Abstract
Pentachlorophenol (PCP), a highly toxic anthropogenic pesticide, can be mineralized by Sphingobium chlorophenolicum, a gram-negative bacterium isolated from PCP-contaminated soil. However, degradation of PCP is slow and S. chlorophenolicum cannot tolerate high levels of PCP. We have used genome shuffling to improve the degradation of PCP by S. chlorophenolicum. We have obtained several strains that degrade PCP faster and tolerate higher levels of PCP than the wild-type strain. Several strains obtained after the third round of shuffling can grow on one-quarter-strength tryptic soy broth plates containing 6 to 8 mM PCP, while the original strain cannot grow in the presence of PCP at concentrations higher than 0.6 mM. Some of the mutants are able to completely degrade 3 mM PCP in one-quarter-strength tryptic soy broth, whereas no degradation can be achieved by the wild-type strain. Analysis of several improved strains suggests that the improved phenotypes are due to various combinations of mutations leading to an enhanced growth rate, constitutive expression of the PCP degradation genes, and enhanced resistance to the toxicity of PCP and its metabolites.
Thousands of anthropogenic chemicals are released into the environment as a result of agricultural, industrial, military, and domestic activities. Biodegradation of anthropogenic compounds is often inefficient because bacteria in soil and water have had little time to evolve the metabolic pathways required to convert such compounds into readily metabolized products. We are studying the biodegradation of pentachlorophenol (PCP), an anthropogenic pesticide first introduced into the environment in 1936 (4). Biodegradation of PCP is particularly challenging because it uncouples oxidative phosphorylation and alters membrane fluidity. Despite its toxicity and recent introduction into the environment, PCP can be completely degraded by Sphingobium chlorophenolicum, several strains of which (3, 17, 18, 20) have been isolated from PCP-contaminated soil. Although the ability of S. chlorophenolicum to mineralize PCP is remarkable, the inefficiency of the metabolic pathway has limited the potential for its use in bioremediation.
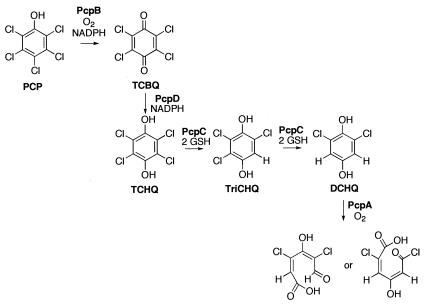
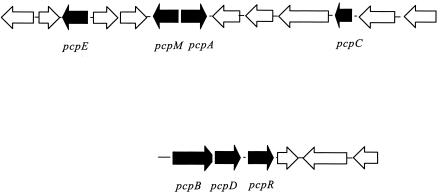
The initial steps of the pathway for degradation of PCP in S. chlorophenolicum (5, 14) are shown in Fig. 1. The pathway begins with hydroxylation of PCP by PCP hydroxylase (PcpB) to produce tetrachlorobenzoquinone (TCBQ). TCBQ is subsequently reduced by a reductase (PcpD) to tetrachlorohydroquinone (TCHQ). Two successive reductive dehalogenation reactions are catalyzed by TCHQ dehalogenase (PcpC). 2,6-Dichlorohydroquinone (2,6-DCHQ) is cleaved by an Fe(II)-dependent extradiol dioxygenase (PcpA). A plausible pathway for further degradation to produce β-ketoadipate has been proposed (2). The genes involved in the degradation of PCP have been found on two different gene fragments (Fig. 2) (2). Expression of pcpA, pcpB, pcpD, and pcpE is induced by PCP, while expression of pcpC is constitutive.
FIG. 1.
Pathway for degradation of PCP in S. chlorophenolicum strain ATCC 39723. PcpB, PCP hydroxylase; PcpD, TCBQ reductase; PcpC, TCHQ dehalogenase; PcpA, 2,6-DCHQ dioxygenase; GSH, glutathione.
FIG. 2.
Organization of genes involved in the degradation of PCP on two chromosomal fragments from S. chlorophenolicum ATCC 39723 (2). pcpA encodes DCHQ dioxygenase, pcpB encodes PCP hydroxylase, pcpC encodes TCHQ dehalogenase, pcpD encodes TCBQ reductase, pcpE encodes a putative maleylacetate reductase, and pcpM and pcpR encode putative LysR transcriptional regulators.
