Abstract
Previous studies have shown that membrane-aerated biofilm (MAB) reactors can simultaneously remove carbonaceous and nitrogenous pollutants from wastewater in a single reactor. Oxygen is provided to MABs through gas-permeable membranes such that the region nearest the membrane is rich in oxygen but low in organic carbon, whereas the outer region of the biofilm is void of oxygen but rich in organic carbon. In this study, MABs were grown under similar conditions but at two different fluid velocities (2 and 14 cm s−1) across the biofilm. MABs were analyzed for changes in biomass density, respiratory activity, and bacterial community structure as functions of biofilm depth. Biomass density was generally highest near the membrane and declined with distance from the membrane. Respiratory activity exhibited a hump-shaped profile, with the highest activity occurring in the middle of the biofilm. Community analysis by PCR cloning and PCR-denaturing gradient gel electrophoresis of 16S rRNA genes demonstrated substantial stratification of the community structure across the biofilm. Population profiles were also generated by competitive quantitative PCR of gene fragments specific for ammonia-oxidizing bacteria (AOB) (amoA) and denitrifying bacteria (nirK and nirS). At a flow velocity of 14 cm s−1, AOB were found only near the membrane, whereas denitrifying bacteria proliferated in the anoxic outer regions of the biofilm. In contrast, at a flow velocity of 2 cm s−1, AOB were either not detected or detected at a concentration near the detection limit. This study suggests that, under the appropriate conditions, both AOB and denitrifying bacteria can coexist within an MAB.
The aeration of bioreactors is one of the most significant operating costs incurred during the treatment of municipal and industrial wastewaters. One technological alternative to conventional coarse and fine bubble diffusers is the use of microporous membranes that contain small (<0.1 μm) hydrophobic pores for bubbleless oxygen transfer to the wastewater (1). Biofilms actively grow on these membranes, such that membrane-aerated biofilm (MAB) reactors can treat wastewater at rates comparable to conventional suspended-growth processes (6, 33, 37). MAB reactors have lower operating costs, because oxygen transfer efficiency approaches 100% (5, 26) and because expensive air compression is no longer required (9). In addition, MAB reactors generate fewer odors, because volatile pollutants are not released into the atmosphere (5, 6).
MABs are unique compared to biofilms grown on inert substrata. Nutrient concentrations in MABs are highest at the biofilm-liquid boundary and decrease with depth into the biofilm; in contrast, oxygen concentrations are highest at the membrane and decrease toward the outside of the biofilm (Fig. 1). In thick MABs, oxygen partially penetrates the biofilm, generating an anoxic zone at the biofilm-liquid boundary (6). In such biofilms, the region of greatest microbial activity is variable and depends on the relative availability of oxygen and substrate (8, 12). Thick MABs also simultaneously provide favorable conditions for both nitrification (near the membrane) and denitrification (near the biofilm-liquid boundary) within a single biofilm.
FIG. 1.

Hypothetical structure of an MAB treating wastewater.
The exceptional ability of MABs to achieve both nitrification and denitrification within a single biofilm has substantial potential for improving existing wastewater treatment processes. Numerous researchers have previously reported that ammonia-oxidizing bacteria (AOB) are concentrated near the membrane in MABs (16, 32, 36). Additional research, however, is needed to understand the stratification of the denitrifying bacterial populations as well as the pertinent factors (e.g., nutrient loading, fluid shear, etc.) that control the development of both the nitrifying and denitrifying bacterial populations within a single MAB.
The purpose of this study, therefore, was to characterize the unique stratification of MABs with respect to bacterial community structure and function. We hypothesized that these biofilms would vary substantially in biomass density and respiratory activity as a function of biofilm depth. More importantly, AOB should be found near the membrane, where oxygen concentrations are high and organic concentrations are low. Denitrifying bacteria should preferentially grow in the outer anoxic region of the biofilm, where organic concentrations are high. MABs were grown under well-defined conditions of fluid flow and substrate concentrations. Biofilms were separately grown at different fluid velocities (2 and 14 cm s−1) to compare the effect of fluid shear on MAB characteristics. The bacterial community structure was examined by PCR-denaturing gradient gel electrophoresis (PCR-DGGE) and PCR cloning of 16S rRNA genes and by competitive quantitative PCR of genes encoding enzymes required for ammonia oxidation (amoA) and for denitrification (nirS and nirK).
MATERIALS AND METHODS
Biofilm reactor.
MABs were grown under well-defined conditions of fluid flow and substrate concentrations in a rectangular, closed flow-cell reactor (6 cm wide, 2.5 cm high, 2 m long). Biofilms grew on flat-sheet microporous membranes (20 cm by 1 cm) set into the base of the reactor, flush with the bottom of the tank. Each membrane was exposed to a separate gas chamber so that oxygen addition could be independently controlled. Honeycombs and 1.5 m of tank length combined to allow complete dissipation of hydraulic entrance effects prior to contact with the membranes. The reactor hydraulic retention time was 6 h, to help control substrate concentrations in the reactor. A centrifugal pump recirculated the water in the reactor to control the fluid velocity past the membranes independent of hydraulic residence time. Biofilms were grown separately at average fluid velocities of 2 or 14 cm s−1.
For each experiment, the reactor was inoculated with 1 ml of cryopreserved (15% [vol/vol] glycerol) activated sludge collected from the aeration tanks at the St. Paul, Minn., metropolitan wastewater treatment facility. The reactor was fed a mixture containing sodium acetate and ammonium chloride dissolved in dechlorinated tap water. Bulk reactor conditions were monitored daily and maintained at approximately 100 mg of chemical oxygen demand liter−1 (mean = 102; standard deviation = 21) and 25 mg of ammonia nitrogen liter−1 (mean = 25; standard deviation = 6). Each membrane was supplied with 30 ml of atmospheric air liter−1 (pO2 = 0.21 atm) or an oxygen-air mixture (pO2 = 0.75 atm).
