubMed] [Google Scholar]
- 13.Henrich B, Bergamaschi A, Broennimann C, Dinapoli R, Eikenberry EF, Johnson I, Kobas M, Kraft P, Mozzanica A, Schmitt B. PILATUS: A single photon counting pixel detector for X-ray applications. **Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment. 2009;607:247–249. [Google Scholar]
- 14.Moukhametzianov R, Burghammer M, Edwards PC, Petitdemange S, Popov D, Fransen M, McMullan G, Schertler GFX, Riekel C. Protein crystallography with a micrometre-sized synchrotron-radiation beam. Acta Cryst. 2008;D64:158–166. doi: 10.1107/S090744490705812X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caffrey M. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Ann Rev Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- 16.Tate CG, Schertler GF. Engineering G protein-coupled receptors to facilitate their structure determination. Curr Opin Struct Biol. 2009;19:386–395. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Blois TM, Bowie JU. G-protein-coupled receptor structures were not built in a day. Protein Sci. 2009;18:1335–1342. doi: 10.1002/pro.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen S, Choi H, Rosenbaum D, Kobilka T, Thian F, Edwards P, Burghammer M, Ratnala V, Sanishvili R, Fischetti R, et al. Crystal structure of the human beta(2) adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 19.Ostermeier C, Harrenga A, Ermler U, Michel H. Structure at 2.7 Å resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody FV fragment. Proc Natl Acad Sci USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang Y, Jayaram H, Shane T, Kolmakova-Partensky L, Wu F, Williams C, Xiong Y, Miller C. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature. 2009;460:1040–1043. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen BP, Kumar H, Waight AB, Risenmay AJ, Roe-Zurz Z, Chau BH, Schlessinger A, Bonomi M, Harries W, Sali A, et al. Crystal structure of a eukaryotic phosphate transporter. Nature. 2013;496:533–536. doi: 10.1038/nature12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan H, Huang W, Yan C, Gong X, Jiang S, Zhao Y, Wang J, Shi Y. Structure and mechanism of a nitrate transporter. Cell Rep. 2013;3:716–723. doi: 10.1016/j.celrep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Gourdon P, Liu XY, Skjørringe T, Morth JP, Møller LB. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475:59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- 24.Long F, Su CC, Zimmermann MT, Boyken SE. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;476:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen BP, Morth JP, Nissen P. Structure determination using poorly diffracting membrane-protein crystals: the H +-ATPase and Na +, K +-ATPase case history. Acta Cryst. 2010;D66:309–313. doi: 10.1107/S0907444909053244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