The flux through the pathway appears to be limited by the poor catalytic activity of the first enzyme, PCP hydroxylase (9). This enzyme has a kcat of only 0.02 s−1 (P. M. Kiefer and S. D. Copley, unpublished results); flavin monooxygenases typically have kcat values of 25 to 50 s−1 (21). The ability of S. chlorophenolicum to evolve a more effective PCP hydroxylase is limited because the enzyme converts PCP to an extremely toxic product, TCBQ (5). TCBQ is a potent electrophile and reacts with nucleophiles in proteins (23), DNA (8), and glutathione (22). Thus, substantial improvement of the activity of PCP hydroxylase would expose the cells to higher levels of TCBQ. Consequently, we expect that improvement of flux through the pathway will require simultaneous improvement of both PCP hydroxylase and the downstream enzyme TCBQ reductase, which converts TCBQ to the less toxic TCHQ. This is unlikely to occur either in nature or during efforts to improve the strain by classic strain selection in the laboratory.
We have explored the use of genome shuffling to improve the degradation of PCP by S. chlorophenolicum. Genome shuffling involves generation of mutant strains that have an improved phenotype, followed by multiple rounds of protoplast fusion to allow recombination between genomes. This approach has recently been used to improve the production of the polyketide antibiotic tylosin in Streptomyces fradiae (26) and to improve acid tolerance in Lactobacillus (16). Genome shuffling is useful for engineering of multitrait phenotypes that would be difficult to engineer directly because it may be impossible to anticipate all of the mutations needed to improve a complex trait while still maintaining robust growth. The improvement of PCP degradation is an excellent problem to address by this method. Because we expect that it may be difficult to improve flux through the pathway, adaptations that reduce the toxic effects of PCP and its metabolites are likely to be particularly important, and these are likely to require multiple mutations. We report here that successive rounds of protoplast fusion alternating with selection for improved growth in the presence of PCP result in substantial improvements in both the rate of PCP degradation and the concentration of PCP that can be tolerated. Analysis of several improved strains indicates that various combinations of mutations leading to an enhanced growth rate, constitutive expression of the PCP degradation genes, and enhanced resistance to the toxicity of PCP and its metabolites contribute to the improved phenotypes. These results suggest that enhanced performance can be achieved in a number of different ways, an issue that has not heretofore been explored in reports of genome shuffling experiments.
MATERIALS AND METHODS
Materials.
Restriction endonucleases and Taq DNA polymerase were purchased from Promega. PCP, TCBQ, NADPH, and PEG6000 were obtained from Sigma-Aldrich-Fluka. Tryptic soy broth (TSB) was obtained from Becton Dickinson. Oligonucleotide primers were obtained from IDT Technologic.
Bacterial strains.
S. chlorophenolicum strain ATCC 39723 was obtained from the American Type Culture Collection. This strain was grown routinely on one-quarter-strength TSB containing 50 μM PCP. S. chlorophenolicum cannot tolerate full-strength TSB, and dilution to one-quarter strength was found to support optimal growth. PCP was added to prevent loss of PCP degradation genes, which can occur when the cells grow in the absence of PCP.
Genome shuffling.
S. chlorophenolicum cells were mutagenized with nitrosoguanidine as described by Miller (10) and then spread on plates containing one-quarter-strength TSB and 1.6 mM PCP. The plates were incubated at 25°C for 5 days. Thirty-four colonies were obtained from the plates. Protoplast fusion was carried out as described by Wei et al. (24). Cells were grown in one-quarter-strength TSB until the optical density at 550 nm (OD550) reached 1.0. The cells were harvested by centrifugation at 4°C at 3,000 × g for 10 min. The cells were washed twice with SMM (0.5 M sucrose, 20 mM MgCl2, 20 mM sodium maleate buffer, pH 6.5) and treated with lysozyme (0.4 mg/ml in SMM) for 30 min at 25°C. Sodium EDTA (pH 8.0) was then added to a final concentration of 0.1% (wt/vol), and the cells were shaken gently for 10 min to complete protoplast formation. Protoplasts were fused by suspension in SMM containing 30% PEG 6000, 15% dimethyl sulfoxide, and 10 mM CaCl2. After 30 min of gentle shaking at 25°C, the suspension was diluted 10-fold with regeneration medium (2.9 g of Na2HPO4 per liter, 0.3 g of KH2PO4 per liter, 0.1 g of NH4NO3 per liter, 0.02 g of MgSO4 · 7H2O per liter, 10 mg of FeSO4 · 7H2O per liter, 4 g of sodium glutamate per liter, 0.5 M sucrose) and protoplasts were harvested by centrifugation at 2,000 × g for 20 min at 20°C. The protoplasts were resuspended in regeneration medium and shaken at 100 rpm for 10 h before plating on plates containing one-quarter-strength TSB and various concentrations of PCP. The plates were incubated for 5 to 8 days at 25°C. Subsequent rounds of genome shuffling were carried out by the same methods.