Biofilm samples (∼1 cm by 1 cm) were removed from the reactor 6 to 8 weeks after inoculation. Samples were excised with a razor blade and immediately frozen at −15°C. Precise dimensions of each biofilm core were measured with a vernier caliper. Biofilms were then thin sliced (typical thickness, 100 to 300 μm) parallel to the membrane in a combined cryostat-microtome (Richard-Allan Scientific, Kalamazoo, Mich.). Biofilm slices were transferred to sterile microcentrifuge tubes and stored separately at −20°C until further analysis.
Analytical methods.
Dissolved oxygen profiles were obtained using a Clark-type microelectrode (tip diameter, 10 to 15 μm; response time, <5 s; spatial resolution, 20 to 30 μm; Unisense, Aarhus, Denmark). The microelectrode was lowered through an access window in the top of the reactor and into the biofilm using a computer-controlled micromanipulator (Oriel Inst., Stratford, Conn.) capable of maneuvering in 0.1-μm increments. Biofilm thickness was determined utilizing a 250× magnification microscope inspection system (Integrated Endoscopy, Irvine, Calif.). The microscope was focused alternately on the biofilm surface and a reference point at the bottom of the reactor. The distance between the membrane surface and the reference point was determined prior to biofilm growth, and the thickness was determined by the distance traveled by the micromanipulator (35).
Respiratory activity was measured by iodonitrotetrazolium chloride (INT) reduction to INT-formazan (39). Biofilm samples (∼1 cm by 1 cm) were removed from the reactor and placed into 2 ml of INT solution (9.9 mM in 2% [vol/vol] N,N-dimethylformamide) and 1.5 ml of Tris buffer (1.0 M; pH 7.0) and incubated in the dark for 2 h at 40°C prior to freezing and slicing in the cryostat-microtome. INT-formazan was extracted from the biofilm slices using 1.0 ml of extraction solution (50% [vol/vol] N,N-dimethylformamide in ethanol) and incubated at room temperature in the dark for 1 h. INT-formazan concentrations were quantified using a UV-vis spectrophotometer (λ = 464 nm; DU-530; Beckman, Fullerton, Calif.) and calibrated to a formazan standard curve.
Total protein concentrations were quantified by the Lowry method (19) using bovine serum albumin as a standard. Biofilm slices first were dried in a centrifugal drier (Labnet, Edison, N.J.) and resuspended in 1.0 N NaOH prior to analysis. Polysaccharide concentrations were determined by the anthrone method (15) using glucose as a standard. Biofilm slices first were dried in the centrifugal drier and resuspended in deionized water prior to analysis. Dry cell mass concentrations were determined by drying a known volume of biofilm at 103°C overnight and weighing the residue with an analytical balance.
DNA extraction.
Biofilm slices were suspended in 1 ml of lysis buffer (120 mM sodium phosphate buffer [pH 8.0], 5% [wt/vol] sodium dodecyl sulfate). Samples then underwent three successive freeze-thaw cycles followed by an incubation at 70°C for 90 min. Genomic DNA was extracted and purified from these samples using the FastDNA Spin kit per the manufacturer's instructions (QBioGene, Vista, Calif.).
PCR-DGGE.
Partial 16S rRNA genes were amplified from the extracted genomic DNA by PCR using a PTC 100 thermal cycler (MJ Research, Inc., Watertown, Mass.) using primers PRBA338F (18) and PRUN518R (23) with a GC clamp attached to the forward primer (23). The final 50-μl reaction mixture contained 1× PCR buffer (Promega, Madison, Wis.), 175 μmol of MgCl2, 4 nmol of deoxynucleoside triphosphates, 2% bovine serum albumin, 25 pmol of forward and reverse primers, 1.25 U of Taq polymerase (Promega), and ∼1 ng of template DNA. The PCR protocol included a 5-min initial denaturation at 94°C, 30 cycles of 92°C for 30 s, 55°C for 30 s, and 72°C for 30s, and a final extension for 10 min at 72°C.
DGGE was performed on a D-Code apparatus (Bio-Rad, Hercules, Calif.). Samples containing equal amounts of PCR amplicons were loaded onto 8% (wt/vol) polyacrylamide gels (37.5:1, acrylamide-bisacrylamide) in 0.5× Tris-acetate-EDTA (TAE) buffer (31) using a denaturing gradient ranging from 30 to 50% denaturant (100% denaturant contains 7 M urea, 40% [vol/vol] formamide in 0.5× TAE). Electrophoresis was performed at 60°C, initially at 20 V (15 min) and then at 200 V (200 min). Following electrophoresis, the gel was stained with SYBR Green I (Molecular Probes, Eugene, Oreg.; diluted 1:5,000 in 0.5× TAE). Gels were visualized on a UV transilluminator and photographed with a digital charge-coupled device camera (BioChemi system; UVP, Inc., Upland, Calif.). Photographs were enhanced for contrast and brightness using Adobe Photoshop version 6.0. Specific PCR-DGGE bands were manually excised from the gel, suspended in 30 μl of sterile water, and incubated overnight at room temperature. The PCR-DGGE process was then repeated using these samples as template until only a single band was detectable on a denaturing gel. A final PCR step was performed without the GC clamp attached to the forward primer. PCR products were then purified using the GeneClean II kit (Qbiogene) prior to nucleotide sequence determination.
