Abstract
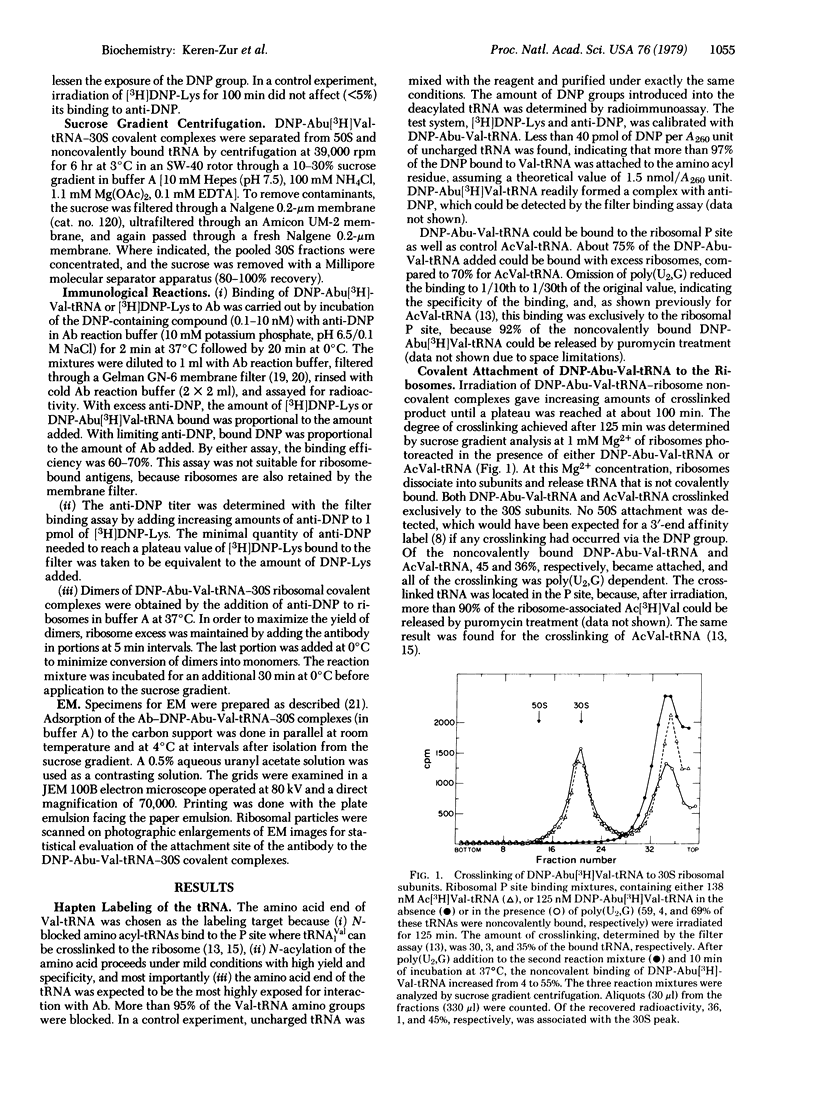
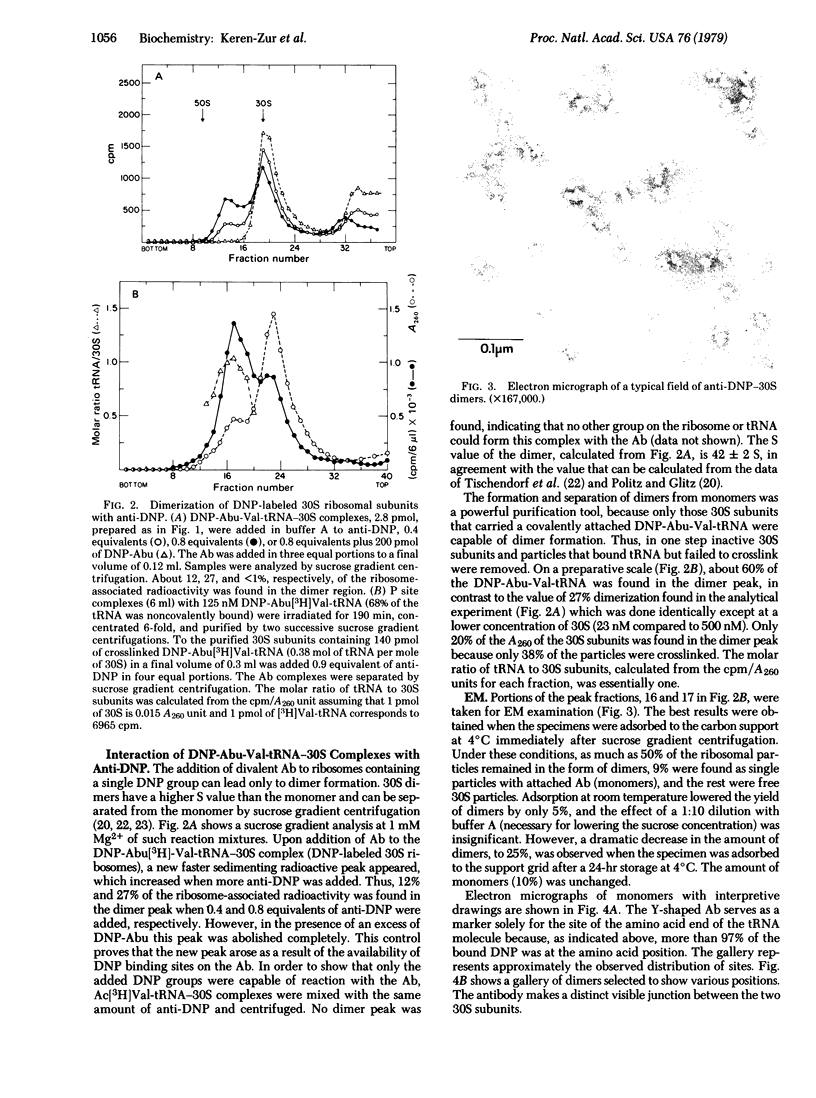
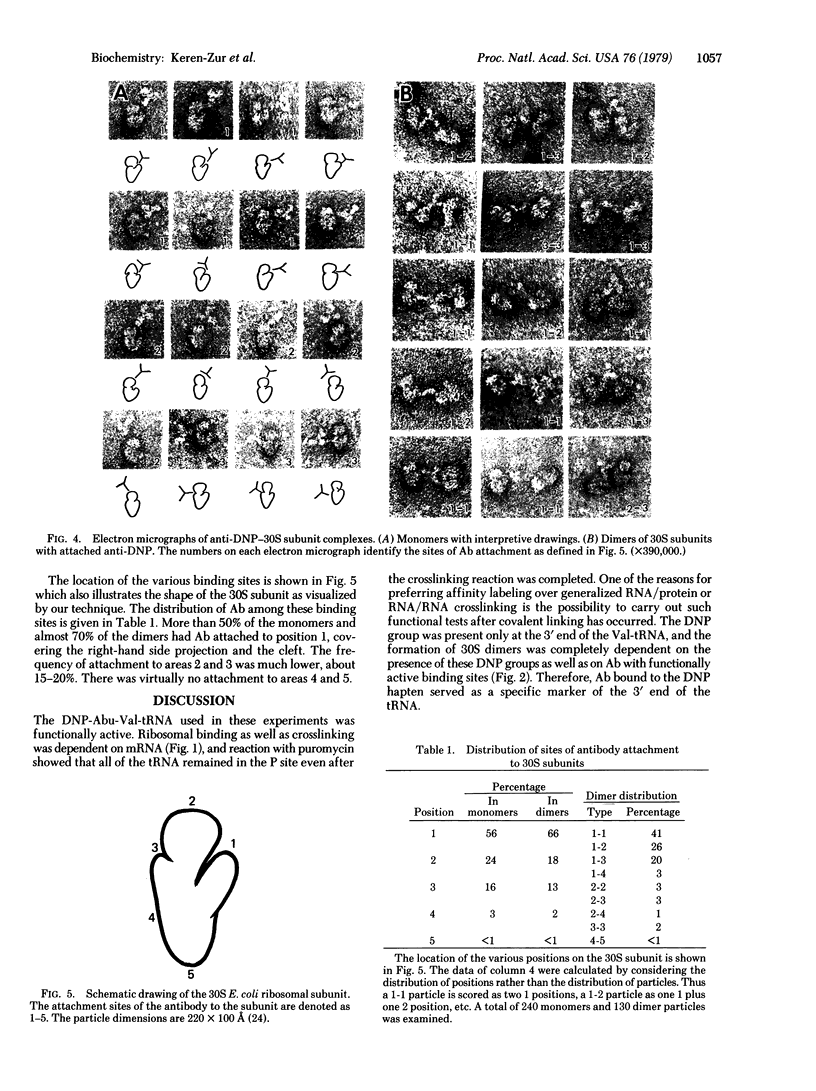
The decoding region of the Escherichia coli ribosome has been localized by affinity immunoelectron microscopy. Valyl-tRNA1Val, derivatized at the alpha-amino end with the dinitrophenyl group, was bound to the ribosomal P site of 70S ribosomes and crosslinked specifically to 16S RNA by 310- to 325-nm irradiation. Previous work has shown that crosslink occurs between the 5' anticodon base of the tRNA and a pyrimidine in the 3' one-third of the 16S RNA. By reaction of the dinitrophenyl group with antibody, dimers of the 30S tRNA covalent complexes were prepared containing one covalently attached tRNA per 30S subunit. Electron microscopic visualization of the antibody attached to the dinitrophenyl group located the position of the 3' end of the tRNA. Several sites, with a strong preference for the large protrusion or cleft region in the upper one-third of the subunit, were found. The multiplicity of sites is likely due to the freedom of orientation of the 3' end of the tRNA which is approximately 80 A from the site of crosslinking. By extrapolating this distance from each of the antigenic sites, we concluded that the anticodon of tRNA when in the P site is probably located in the cleft region of the 30S subunit.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boulik M., Hellmann W. Comparison of Artemia salina and Escherichia coli ribosome structure by electron microscopy. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2829–2833. doi: 10.1073/pnas.75.6.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe R., Nierhaus K. H., Garrett R. A., Wittmann H. G. The ribosome of Escherichia coli. Prog Nucleic Acid Res Mol Biol. 1976;18:1-44, 323-5. doi: 10.1016/s0079-6603(08)60585-1. [DOI] [PubMed] [Google Scholar]