Analysis of cell growth and PCP degradation in liquid cultures.
Wild-type and mutant strains of S. chlorophenolicum were grown in 20 ml of one-quarter-strength TSB in 125-ml flasks. Each strain was grown in at least three different flasks. Each flask was inoculated with 0.5 ml of a starter culture grown in one-quarter-strength TSB (in some cases containing 50 μM PCP) and shaken at 200 rpm and 22°C. PCP was added to a final concentration of either 0.3 or 3 mM when the OD550 reached 0.5. Cell growth was monitored by measurement of the OD550. PCP concentration was determined by high-performance liquid chromatography as previously described (5).
PCR amplification of pcpB, pcpD, pcpR, and the promoter region of pcpB.
Cells were grown on one-quarter-strength TSB in the presence of 50 μM PCP. After an OD550 of 1.2 to 1.5 was reached, genomic DNA was isolated with a QIAGEN DNeasy kit. pcpB, pcpD, pcpR, and the promoter region of pcpB were amplified by PCR. The PCR program consisted of a denaturation step of 5 min at 95°C, followed by 25 cycles of denaturation at 95°C for 50 s, annealing at 52°C for 50 s, and extension at 72°C for 50 s for pcpD, pcpR, and the promoter region of pcpB or 2 min for pcpB, with a final extension step of 72°C for 10 min. The following primers were used: pcpB forward primer, 5′-CGGAATTCTCGGGCAAGACCACC-3′; pcpB reverse primer, 5′-ATCCCAAGCTTAGGCGCACGGCACAAGCA-3′; pcpD forward primer, 5′-GAAGATCTCACAAACCCCGTTTCGA-3′; pcpD reverse primer, 5′-CCCAAGCTTATCAGATGTCCAGCACCAGC-3′; pcpR forward primer, 5′-TAGGAAGATCTAATGATTCCGTATTACCGCT-3′; pcpR reverse primer, 5′-CGGAAGCTTATCACTCCGCCGGCGGATT-3′; promoter region of pcpB forward primer, 5′-ATGGGATCCTTATGCGGCCGGCA-3′; promoter region of pcpB reverse primer, 5′-CGGAAGCTTAATAACAATCTCTCTCCCGA-3′. PCR products from two different amplification reactions were sequenced at the University of Colorado DNA Sequencing Facility.
RT-PCR to detect transcription of pcpA, pcpB, pcpC, and pcpD.
Cells were grown on one-quarter-strength TSB in the presence or absence of 50 μM PCP. After an OD550 of 0.5 was reached, RNA was isolated with a QIAGEN RNeasy kit. RNase-free DNase was added in accordance with the manufacturer's protocol in order to eliminate contaminating DNA. Reverse transcription (RT) was performed with a Qiagen Omniscript reverse transcriptase kit. The resulting cDNA was amplified by PCR with primers specific for each gene. The PCR program consisted of a denaturation step of 5 min at 95°C, followed by 28 cycles of denaturation at 95°C for 50 s, annealing at 52°C for 50 s, and extension at 72°C for 50 s for pcpA, pcpC, and pcpD and 2 min for pcpB, with a final extension step of 72°C for 10 min. The following primers were used: pcpA forward primer, 5′-ATGGAAACGAACCATATCACCA-3′; pcpA reverse primer, 5′-TCATCAAACCACGATGGGATCGT-3′; pcpB forward primer, 5′-CCGGAATTCATGTCGACCTATCCAATCAATG-3′; pcpB reverse primer, 5′-ATCCCAAGCTTAGGCGCACGGCACAAGCA-3′; pcpC forward primer, 5′-AGAGGATCCGGAGCTTATCGACT-3′; pcpC reverse primer, 5′-CCGCTCGAGGATGCCGCCCTTCCAATTG-3′; pcpD forward primer, 5′-GGGGTACCATGACAAACCCCGTTTCGACAAT-3′; pcpD reverse primer, 5′-CCCAAGCTTATCAGATGTCCAGCACCAGC-3′. To ensure that contaminating DNA was not present, the same procedure was followed but without the initial RT step. PCR products were separated by gel electrophoresis and visualized by staining with ethidium bromide.
Preparation of crude extracts for enzyme assays.
