Abstract
A biochemical assay that is designed to detect recombination intermediates formed in vitro is described. The assay measures the fusion of two essentially homologous plasmids, one of which is radioactively labeled and the other of which carries several copies of the lac operator. The fusion product is radioactive and can be bound to a nitrocellulose filter by lac repressor. This assay for genome fusion is rapid and readily applicable to the many fractions that result during enzyme purification. The fused product is not destroyed in the assay and may be recovered from the filter for further analysis by electron microscopy. The product is then seen to consist of figure 8 structures that can be cleaved by the restriction enzyme EcoRI to give chi forms, structures similar to those recovered from recombination-proficient cells. It is expected that this assay will be useful in the purification of the "recombinase-type" activity detected in crude cell lysates. To demonstrate this point, the assay was applied to the protein fractions recovered from a molecular sieve column. The results indicate that the fusion activity has an apparent molecular weight of 50,000--100,000.
Full text
PDF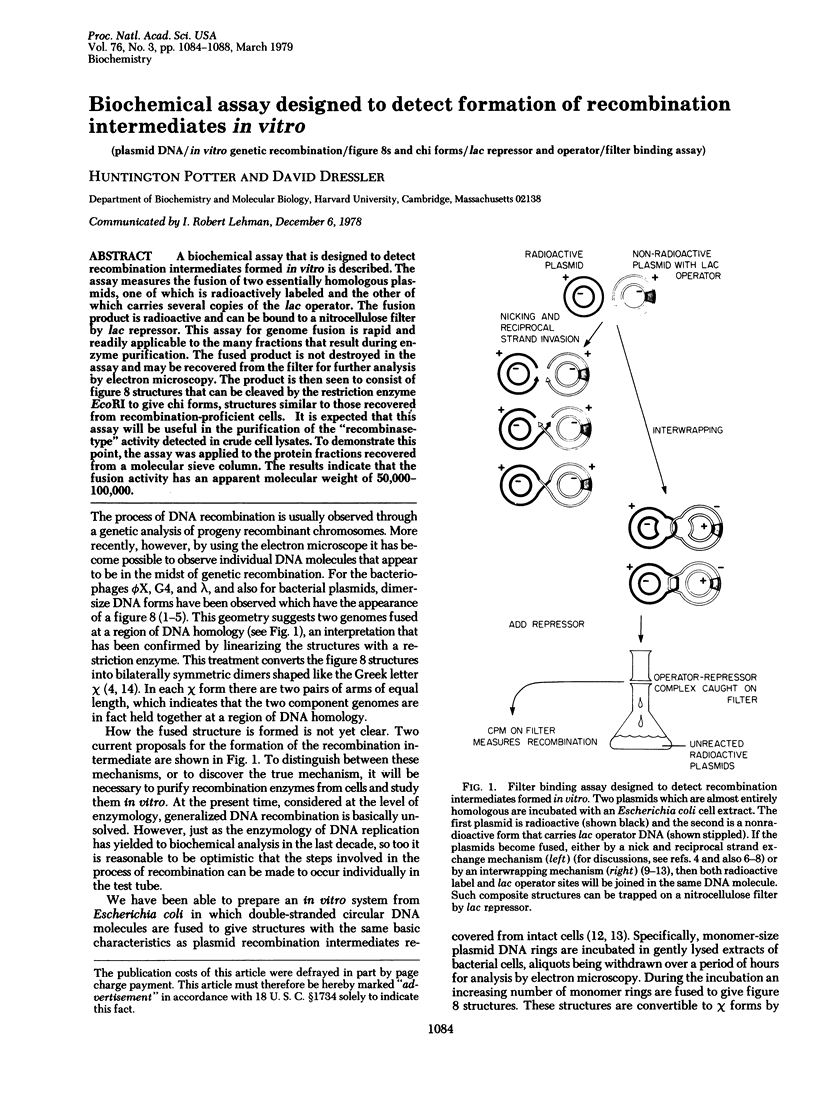

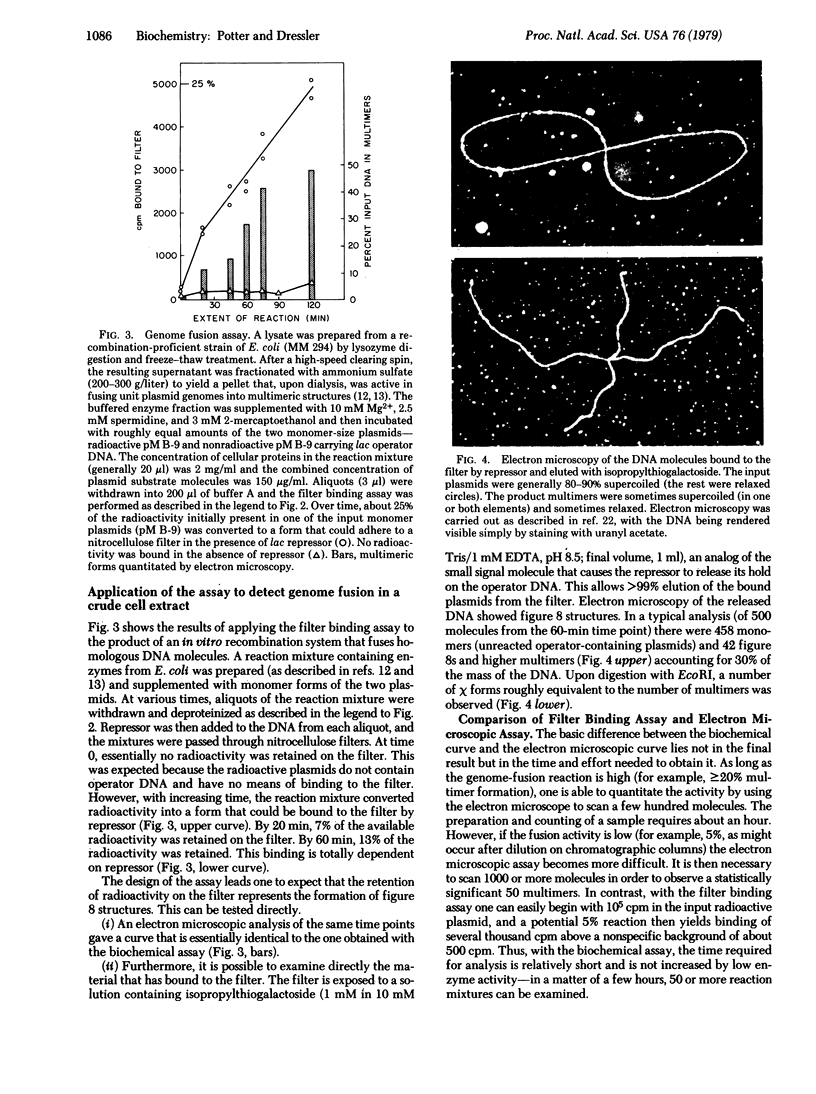
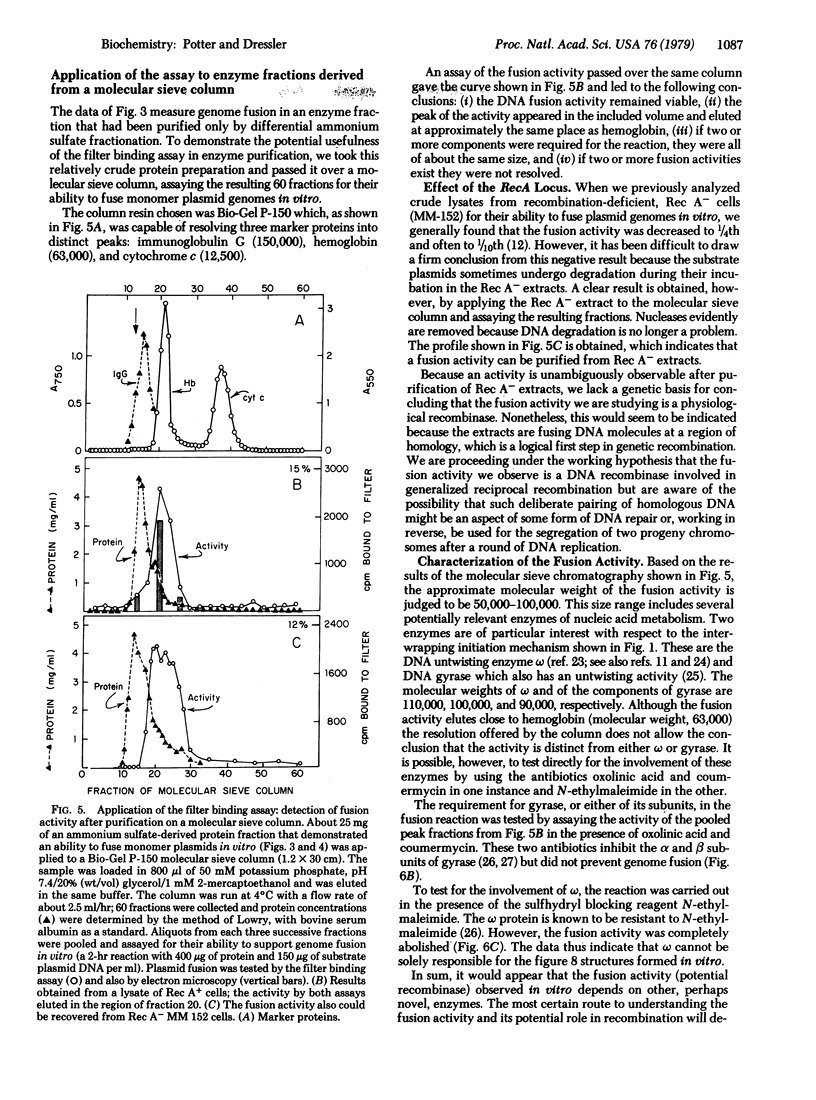

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benbow R. M., Zuccarelli A. J., Sinsheimer R. L. Recombinant DNA molecules of bacteriophage phi chi174. Proc Natl Acad Sci U S A. 1975 Jan;72(1):235–239. doi: 10.1073/pnas.72.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto E., Mursalim J., Howard-Flanders P. Homology-dependent cutting in trans of DNA in extracts of Escherichia coli: an approach to the enzymology of genetic recombination. Proc Natl Acad Sci U S A. 1978 Feb;75(2):620–624. doi: 10.1073/pnas.75.