PCR cloning.
Nearly complete 16S rRNA genes were amplified by PCR as described above except that primers PRBA8F (11) and PRBA1522R (11) were used. PCR products were purified, ligated into the pGEM-T Easy cloning vector (Promega), and transformed into competent Escherichia coli DH5α cells (31). Plasmids were purified by the alkaline lysis procedure (31). Plasmids were then digested with RsaI restriction enzyme, and their fragment lengths were compared by electrophoresis on a 2% agarose gel.
Nucleotide sequence determination.
Nucleotide sequences were determined fully in both directions from purified PCR-DGGE bands and plasmids at the Advanced Genetic Analysis Center at the University of Minnesota using an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.). PCR-DGGE bands were sequenced using primers PRBA338F and PRUN518R. Nucleotide sequences were determined from plasmids using three forward primers (PRBA8F, PRUN518F, and PRUN907F) and three reverse primers (PRUN518R, PRUN907R, and PRBA1522R) (24). All reported sequences are the consensus of bidirectional sequence information. Reported nucleotide sequences do not include the original PCR primer sequences.
cPCR.
Nitrosomonas oligotropha-like populations were quantified in each biofilm slice by competitive PCR (cPCR) of ammonia monoxygenase (amoA) gene fragments (10). A competitor (10) of 100 nucleotides in length was synthesized by AlphaDNA (Montreal, Canada). PCR mixtures were as described above except that primers AMO598F (10) and AMO718R (10) and AmpliTaq polymerase (Roche Diagnostics, Indianapolis, Ind.) were used. The PCR protocol included a 5-min initial denaturation at 94°C, 40 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, and a final extension for 7 min at 72°C. For each biofilm sample slice (target), cPCR amplifications were performed on four mixtures containing a constant target DNA concentration and decreasing competitor copy numbers. Products were resolved on a 3% (wt/vol) NuSieve 3:1 agarose gel (BioWhittaker, Rockland, Maine) in 1× Tris-borate-EDTA buffer (31) stained with ethidium bromide. Band intensities of target DNA and competitor DNA were quantified using LabWorks Image Acquisition software (UVP).
Denitrifying bacterial populations were quantified also by cPCR of two functionally redundant nitrite reductase gene fragments (nirS and nirK). To synthesize competitor templates, partial nirS and nirK genes were amplified by PCR from bacterial isolates known to contain each gene (nirS, Pseudomonas fluorescens ATCC 33512; nirK, Achromobacter cycloclastes ATCC 21921). Based on the nucleotide sequences of these PCR products (data not shown), internal PCR primers (NIRS-INT, 5′-GAC AGC AGC CAT CGC TA-3′; NIRK-INT, 5′-GCG AGC AGG ACT TCT AC-3′) were designed for each gene fragment with the original forward primer attached to the 5′ end. PCR with this primer resulted in a 102-nucleotide deletion from the nirS gene fragment (735 nucleotides) and a 59-nucleotide deletion from the nirK gene fragment (474 nucleotides). PCR amplicons of the synthesized competitor DNA were then ligated into the pGEM-T Easy cloning vector and transformed into competent E. coli DH5α cells (31). DNA concentrations in the plasmid extracts were quantified by staining with Hoechst 33258 dye and measured on a TD-700 fluorometer (Turner Designs, Sunnyvale, Calif.) using calf thymus as a DNA standard.
The PCR mixtures for cPCR of nirS and nirK gene fragments were the same as those described above except that different primer pairs were used (nirS, cd3 F and cd4 R; nirK, F1aCu and R3Cu) (13, 22). The PCR protocol for the amplification of nirS gene fragments included a 5-min initial denaturation at 94°C, 40 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1.5 min, and a final extension for 10 min at 72°C. The PCR protocol for the amplification of nirK gene fragments included a 3-min initial denaturation at 94°C, 35 cycles of 94°C for 0.5 min, 57°C for 1 min, and 72°C for 1 min, and a final extension for 10 min at 72°C. cPCR products were resolved on 2% agarose gels (Bio-Rad) in 1× TAE buffer (31) and stained with ethidium bromide.
Data analysis.
Total respiratory activity was computed by determining the area beneath the plot of INT-formazan concentration versus distance from the membrane by trapezoidal rule. An INT-formazan concentration of 1,000 μg ml−1 was assumed as a baseline for these calculations.
The concentration of target gene was determined from the cPCR results by computing the y intercept of a linear regression of the log of the competitor copy number versus the log of the ratio of target to corrected competitor band intensity. Competitor band intensity was multiplied by a correction factor to account for the difference in the size of the target and the competitor (amoA, 121/100; nirS, 735/633; nirK, 474/415).
Nucleotide sequences were compared with sequences in the GenBank database (3) using the BLASTn program (2). Nucleotide sequences were checked for possible chimeric sequences using the CHECK_CHIMERA program at the Ribosomal Database Project website (20). Putative chimeric sequences were also manually split into subsections and resubmitted to the GenBank database to determine if the segments were from different phylogenetic groups.
Nucleotide sequence accession number.
Nucleotide sequences have been deposited in the GenBank database under accession numbers AY444967 to AY444996.
RESULTS
Physiological profiles.