Wild-type and mutant strains of S. chlorophenolicum were grown at 25°C in 300 ml of one-quarter-strength TSB containing 200 μM PCP and harvested by centrifugation at 3,000 × g at 4°C for 10 min when an OD550 of 0.5 was reached. Cell pellets were resuspended in 10 ml of lysis buffer [50 mM potassium phosphate (pH 7.0) containing 0.1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride], and lysates were prepared by passage two times through a French pressure cell at 12,000 lb/in2. Lysates were subjected to centrifugation at 17,500 × g at 4°C for 20 min. (Since TCBQ reductase is expected to contain an iron-sulfur cluster and such proteins can be sensitive to O2, cell lysates for TCBQ reductase assays were prepared under an N2 atmosphere and then dialyzed under N2 for 4 h against lysis buffer to remove inhibitory small molecules.)
Enzyme assays.
PCP hydroxylase and TCBQ reductase were assayed as previously described (5). Protein concentrations were determined by the method of Bradford with bovine serum albumin as the standard (1).
RESULTS
Improvement of the ability to grow on PCP-containing plates during three rounds of genome shuffling.
S. chlorophenolicum strain ATCC 39723 cannot grow on one-quarter-strength TSB plates containing PCP at concentrations higher than 0.6 mM. In order to obtain mutants with an improved ability to grow in the presence of PCP, S. chlorophenolicum was treated with nitrosoguanidine and then plated on one-quarter-strength TSB containing 1.6 mM PCP. The 34 colonies obtained were used as the starting population for genome shuffling.
Three successive rounds of protoplast fusion were carried out, and after each round, the concentration of PCP in the plates used for selection was increased. After the first fusion, 50 colonies obtained on plates containing 2 mM PCP were used for the second fusion. After the second fusion, 34 colonies obtained on plates containing 3 mM PCP were used for the third fusion. As a control, the population of mutant strains was also spread again on plates containing various levels of PCP without protoplast fusion to determine whether additional mutations caused by exposure to PCP on the plates could lead to improved growth on PCP. No colonies were observed when the mutant population was respread on plates containing 2 mM PCP.
At the conclusion of the shuffling, the ability of strains isolated during all three rounds to grow in the presence of PCP in liquid medium was assessed, and the best two or three strains from each round were selected for further characterization. (These strains are described by three-digit numbers in which the first digit indicates the round of shuffling from which the strain was obtained.) Figure 3 shows the growth of these strains on plates containing one-quarter-strength TSB and PCP and demonstrates progressive improvement in the ability to grow in the presence of PCP. Strains obtained after the first round of shuffling can grow at PCP concentrations of up to 3 mM. Strains from the second round can grow at 4 mM PCP, and strains from the third round can grow at 5 mM PCP. Two strains (316 and 320) were able to grow on plates containing 8 mM PCP (data not shown). Thus, strains obtained after the third round of shuffling were able to grow in the presence of PCP at levels 13-fold higher than those that can be tolerated by wild-type cells.
FIG. 3.
Growth of wild-type and mutant strains of S. chlorophenolicum on plates containing one-quarter-strength TSB and PCP at 0.4 mM (a), 3 mM (b), 4 mM (c), and 5 mM (d). Strains 108 and 130 were obtained from the first round of genome shuffling, strains 206 and 215 were obtained from the second round, and strains 307, 316, and 320 were obtained from the third round.
Characterization of PCP degradation by selected strains from three rounds of genome shuffling.
Degradation of PCP in liquid cultures was analyzed for the seven selected strains. The original strain cannot grow in one-quarter-strength TSB containing more than 0.4 mM PCP. Figure 4 shows the degradation of PCP in cultures containing one-quarter-strength TSB and 0.3 mM PCP by cells that were not previously exposed to PCP (Fig. 4a) or cells that had been grown initially with 50 μM PCP (Fig. 4c). In both cases, the strains obtained after genome shuffling degraded PCP more quickly than the wild-type strain, primarily because of a shortened lag phase before degradation begins. All strains, including the wild type, degraded PCP more quickly when the cells had been pre-exposed to PCP.
FIG. 4.
Degradation of PCP by and cell growth of wild-type and mutant strains of S. chlorophenolicum. Degradation of 0.3 mM PCP by (a) and growth of (b) cells not previously exposed to PCP, degradation of 0.3 mM PCP by (c) and growth of (d) cells previously exposed to 50 μM PCP, and degradation of 3 mM PCP by (e) and growth of (f) cells previously exposed to 50 μM PCP are shown. Symbols: ▪, wild type; ♦, strain 108; •, strain 130; ♦, strain 206; •, strain 215; ♦, strain 307; •, strain 316; ▪, strain 320. For the experiments in panels c to f, cells were grown in the presence of 50 μM PCP and additional PCP was added when the OD550 reached 0.6.