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. Renaturation of complementary single-stranded DNA circles: complete rewinding facilitated by the DNA untwisting enzyme. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5328–5332. doi: 10.1073/pnas.74.12.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R. A., Lieb M. Heat-inducible lambda phage. V. Induction of prophages with mutations in genes O, P, and R. Genetics. 1967 Nov;57(3):549–560. doi: 10.1093/genetics/57.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Müller-Hill B. Isolation of the lac repressor. Proc Natl Acad Sci U S A. 1966 Dec;56(6):1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. The lac repressor and the lac operator. Ciba Found Symp. 1972;7:245–259. doi: 10.1002/9780470719909.ch14. [DOI] [PubMed] [Google Scholar]
- Holliday R. Molecular aspects of genetic exchange and gene conversion. Genetics. 1974 Sep;78(1):273–287. doi: 10.1093/genetics/78.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones O. W., Berg P. Studies on the binding of RNA polymerase to polynucleotides. J Mol Biol. 1966 Dec 28;22(2):199–209. doi: 10.1016/0022-2836(66)90126-4. [DOI] [PubMed] [Google Scholar]
- LEDER P., NIRENBERG M. RNA CODEWORDS AND PROTEIN SYNTHESIS. II. NUCLEOTIDE SEQUENCE OF A VALINE RNA CODEWORD. Proc Natl Acad Sci U S A. 1964 Aug;52:420–427. doi: 10.1073/pnas.52.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T., Files J. G., Weber K. Lac repressor. Specific proteolytic destruction of the NH 2 -terminal region and loss of the deoxyribonucleic acid-binding activity. J Biol Chem. 1973 Jan 10;248(1):110–121. [PubMed] [Google Scholar]
- Potter H., Dressler D. In vitro system from Escherichia coli that catalyzes generalized genetic recombination. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3698–3702. doi: 10.1073/pnas.75.8.