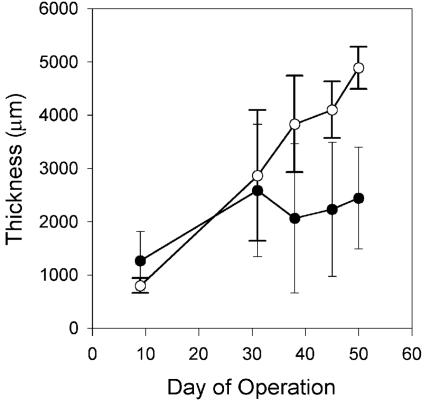
Biofilms grew rapidly after inoculation, fully covering the membranes within 1 week. Biofilm thickness increased over time (Fig. 2), although thickness could vary by as much as twofold along the length of each biofilm. Comparing the two MABs grown at 14 cm s−1, the oxygen-fed biofilm grew more rapidly and was twofold thicker than the air-fed biofilm at the time of harvesting. In contrast, the air-fed and oxygen-fed biofilms grown at 2 cm s−1 were very similar in thickness to each other throughout the growth period (Table 1).
FIG. 2.
MAB thickness as a function of time for biofilms grown at a fluid velocity of 14 cm s−1. •, air-fed biofilm; ○, oxygen-fed biofilm. Error bars represent the standard deviations of the means (n = 9).
TABLE 1.
Summary of pertinent MAB characteristicsa
| Biofilm | O2 at membrane (mg liter−1) | O2 penetration (μm) | Final thickness (μm) | Avg protein (mg ml−1) | Avg carbohydrate (mg ml−1) | Bulk dry cell mass (mg ml−1) | Location of maximum respiratory activity (μm) | Total respiratory activity (μg mm−2) |
|---|---|---|---|---|---|---|---|---|
| Air (2 cm s−1) | 10.4 ± 0.5 | 1,000 ± 100 | 3,600 ± 900 | 3.2 | 2.8 | 12 | 900 | 2.5 |
| O2 (2 cm s−1) | 34.1 ± 1.9 | 2,100 ± 300 | 3,200 ± 1,000 | 4.9 | 2.0 | 12 | 1,100 | 2.9 |
| Air (14 cm s−1) | 9.1 ± 0.3 | 600 ± 100 | 2,400 ± 960 | 6.8 | 5.0 | 70 | 500 | 4.4 |
| O2 (14 cm s−1) | 32.8 ± 2.9 | 1,600 ± 400 | 4,900 ± 400 | 4.5 | 4.0 | 66 | 1,300 | 3.4 |
Grown at fluid velocities of 2 cm and 14 cm s−1. Distances of oxygen penetration and maximum respiratory activity are relative to the membrane.
Dissolved oxygen concentrations in each of the biofilms were at saturation levels near the membrane (Table 1) and then declined approximately linearly as a function of distance from the membrane (Fig. 3). The depth of oxygen penetration generally increased during the first 2 weeks of biofilm growth but then remained relatively constant until biofilms were harvested. The average depth of dissolved oxygen penetration decreased with increasing fluid velocity by 60% for the air-fed biofilm and by 75% for the oxygen-fed biofilm (Table 1).
FIG. 3.
Dissolved oxygen profiles of air-fed (a) and oxygen-fed (b) biofilms grown at a fluid velocity of 14 cm s−1. Different profiles represent data collected on different days.
After the MABs had been removed from the reactor and thin sliced, biomass densities were quantified as cellular protein as a function of depth (Fig. 4). For the biofilms grown at a fluid velocity of 2 cm s−1, protein concentrations either gradually decreased from the membrane towards the outer region of the biofilm (air fed) or exhibited a hump-shaped pattern in which the highest concentration of protein occurred in the middle of the biofilm (oxygen fed) (Fig. 4a). For both of the biofilms grown at a fluid velocity of 14 cm s−1, the highest protein concentration occurred near the membrane and concentrations declined as a function of distance from the membrane (Fig. 4b).
FIG. 4.
Protein concentration profiles within MABs grown at fluid velocities of 2 cm s−1 (a) and 14 cm s−1 (b). •, air-fed biofilm; ○, oxygen-fed biofilm. Error bars represent the standard deviations of the means (n = 3).
The dry cell mass concentrations of entire biofilms were also determined (Table 1). The dry cell mass concentration of the biofilm grown at 14 cm s−1 was fivefold higher than that of the biofilm grown at 2 cm s−1. The ratio of cellular protein to the dry cell mass was 25 to 45% and 7 to 10% at fluid velocities of 2 and 14 cm s−1, respectively. Because cell protein is normally about 50% the dry cell mass of bacterial cells (25), we hypothesized that a substantial fraction of the dry cell mass was complex polysaccharides responsible for the cohesive strength of the biofilm. The concentration profiles of carbohydrates in the biofilms exhibited similar patterns (data not shown) and concentrations (Table 1) as the cell protein as a function of depth.
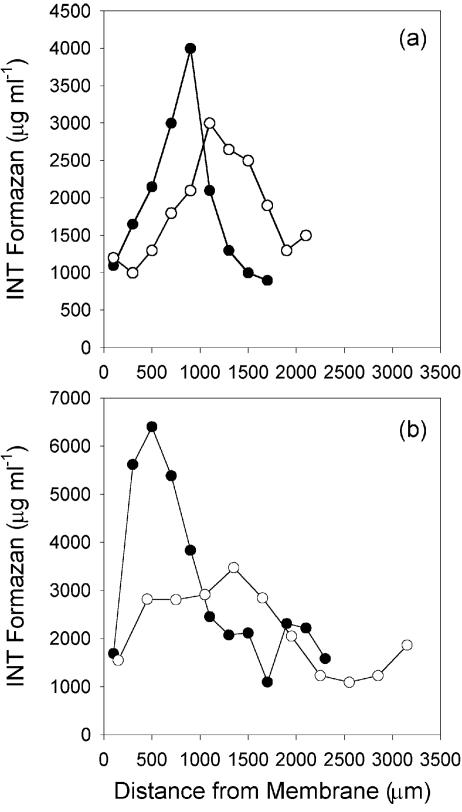
Respiratory activity was also quantified in the biofilms (Fig. 5). For each biofilm, respiratory activity exhibited a hump-shaped pattern as a function of depth. Respiratory activity was low near the membrane and at the biofilm-liquid interface, with the highest activities occurring at an intermediate depth. The location of highest respiratory activity was closer to the membrane for the air-fed biofilms than the oxygen-fed biofilms at both fluid velocities (Table 1). The total respiratory activities were 40 to 80% higher in the biofilms grown at a fluid velocity of 14 cm s−1 than in the MABs grown at 2 cm s−1 (Table 1).