Figure 4 shows the degradation of PCP (Fig. 4e) and growth of cells (Fig. 4f) in cultures containing one-quarter-strength TSB and 3 mM PCP by cells that had been preinduced with 50 μM PCP. The differences between the original and mutant strains are much more dramatic under these circumstances. The original strain can neither grow nor degrade PCP. In contrast, all of the mutant strains grow and degrade PCP. The extent of degradation varies. Five of the strains stop degrading PCP when 30 to 50% of the PCP remains. Under these conditions, the toxicity of PCP eventually overwhelms the cells, and the cessation of degradation is due to cell death. However, two strains can completely degrade 3 mM PCP within 48 h.
Analysis of growth rates of the original and mutant strains.
The growth of the original and mutant strains in one-quarter-strength TSB in the presence and absence of 0.3 mM PCP was followed. Figure 4b and d show that the mutant strains grew faster than the wild type in the presence of PCP, regardless of whether the cells had been pre-exposed to PCP. Under both conditions, the growth rate was closely correlated with the rate of removal of PCP from the medium. The relative growth rates of the mutant strains differed, however, depending on whether the cells had been pre-exposed to PCP. Additional experiments showed that the mutant strains also grew faster in one-quarter-strength TSB lacking PCP (data not shown). In the absence of PCP, the mutant strains had a shorter lag phase (10 h versus 15 h for the original strain) and grew to a cell density greater (up to 45% greater) than that of the wild-type strain.
Analysis of protein levels in crude extracts of wild-type and mutant strains.
Aliquots of crude extracts prepared from wild-type and mutant strains grown in the presence of 0.2 mM PCP were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in order to determine whether higher levels of the PCP degradation enzymes were apparent in the mutant strains. No discernible differences in protein expression levels were observed (data not shown).
Analysis of the expression of PCP degradation genes in cells grown in the absence of PCP by RT-PCR.
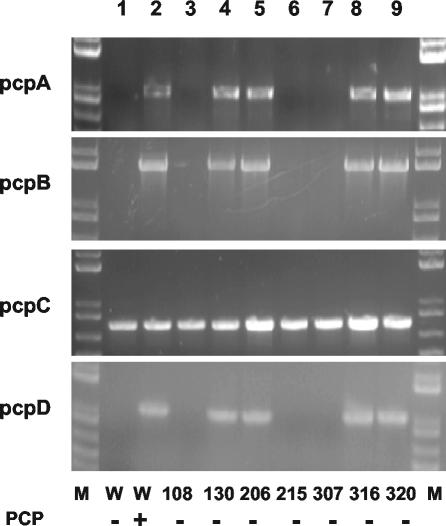
Expression of several genes involved in PCP degradation was analyzed in the selected strains. Mutant strains were grown in the absence of PCP, and the original strain was grown in the absence and presence of PCP. Total RNA was isolated, and the presence of transcripts for pcpA, pcpB, pcpC, and pcpD was assessed by RT-PCR. These genes encode dichlorohydroquinone dioxygenase, PCP hydroxylase, TCHQ dehalogenase, and TCBQ reductases, respectively (Fig. 1). In the wild-type strain, expression of pcpA (25), pcpB (15), and pcpD (5) has been reported to be induced by PCP, while expression of pcpC has been reported to be constitutive (13). Our results (Fig. 5) confirm these findings for the wild-type strain and demonstrate that in four of the seven strains under investigation (130, 206, 316, and 320), expression of pcpA, pcpB, and pcpD is constitutive.
FIG. 5.
RT-PCR analysis of the presence of transcripts for genes involved in PCP degradation. Left- and right-most lanes, molecular size markers (M); lane 1, wild-type (W) cells grown in the absence (−) of PCP; lane 2, wild-type cells grown in the presence (+) of 50 μM PCP; lanes 3 to 9, mutant strains (as indicated at the bottom) grown in the absence (−) of PCP. pcpA, pcpB, pcpC, and pcpD encode DCHQ dioxygenase, PCP hydroxylase, TCHQ dehalogenase, and TCBQ reductase, respectively.
Sequencing of pcpB, pcpD, pcpR, and the pcpB promoter region in selected mutant strains.