- Fanning T. G., Cantrell M., Shih C. Y., Craven G. R. Evidence that proteins S1, S11 and S21 directly participates in the binding of transfer RNA to the 30S ribosome. Nucleic Acids Res. 1978 Mar;5(3):933–950. doi: 10.1093/nar/5.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser I., Scheit K. H., Kuechler E. Poly(4-thiouridylic acid) as messenger RNA and its application for photoaffinity labelling of the ribosomal mRNA binding site. Eur J Biochem. 1977 Apr 15;74(3):447–456. doi: 10.1111/j.1432-1033.1977.tb11411.x. [DOI] [PubMed] [Google Scholar]
- Ginzburg I., Miskin R., Zamir A. N-ethyl maleimide as a probe for the study of functional sites and conformations of 30 S ribosomal subunits. J Mol Biol. 1973 Sep 25;79(3):481–494. doi: 10.1016/0022-2836(73)90400-2. [DOI] [PubMed] [Google Scholar]
- Humayun M. Z., Jacob T. M. Immunologic studies on nucleic acids and their components. I. An analysis of the specificity of anti-deoxyadenylate antibodies by a membrane-binding technique. Biochim Biophys Acta. 1973 Nov 26;331(1):41–53. doi: 10.1016/0005-2787(73)90417-6. [DOI] [PubMed] [Google Scholar]
- Kim S. H. Three-dimensional structure of transfer RNA. Prog Nucleic Acid Res Mol Biol. 1976;17:181–216. doi: 10.1016/s0079-6603(08)60070-7. [DOI] [PubMed] [Google Scholar]
- Lake J. A. Aminoacyl-tRNA binding at the recognition site is the first step of the elongation cycle of protein synthesis. Proc Natl Acad Sci U S A. 1977 May;74(5):1903–1907. doi: 10.1073/pnas.74.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A., Pendergast M., Kahan L., Nomura M. Localization of Escherichia coli ribosomal proteins S4 and S14 by electron microscopy of antibody-labeled subunits. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4688–4692. doi: 10.1073/pnas.71.12.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritella P., Kuechler E. Specificity of the photo-crosslinking reaction between poly(U) and protein S1 on the Escherichia coli ribosome. FEBS Lett. 1978 Apr 1;88(1):131–134. doi: 10.1016/0014-5793(78)80624-3. [DOI] [PubMed] [Google Scholar]