FIG. 5.
Profiles of respiratory activity as measured by the reduction of INT to INT-formazan within MABs grown at fluid velocities of 2 cm s−1 (a) and 14 cm s−1 (b). •, air-fed biofilm; ○, oxygen-fed biofilm.
16S rRNA gene-based population profiles.
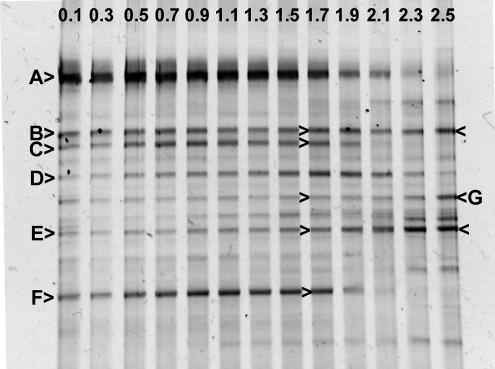
The bacterial community structure was investigated as a function of depth in the oxygen-fed biofilm grown at a fluid velocity of 2 cm s−1 by PCR-DGGE of 16S rRNA gene fragments (Fig. 6). Community profiles were reasonably complex (10 to 13 bands per lane) and indicated a substantial shift in community structure from the membrane to the outer anoxic zone of the biofilm. Several bands were specific to the aerobic zone of the biofilm (<1,700 μm from the membrane; for example, bands A, C, and F), whereas other bands were detected throughout the entire depth of the biofilm (for example, bands B, D, and E).
FIG. 6.
DGGE of PCR-amplified 16S rRNA gene fragments of an oxygen-fed biofilm grown at a fluid velocity of 2 cm s−1. Individual gel lanes are identified by a number indicating the distance (in millimeters) from the membrane where the sample was collected. Letters and arrows identify specific bands that were excised from the gel and sequenced. The results of the nucleotide sequence analysis for these PCR-DGGE bands are shown in Table 2.
Fourteen PCR-DGGE bands, representing seven putatively different vertical band positions, were excised from three different lanes of the gel and their nucleotide sequences were determined (Table 2). In each case, when vertically comigrating bands were analyzed from different gel lanes, nucleotide sequences had a 100% identity to the comigrating band from a different lane. Nucleotide sequence analysis of band E failed for each of the three lanes from which it was excised, likely due to the comigration of multiple 16S rRNA gene fragments. The sequences of the other six bands indicated that these organisms were from a relatively broad phylogenetic distribution, including Bacteriodetes, the alpha subdivision of Proteobacteria (α-Proteobacteria), β-Proteobacteria, and γ-Proteobacteria.
TABLE 2.
Sequence length of the PCR-DGGE bands and clones analyzed in this study and their closest phylogenetic match found in the GenBank databasea
| PCR-DGGE band or clone | Sequence length (bases) | Phylogenetic relationship
|
||
|---|---|---|---|---|
| Most closely related sequence (GenBank Accession no.) | Bacterial division | % Identity | ||
| A | 155 | Uncultivated bacterium (AF532194) | Bacteriodetes | 99.3 |
| B | 160 | Pseudomonas sp. strain C81E ((AF408915) | γ-Proteobacteria | 99.4 |
| C | 160 | Variovorax paradoxus strain Isol ((AY127900) | α-Proteobacteria | 100 |
| D | 156 | Isolate CAGY1 ((AF538743) | Bacteriodetes | 96.8 |
| E | Sequencing unsuccessful | |||
| F | 135 | Clone LED12 ((AF392632) | α-Proteobacteria | 99.3 |
| G | 160 | Acidovorax delafieldii ((AJ420323) | β-Proteobacteria | 100 |
| M5 | 1,487 | Variovorax paradoxus strain Iso1 ((AY127900) | β-Proteobacteria | 99.7 |
| M8 | 1,484 | Pseudomonas sp. strain R1 ((AJ002805) | γ-Proteobacteria | 98.6 |
| M11 | 1,490 | Dechloromonas sp. strain JM ((AF323489) | β-Proteobacteria | 99.9 |
| M12 | 1,438 | Rhizobium sp. strain RM1-2001 ((AF331662) | α-Proteobacteria | 99.9 |
| M15 | 1,436 | Caulobacter sp. strain DNA ((AJ227777) | α-Proteobacteria | 99.7 |
| M16 | 1,437 | Afipia genosp. 6 ((U87772) | α-Proteobacteria | 99.3 |
| M17 | 1,441 | Uncultured sludge bacterium S43 ((AF234739) | α-Proteobacteria | 97.7 |
| M18 | 1,441 | Uncultured bacterium DSSD33 ((AY328732) | α-Proteobacteria | 97.9 |
| M26 | 1,459 | Roseomonas fauriae ((AY150046) | α-Proteobacteria | 94.6 |
| M32 | 1,485 | Acidovorax sp. strain BSB421 ((Y18617) | β-Proteobacteria | 99.6 |
| M35 | 1,437 | Uncultured α-proteobacterium 34626 ((AF288303) | α-Proteobacteria | 98.8 |
| M43 | 1,493 | Uncultured Crater Lake bacterium ((AF316731) | Verrucomicrobia | 94.5 |
| M45 | 1,485 | Uncultured bacterium HOCICi59 ((AY328608) | β-Proteobacteria | 99.9 |
| L2 | 1,458 | Azospirillum sp. ((AF170353) | α-Proteobacteria | 98.3 |
| L5 | 1,503 | Desulfomicrobium norvegicum ((AJ277897) | δ-Proteobacteria | 98.7 |
| L7 | 1,489 | Pseudomonas mosselii ((AF072688) | γ-Proteobacteria | 99.0 |
| L8 | 1,490 | Acinetobacter johnsonii ((AB099655) | γ-Proteobacteria | 98.3 |
| L9 | 1,490 | Dechloromonas sp. strain JM ((AF323489) | β-Proteobacteria | 99.8 |
| L10 | 1,511 | Uncultured eubacterium WCHB1-67 ((AF050536) | δ-Proteobacteria | 97.4 |
| L11 | 1,481 | Acidovorax temperans ((AF078766) | β-Proteobacteria | 99.7 |
| L15 | 1,470 | Uncultured bacterium KD6-12 ((AY188321) | Bacteriodetes | 97.1 |
| L17 | 1,487 | Variovorax paradoxus strain Iso1 ((AY127900) | β-Proteobacteria | 99.8 |