The genes encoding PcpB (PCP hydroxylase), PcpD (TCBQ reductase), and PcpR (the LysR transcriptional regulator responsible for induction of expression in the presence of PCP), as well as the promoter region upstream of pcpB, were amplified from several mutant strains by PCR and sequenced at the University of Colorado Microsequencing Facility. The mutations found in pcpB and pcpD are summarized in Table 1. No mutations were found in pcpR or in the pcpB promoter region.
TABLE 1.
Mutations found in pcpB and pcpD in mutant strains of S. chlorophenolicum produced by genome shuffling
| Strain | pcpB mutation(s) | Amino acid change(s) in PcpB | pcpD mutation | Amino acid change in PcpD |
|---|---|---|---|---|
| 108 | None | None | None | None |
| 130 | C106T, G846A | None, Arg283Gln | None | None |
| 206 | C134T, C220T, C759A | Arg45Cys, Arg74Cys, none | None | None |
| 215 | C1270A | Ala424Val | G721A | Val241Met |
| 307 | G485A | None | None | None |
| 316 | None | None | None | None |
| 320 | C30T | None | None | None |
Analysis of PCP hydroxylase and TCBQ reductase in crude extracts of wild-type and mutant strains.
Levels of PCP hydroxylase and TCBQ reductase in crude extracts from wild-type and mutant strains were measured (data not shown). The level of PCP hydroxylase activity was equivalent to that in the wild-type strain in six of the seven strains. The specific activity in strain 206 was 63% of that in the original strain. The level of TCBQ reductase activity in extracts of the seven mutant strains was essentially identical to that in extracts of the wild-type strain.
DISCUSSION
Mutant strains obtained after genome shuffling show substantial improvement in the ability to grow in the presence of PCP on solid medium. Several strains obtained after the third round of shuffling can grow on plates containing 6 to 8 mM PCP, while the original strain cannot grow in the presence of concentrations higher than 0.6 mM. Characterization of selected mutant strains in liquid medium shows that the improvement in the ability to grow in the presence of PCP is correlated with increased degradation of PCP and so is not simply due to mutations that result in exclusion of PCP from the cell.
A clue to the mechanisms underlying the improved performance of the mutant strains is provided by the data in Fig. 4, which demonstrate that the growth rate of the mutant strains is closely correlated with the ability to remove PCP from the medium. Thus, an important factor in the rate of PCP degradation is simply the cell mass that can be achieved. The enhanced growth rate of the mutant strains might be due to mutations that allow the cells to use the nutrients available in one-quarter-strength TSB more effectively. Indeed, the mutant strains grow faster than the wild type on one-quarter-strength TSB, even in the absence of PCP. However, mutations that allow faster removal of PCP or that ameliorate the adverse effects of PCP or its metabolites must also be involved, since the mutant strains were able to grow in the presence of high levels of PCP that completely inhibited the growth of the original strain.
Constitutive expression of pcpB, pcpA, and pcpD was found in four of the mutant strains. These strains degraded PCP most quickly when the cells were not preinduced with PCP (Fig. 4a). Constitutive expression of PCP degradation genes may protect cells from the initial shock of exposure to PCP by allowing degradation to begin immediately. Expression of the PCP degradation genes is regulated by pcpR, a LysR family transcriptional regulator (2). LysR transcriptional regulators appear to function as tetramers (11) that bind to promoter DNA and induce a sharp bend of about 78° (12). Upon binding of inducer to the regulatory domain, the angle is relaxed to about 54° (12) and transcription is activated. Previous studies of LysR transcriptional regulators have shown that inducer-independent expression can result from mutations in the regulatory domain of the protein (7, 19). However, we found no mutations in pcpR in any of the strains, including those that constitutively express pcpB, pcpD, and pcpA. It is possible that mutations in the promoter regions upstream of pcpB and pcpA might allow recognition by another transcriptional regulator capable of directing the expression of these genes in the absence of PCP. This possibility can be ruled out because the promoter region upstream of pcpB was not mutated in any of the strains. Thus, the most likely explanation is that a mutation in a gene encoding another transcriptional regulator allowed it to bind to the promoter regions of pcpB and pcpA and direct expression of the PCP degradation genes.