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter H., Dressler D. On the mechanism of genetic recombination: electron microscopic observation of recombination intermediates. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3000–3004. doi: 10.1073/pnas.73.9.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter H., Dressler D. On the mechanism of genetic recombination: the maturation of recombination intermediates. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4168–4172. doi: 10.1073/pnas.74.10.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. A mechanism to activate branch migration between homologous DNA molecules in genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):279–283. doi: 10.1073/pnas.72.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Camien M. N., Warner R. C. Kinetics of branch migration in double-stranded DNA. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2299–2303. doi: 10.1073/pnas.73.7.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Escarmis C., Parker B., Slater W. C., Doniger J., Tessman I., Warner R. C. Figure-8 configuration of dimers of S13 and phiX174 replicative form DNA. J Mol Biol. 1975 Feb 5;91(4):409–419. doi: 10.1016/0022-2836(75)90269-7. [DOI] [PubMed] [Google Scholar]
- Valenzuela M. S., Inman R. B. Visualization of a novel junction in bacteriophage lambda DNA. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3024–3028. doi: 10.1073/pnas.72.8.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- Wolfson J., Dressler D., Magazin M. Bacteriophage T7 DNA replication: a linear replicating intermediate (gradient centrifugation-electron microscopy-E. coli-DNA partial denaturation). Proc Natl Acad Sci U S A. 1972 Feb;69(2):499–504. doi: 10.1073/pnas.69.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M., Berg P. Recognition of tRNA by aminoacyl tRNA synthetases. J Mol Biol. 1967 Sep 28;28(3):479–490. doi: 10.1016/s0022-2836(67)80098-6. [DOI] [PubMed] [Google Scholar]