| L19 | 1,477 | Unidentified bacterium ((Z93973) | β-Proteobacteria | 99.7 |
| L22 | 1,490 | Zoogloea ramigera ((X74913) | β-Proteobacteria | 97.9 |
PCR-DGGE band letters are also shown in Fig. 6. Clones are identified by an abbreviation for the location from which they were detected (M = nearest the membrane; L = furthest from the membrane) and a number.
The bacterial community structure was also investigated within two biofilm slices from the air-fed biofilm grown at 2 cm s−1 by PCR cloning of nearly complete 16S rRNA genes (Table 2). From the biofilm slice nearest the membrane, 34 clones were screened to give 13 different DNA sequences. Nucleotide sequence analysis revealed that seven and four of these clones were from the α-Proteobacteria and β-Proteobacteria, respectively. In the biofilm slice furthest from the membrane, 29 clones were screened to reveal 11 different DNA sequences. Five of these clones were from the β-Proteobacteria; other clones were from Bacteriodetes, α-Proteobacteria, γ-Proteobacteria, and δ-Proteobacteria. Although there were substantial differences between these two bacterial communities, there were two instances (clones M11 and L9 and clones M32 and L11) in which highly similar clones (>99.5% identical) were detected both near the membrane and at the biofilm-liquid interface.
Functional gene-based population profiles.
The bacterial community structure in the MABs was also analyzed by competitive quantitative PCR of genes coding for ammonia oxidation (amoA) or denitrification (nirK and nirS). In the air-fed biofilm grown at a fluid velocity of 2 cm s−1, amoA gene fragments were detected only in the biofilm slice closest to the membrane, at a concentration near the detection limit (∼400 gene copies, or 2 × 104 gene copies per ml of biofilm). Similarly, amoA genes were not detectable throughout the oxygen-fed biofilm. In both the air-fed and oxygen-fed biofilms grown at a fluid velocity of 2 cm s−1, the concentrations of nirS gene fragments exhibited a hump-shaped pattern as a function of depth, with the highest concentration occurring in the middle of the biofilm (data not shown). The nirK gene fragment was difficult to quantify in either biofilm due to the coamplification of a similar-sized fragment.
In contrast, the population profiles in the biofilms grown at a fluid velocity of 14 cm s−1 were substantially different (Fig. 7) than those from the MABs grown at 2 cm s−1. In both the air-fed and oxygen-fed biofilms, the highest concentration of amoA gene fragments occurred near the membrane in the aerobic region of the biofilm. These concentrations gradually declined with distance from the membrane. In contrast, the concentrations of nirK gene fragments exhibited a hump-shaped pattern as a function of depth, with the highest concentration occurring in the middle of the biofilm. The nirS populations were not detectable near the membrane (air-fed biofilm, <1,000 μm; oxygen-fed biofilm, <1,900 μm) but increased rapidly as a function of distance from the membrane. The profile of nirS populations in the air-fed biofilm was such that the highest concentration of genes occurred near the biofilm-liquid interface. In contrast, the nirS population profile in the oxygen-fed biofilm was hump shaped, with the maximum concentration occurring in the middle of the biofilm. The concentrations of nirS populations were approximately 10-fold higher in the air-fed biofilm than in the oxygen-fed biofilm.
FIG. 7.
Population profiles of ammonia-oxidizing (amoA) and denitrifying (nirS and nirK) bacterial populations as determined by competitive quantitative PCR within air-fed (a) and oxygen-fed (b) biofilms grown at a fluid velocity of 14 cm s−1. •, nirS; ○, nirK; ▴, amoA.
DISCUSSION
An understanding of the factors controlling the stratification of the bacterial physiology and community structure in MABs is important to optimize their application for biological wastewater treatment. Previous studies have used fluorescent in situ hybridization to detect AOB near the membrane of MABs (16, 32, 36). Our study, however, is the first to simultaneously detect AOB and denitrifying bacteria within a single MAB. Our results are also consistent with previous studies that reported complete nitrogenous pollutant removal in MAB reactors (16, 26, 33, 36). In contrast, AOB and denitrifying bacterial populations did not coexist in MABs grown under conditions of low fluid velocity (2 cm s−1). This suggests that fluid velocity, which controls the rate of substrate mass transfer into the biofilm (7, 28, 29, 34, 38), has a profound effect on the stratification of bacterial activity and community structure in MABs.