Strains containing mutations leading to constitutive expression of PCP degradation genes did not necessarily perform better when the cells were pretreated with PCP (Fig. 4c), suggesting that other types of mutations also conferred enhanced degradative abilities. An obvious mechanism by which PCP degradation might be improved would be up-regulation of the enzymes in the metabolic pathway. Surprisingly, analysis of crude extracts showed that there were no discernible differences in the pattern of proteins produced by the wild-type and mutant strains. Furthermore, assays of PCP hydroxylase and TCBQ reductase in crude extracts of cells grown in the presence of PCP showed no significant differences between the wild-type and mutant strains, with a single exception. The PCP hydroxylase activity in strain 206 was only 63% of that of the wild-type strain. The gene encoding PCP hydroxylase in this strain was sequenced and found to have mutations resulting in changes of Arg45 and Arg74 to Cys. On the basis of sequence comparisons with the homologous and structurally characterized phenol hydroxylase (6), Arg45 corresponds to a residue involved in binding flavin (Lys43), while Arg74 corresponds to a residue located at the subunit interface (Lys72). Mutation of either or both of these residues could lead to the observed diminution of activity in the strain 206 enzyme.
We suspect that multiple mutations are responsible for the improved phenotypes exhibited by the mutant strains, as the performance of the mutants increased with successive rounds of shuffling, presumably as a result of the combination of different beneficial mutations. Support for the hypothesis that the improved phenotypes were achieved by genome shuffling rather than by accumulation of successive mutations within strains is provided by control experiments in which the initial mutant population was replated without protoplast fusion. No clones capable of growth on 2 mM PCP were found, while plating of a comparable number of cells after protoplast fusion gave 50 colonies on plates containing 2 mM PCP.
Taken together, the different rates of growth and PCP degradation among the mutants when they are grown under different conditions (compare Fig. 3a and c and 4), the differences in the regulation of the PCP degradation genes (Fig. 5), and the mutations in pcpB and pcpD (Table 1) demonstrate that the seven mutant strains are genetically distinct. This finding suggests that improved performance can be achieved in a number of different ways. The mutations leading to improved performance appear to confer enhanced growth under the selection conditions and enhanced resistance to the toxicity of PCP and TCBQ. Notably, mutations leading to higher activities of the first two enzymes in the pathway, which limit flux through the pathway and detoxify a highly toxic intermediate, respectively, were not found. This is not surprising, since the experimental design involves initial selection for mutants that have improved growth in the presence of PCP. Mutations resulting in substantial improvement of PCP hydroxylase activity would likely result in decrease fitness because of the increased production of toxic TCBQ. Furthermore, if the existing TCBQ reductase activity is already sufficient to protect the cells from the toxic effects of the small amount of TCBQ formed by PCP hydroxylase, then substantial improvements in this activity would also not enhance fitness. Thus, the initial pool of mutants that displayed improved growth on plates containing PCP would not be expected to include mutants with improved versions of either PCP hydroxylase or TCBQ reductase.
The findings reported here have important implications. It is remarkable how much improvement in PCP degradation could be achieved without altering the key enzymes in the degradative pathway. These improvements apparently resulted from mutations that enhanced growth under these particular growth conditions and enhanced resistance to the toxicity of PCP. Thus, we expect that genome shuffling procedures should be most successful when they are tailored for particular applications by matching the selection conditions to the conditions of the process for which the bacteria will be used. In addition, these results suggest that genome shuffling will provide an excellent experimental tool for examination of fitness landscapes by generating a number of mutant strains that have reached the same phenotype via different routes.
Acknowledgments
This work was supported by the U.S. Army Research Office under grant DAAD19-99-1-0301.
We thank Ryan Gill for helpful discussion.