- Noll M., Hapke B., Schreier M. H., Noll H. Structural dynamics of bacterial ribosomes. I. Characterization of vacant couples and their relation to complexed ribosomes. J Mol Biol. 1973 Apr 5;75(2):281–294. doi: 10.1016/0022-2836(73)90021-1. [DOI] [PubMed] [Google Scholar]
- Ofengand J., Schwartz I., Chinali G., Hixson S. S., Hixson S. H. Photoaffinity-probe-modified tRNA for the analysis of ribosomal binding sites. Methods Enzymol. 1977;46:683–702. doi: 10.1016/s0076-6879(77)46086-5. [DOI] [PubMed] [Google Scholar]
- Politz S. M., Glitz D. G. Ribosome structure: localization of N6,N6-dimethyladenosine by electron microscopy of a ribosome-antibody complex. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1468–1472. doi: 10.1073/pnas.74.4.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongs O., Nierhaus K. H., ERDMANN V. A., Wittmann H. G. Active sites in Escherichia coli ribosomes. FEBS Lett. 1974 Mar 23;40(0):suppl–suppl:S37. [PubMed] [Google Scholar]
- Schofield P., Zamecnik P. C. Cupric ion catalysis in hydrolysis of aminoacyl-tRNA. Biochim Biophys Acta. 1968 Feb 26;155(2):410–416. doi: 10.1016/0005-2787(68)90185-8. [DOI] [PubMed] [Google Scholar]
- Schwartz I., Gordon E., Ofengand J. Photoaffinity labeling of the ribosomal A site with S-(p-azidophenacyl)valyl-tRNA. Biochemistry. 1975 Jul;14(13):2907–2914. doi: 10.1021/bi00684a018. [DOI] [PubMed] [Google Scholar]
- Schwartz I., Ofengand J. Photo-affinity labeling of tRNA binding sites in macromolecules. I. Linking of the phenacyl-p-azide of 4-thiouridine in (Escherichia coli) valyl-tRNA to 16S RNA at the ribosomal P site. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3951–3955. doi: 10.1073/pnas.71.10.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz I., Ofengand J. Photochemical cross-linking of unmodified acetylvalyl-tRNA to 16S RNA at the ribosomal P site. Biochemistry. 