Our results also suggest a niche differentiation between the nirS and nirK denitrifying bacterial populations. In the biofilms grown at 14 cm s−1, the nirK populations reached their maximum population density in the middle of the biofilm, whereas the nirS populations reached their maximum in the outer region of the biofilm. The conditions at these locations differed substantially in that the middle of the biofilm was nearer the aerobic zone and should have contained lower concentrations of organic nutrients, whereas the outer region of the biofilm was further from the aerobic zone but contained higher concentrations of organic nutrients. We therefore infer that these conditions are pertinent factors in the competition between nirS and nirK bacterial populations, although other factors that we did not quantify (e.g., nitrite and/or nitrate concentrations) could also be important.
PCR-DGGE and PCR cloning of 16S rRNA genes provided further evidence for the stratification of the bacterial community structure in MABs. These data also supported our cPCR results of amoA gene fragments in that none of the known AOB were detected in the biofilms grown at a fluid velocity of 2 cm s−1. Previous researchers demonstrated that the phylogenies of the known AOB give similar results whether amoA or 16S rRNA gene sequences are analyzed (30). The results from the 16S rRNA approach, however, cannot be compared to the cPCR results for nirK and nirS gene fragments, because denitrifying bacteria are of broad phylogenetic distribution (4, 27).
Previous researchers have discussed the possible biases of cultivation-independent community analysis that can occur during nucleic acid extraction (40) and PCR (14, 40). In addition to these biases, our competitive quantitative PCR approach is limited by two other factors. First, gene copy numbers do not necessarily correlate to the number of cells comprising a particular population, because individual cells can have variable copies of a gene within their genome (17, 21). This complicates the comparison of gene copy number of the different population types (amoA, nirS, and nirK) detected herein. Second, all PCR-based assays for community analysis simultaneously must balance generality (i.e., are all of the specific gene targets amplified?) versus specificity (i.e., are all of the nontargeted genes excluded?). The cPCR technique used herein for AOB was selected because previous researchers have demonstrated that it had a high specificity for N. oligotropha-like populations (10). This PCR protocol, however, does not amplify all of the known AOB (e.g., Nitrosomonas europaea ATCC 19718 [data not shown]), and thus it is possible that our cPCR underestimated the number of physiologically relevant AOB in our biofilms. Similarly, the quantification of nirK and nirS genes by cPCR could have overestimated the abundance of these populations if genes other than those coding for nitrite reductase were amplified. The researchers who developed the PCR protocols used herein to amplify the nirK and nirS gene fragments emphasized generality (13) and specificity (22), respectively. Conversely, cPCR could have underestimated the nirK and nirS populations if the PCR amplifications failed to incorporate all of the diversity of nitrite reductase genes that existed in our biofilms. This concern over PCR generality and specificity is particularly pertinent because relatively few studies have performed PCR analysis of nirK and nirS populations.
In addition to the differences in bacterial community structure, substantial differences in bacterial physiology and activity were detected in MABs as functions of depth, flow velocity, and oxygen partial pressure. All of the biofilms in this study were sufficiently thick such that partial oxygen penetration was achieved, resulting in the formation of an anoxic region that ranged in size from 30 to 75% of the total MAB thickness. Although an anoxic region is necessary for anaerobic denitrification to occur, thick MABs can be detrimental to reactor performance (33). MAB thickness and oxygen penetration must therefore be optimized. Oxygen did not penetrate as far at the higher fluid velocity, which was linked to a shift in respiratory activity towards the membrane (i.e., the source of oxygen). This increase in oxygen penetration was possibly due to an increase in biomass density or to an increase in substrate penetration resulting from improved mass transfer efficiency. Our ongoing research will help elucidate the effects of fluid velocity and other reactor conditions on biofilm characteristics, with the goal of optimizing the performance of MAB reactors.
In conclusion, our research demonstrates that the bacterial community structure and activity in MABs is substantially stratified as a function of biofilm depth. Under the proper conditions, this stratification can lead to the simultaneous existence of both nitrifying and denitrifying bacterial populations within a single biofilm. MAB reactors, therefore, have potential utility for the biological treatment of wastewaters containing both carbonaceous and nitrogenous pollutants. Our research also has demonstrated that fluid velocity, which controls the mass transfer of substrate into biofilms, can be an important factor affecting the simultaneous development of both AOB and denitrifying bacterial populations in MABs.
Acknowledgments
This research was funded by National Science Foundation grant BES 0123394, Water Environment Research Foundation grant WERF/00-CTS-11, and a Graduate Assistantship in Areas of National Need fellowship to A.C.C. from the U.S. Department of Education.