REFERENCES
- 1.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Cai, M., and L. Xun. 2002. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 184:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassidy, M. B., H. Lee, J. T. Trevors, and R. B. Zablotowicz. 1999. Chlorophenol and nitrophenol metabolism by Sphingomonas sp. UG30. J. Ind. Microbiol. Biotechnol. 23:232-241. [DOI] [PubMed] [Google Scholar]
- 4.Cline, R. E., R. H. Hill, D. L. Phillips, and L. L. Needham. 1989. Pentachlorophenol measurements in body fluids of people in log homes and workplaces. Arch. Environ. Contam. Toxicol. 18:475-481. [DOI] [PubMed] [Google Scholar]
- 5.Dai, M., J. Bull Rogers, J. R. Warner, and S. D. Copley. 2003. A previously unrecognized step in pentachlorophenol degradation in Sphingobium chlorophenolicum is catalyzed by tetrachlorobenzoquinone reductase (PcpD). J. Bacteriol. 185:302-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enroth, C., H. Neujahr, G. Schneider, and Y. Lindqvist. 1998. The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis. Structure 6:605-617. [DOI] [PubMed] [Google Scholar]
- 7.Huang, J., and M. A. Schell. 1991. In vivo interactions of the NahR transcriptional activator with its target sequences: inducer-mediated changes resulting in transcription activation. J. Biol. Chem. 266:10830-10838. [PubMed] [Google Scholar]
- 8.Lin, P.-H., J. Nakamura, S. Yamaguchi, P. B. Upton, D. K. La, and J. A. Swenberg. 2001. Oxidative damage and direct adducts in calf thymus DNA induced by the pentachlorophenol metabolites, tetrachlorohydroquinone and tetrachloro-1,4-benzoquinone. Carcinogenesis 22:627-634. [DOI] [PubMed] [Google Scholar]
- 9.McCarthy, D. L., A. Claude, and S. D. Copley. 1997. In vivo levels of chlorinated hydroquinones in a pentachlorophenol-degrading bacterium. Appl. Environ. Microbiol. 63:1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller, J. H. 1972. Experiments in molecular genetics, p. 125-129. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Muraoka, S., R. Okumura, N. Ogawa, T. Nonaka, K. Miyashita, and T. Senda. 2003. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol. 328:555-566. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa, N., S. M. McFall, T. J. Klem, K. Miyashita, and A. M. Chakrabarty. 1999. Transcriptional activation of the chlorocatechol degradative genes of Ralstonia eutropha NH9. J. Bacteriol. 181:6697-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orser, C. S., J. Dutton, C. Lange, P. Jablonski, L. Xun, and M. Hargis. 1993. Characterization of a Flavobacterium glutathione S-transferase gene involved in reductive dehalogenation. J. Bacteriol. 175:2640-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orser, C. S., and C. C. Lange. 1994. Molecular analysis of pentachlorophenol degradation. Biodegradation 5:277-288. [DOI] [PubMed] [Google Scholar]
- 15.Orser, C., C. C. Lange, L. Xun, T. C. Zahrt, and B. J. Schneider. 1993. Cloning, sequence analysis, and expression of the Flavobacterium pentachlorophenol-4-monooxygenase gene in Escherichia coli. J. Bacteriol. 175:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patnaik, R., S. Louie, V. Gavrilovic, K. Perry, W. P. C. Stemmer, C. M. Ryan, and S. del Cardayré. 2002. Genome shuffling of Lactobacillus for improved acid tolerance. Nat. Biotechnol. 20:707-712. [DOI] [PubMed] [Google Scholar]
- 17.Radehaus, P. M., and S. K. Schmidt. 1992. Characterization of a novel Pseudomonas sp. that mineralizes high concentrations of pentachlorophenol. Appl. Env. Microbiol. 58:2879-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saber, D. L., and R. L. Crawford. 1985. Isolation and characterization of Flavobacterium strains that degrade pentachlorophenol. Appl. Environ. Microbiol. 50:1512-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schell, M. A. 1992. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi, M., K. Hamana, and H. Akira. 2001. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 51:1405-1417. [DOI] [PubMed] [Google Scholar]
- 21.van Berkel, W. J. H., and F. Müller. 1991. Flavin-dependent monooxygenases with special reference to p-hydroxybenzoate hydroxylase, p. 1-29. In F. Müller (ed.), Chemistry and biochemistry of flavoenzymes, vol. II. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 22.van Ommen, B., A. E. P. Adang, L. Brader, M. A. Posthumus, F. Mueller, and P. J. van Bladeren. 1986. The microsomal metabolism of hexachlorobenzene. Biochem. Pharmacol. 35:3233-3238. [DOI] [PubMed] [Google Scholar]
- 23.van Ommen, B., J. W. Voncken, F. Müller, and P. J. van Bladeren. 1988. The oxidation of tetrachloro-1,4-hydroquinone by microsomes and purified cytochrome P-450b: implications for covalent binding to protein and involvement of reactive oxygen species. Chem.-Biol. Interact. 65:247-259. [DOI] [PubMed] [Google Scholar]
- 24.Wei, W., K. Wu, Y. Qin, Z. Xie, and X. Zhu. 2001. Intergeneric protoplast fusion between Kluyveromyces and Saccharomyces cerevisiae to produce sorbitol from Jerusalem artichokes. Biotechnol. Lett. 23:799-803. [Google Scholar]
- 25.Xun, L., and C. S. Orser. 1991. Purification of a Flavobacterium pentachlorophenol-induced periplasmic protein (PcpA) and nucleotide sequence of the corresponding gene (pcpA). J. Bacteriol. 173:2920-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, Y.-X., K. Perry, V. A. Vinci, K. Powell, W. P. C. Stemmer, and S. B. del Cardayré. 2002. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644-646. [DOI] [PubMed] [Google Scholar]