1978 Jun 27;17(13):2524–2530. doi: 10.1021/bi00606a011. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Craven G. R. Chemical inactivation of Escherichia coli 30-S ribosomes by iodination. Identification of proteins involved in tRNA binding. Eur J Biochem. 1976 Jan 2;61(1):307–315. doi: 10.1111/j.1432-1033.1976.tb10023.x. [DOI] [PubMed] [Google Scholar]
- Thomas G., Sweeney R., Chang C., Noller H. F. Identification of proteins functionally altered by chemical modification of the transfer RNA and polyuridylic acid binding sites of 30 S ribosomal subunits. J Mol Biol. 1975 Jun 15;95(1):91–102. doi: 10.1016/0022-2836(75)90338-1. [DOI] [PubMed] [Google Scholar]
- Tischendorf G. W., Zeichhardt H., Stöffler G. Location of proteins S5, S13 and S14 on the surface of the 3oS ribosomal subunit from Escherichia coli as determined by immune electron microscopy. Mol Gen Genet. 1974;134(3):209–223. doi: 10.1007/BF00267716. [DOI] [PubMed] [Google Scholar]
- Towbin H., Elson D. A photoaffinity labelling study of the messenger RNA-binding region of Escherichia coli ribosomes. Nucleic Acids Res. 1978 Sep;5(9):3389–3407. doi: 10.1093/nar/5.9.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Gassen H. G. Identification of a 16S rna sequence located in the decoding site of 30S ribosomes. FEBS Lett. 1976 Sep 1;67(3):312–315. doi: 10.1016/0014-5793(76)80554-6. [DOI] [PubMed] [Google Scholar]
- Willan K. J., Marsh D., Sunderland C. A., Sutton B. J., Wain-Hobson S., Dwek R. A., Givol D. Comparison of the dimensions of the combining sites of the dinitrophenyl-binding immunoglobulin A myeloma proteins MOPC 315, MOPC 460 and XRPC 25 by spin-label mapping. Biochem J. 1977 Aug 1;165(2):199–206. doi: 10.1042/bj1650199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir A. Affinity labeling of ribosomal functional sites. Methods Enzymol. 1977;46:621–637. doi: 10.1016/s0076-6879(77)46077-4. [DOI] [PubMed] [Google Scholar]