REFERENCES
- 1.Ahmed, T., and M. J. Semmens. 1992. Use of sealed end hollow fibers for bubbleless membrane aeration: experimental studies. J. Membr. Sci. 69:1-10. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2002. GenBank. Nucleic Acids Res. 30:17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bothe, H., G. Jost, M. Schloter, B. B. Ward, and K.-P. Witzel. 2000. Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol. Rev. 24:673-690. [DOI] [PubMed] [Google Scholar]
- 5.Brindle, K., and T. Stephenson. 1996. The application of membrane biological reactors for the treatment of wastewaters. Biotechnol. Bioeng. 49:601-610. [DOI] [PubMed] [Google Scholar]
- 6.Casey, E., B. Glennon, and G. Hamer. 1999. Review of membrane aerated biofilm reactors. Resource Conserv. Recycl. 27:203-215. [Google Scholar]
- 7.Casey, E., B. Glennon, and G. Hamer. 2000. Biofilm development in a membrane-aerated biofilm reactor: effect of flow velocity on performance. Biotechnol. Bioeng. 67:476-486. [PubMed] [Google Scholar]
- 8.Casey, E., B. Glennon, and G. Hamer. 2000. Biofilm development in a membrane-aerated biofilm reactor: effect of intra-membrane oxygen pressure on performance. Bioproc. Eng. 23:457-465. [Google Scholar]
- 9.Côté, P., J.-L. Bersillon, A. Huyard, and G. Faup. 1988. Bubble-free aeration using membranes: process analysis. J. Water Pollut. Control Fed. 60:1986-1992. [Google Scholar]
- 10.Dionisi, H. M., A. C. Layton, G. Harms, I. R. Gregory, K. G. Robinson, and G. S. Sayler. 2002. Quantification of Nitrosomonas oligotropha-like ammonia-oxidizing bacteria and Nitrospira spp. from full-scale wastewater treatment plants by competitive PCR. Appl. Environ. Microbiol. 68:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Essila, N. J., M. J. Semmens, and V. R. Voller. 2000. Modeling biofilms on gas-permeable supports: concentration and activity profiles. J. Environ. Eng. 126:250-257. [Google Scholar]
- 13.Hallin, S., and P.-E. Lindgren. 1999. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl. Environ. Microbiol. 65:1652-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen, M. C., T. Tolkernielsen, M. Givskov, and S. Molin. 1998. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol. Ecol. 26:141-149. [Google Scholar]
- 15.Herbert, D., P. J. Phipps, and R. E. Strange. 1971. Chemical analysis of microbial cells. Methods Microbiol. 5B:209-234. [Google Scholar]
- 16.Hibiya, K., A. Terada, S. Tsuneda, and A. Hirata. 2002. Simultaneous nitrification and denitrification by controlling vertical and horizontal microenvironment in a membrane-aerated biofilm reactor. J. Biotechnol. 100:23-32. [DOI] [PubMed] [Google Scholar]
- 17.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., West Sussex, United Kingdom.
- 19.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 20.Maidak, B. L., J. R. Cole, C. T. Parker, G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McTavish, H., J. A. Fuchs, and A. B. Hooper. 1993. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J. Bacteriol. 175:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michotey, V., V. Méjean, and P. Bonin. 2000. Comparison of method for quantification of cytochrome cd1-denitrifying bacteria in environmental marine samples. Appl. Environ. Microbiol. 66:1564-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis—analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 25.Neidhardt, F. C., et al. (ed.). 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 26.Pankhania, T., T. Stephenson, and M. J. Semmens. 1994. Hollow fiber bioreactor for wastewater treatment using bubbleless membrane aeration. Water Res. 28:2233-2236. [Google Scholar]
- 27.Philippot, L., and O. Højberg. 1999. Dissimilatory nitrate reductases in bacteria. Biochim. Biophys. Acta 1446:1-23. [DOI] [PubMed] [Google Scholar]
- 28.Picioreanu, C., M. C. M. van Loosdrecht, and J. J. Heijnen. 2000. A theoretical study on the effect of surface roughness on mass transport and transformation in biofilms. Biotechnol. Bioeng. 68:355-369. [DOI] [PubMed] [Google Scholar]
- 29.Picioreanu, C., M. C. M. van Loosdrecht, and J. J. Heijnen. 2000. Effect of diffusive and convective transport on biofilm structure formation: a two-dimensional modeling study. Biotechnol. Bioeng. 69:504-515. [DOI] [PubMed] [Google Scholar]
- 30.Purkhold, U., A. Pommerening-Röser, S. Juretchko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Schramm, A., D. De Beer, A. Gieseke, and R. Amann. 2000. Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ. Microbiol. 2:680-686. [DOI] [PubMed] [Google Scholar]
- 33.Semmens, M. J., K. Dahm, J. Shanahan, and A. Christianson. 2003. COD and nitrogen removal by biofilms growing on gas permeable membranes. Water Res. 37:4343-4350. [DOI] [PubMed] [Google Scholar]
- 34.Semmens, M. J., and N. J. Essila. 2001. Modeling biofilms on gas-permeable supports: flux limitations. J. Environ. Eng. 127:126-133. [Google Scholar]
- 35.Siegrist, H., and W. Gujer. 1985. Mass transfer mechanisms in a heterotrophic biofilm. Water Res. 19:1369-1378. [Google Scholar]
- 36.Terada, A., K. Hibiya, J. Nagai, S. Tsuneda, and A. Hirata. 2003. Nitrogen removal characteristics and biofilm analysis of a membrane-aerated biofilm reactor applicable to high-strength nitrogenous wastewater treatment. J. Biosci. Bioeng. 95:170-178. [DOI] [PubMed] [Google Scholar]
- 37.Timberlake, D. L., S. E. Strand, and K. J. Williamson. 1988. Combined aerobic heterotrophic oxidation, nitrification in a permeable support biofilm. Water Res. 22:1513-1517. [Google Scholar]
- 38.van Loosdrecht, M. C. M., J. J. Heijnen, H. Eberl, J. Kreft, and C. Picioreanu. 2002. Mathematical modeling of biofilm structures. Antonie Leeuwenhoek 81:245-256. [DOI] [PubMed] [Google Scholar]
- 39.von Mersi, W., and F. Schinner. 1991. An improved and accurate method for determining the dehydrogenase activity of soils with iodonitrotetrazolium chloride. Biol. Fertil. Soils 11:216-220. [Google Scholar]
- 40.von Witzengerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]