Significance
Dengue virus (DENV) infects nearly 400 million people annually, and approximately 40% of the world’s population lives at risk for infection. Without a therapeutic or vaccine available, DENV remains a major public health burden. Our studies provide a comprehensive structure–function analysis of the DENV Envelope protein, integrating information from numerous Envelope structures with our functional data into a cohesive mechanistic model. This model describes the dynamic processes by which specific residue interactions within Envelope mediate infectivity—from fusion-loop triggering to hinge movement to membrane fusion—and may also apply to related class II viral fusion proteins. Because of the importance of the specific amino acids identified, our results also identify new functional residue targets for DENV vaccines and therapeutics.
Abstract
A number of structures have been solved for the Envelope (E) protein from dengue virus and closely related flaviviruses, providing detailed pictures of the conformational states of the protein at different stages of infectivity. However, the key functional residues responsible for mediating the dynamic changes between these structures remain largely unknown. Using a comprehensive library of functional point mutations covering all 390 residues of the dengue virus E protein ectodomain, we identified residues that are critical for virus infectivity, but that do not affect E protein expression, folding, virion assembly, or budding. The locations and atomic interactions of these critical residues within different structures representing distinct fusogenic conformations help to explain how E protein (i) regulates fusion-loop exposure by shielding, tethering, and triggering its release; (ii) enables hinge movements between E domain interfaces during triggered structural transformations; and (iii) drives membrane fusion through late-stage zipper contacts with stem. These results provide structural targets for drug and vaccine development and integrate the findings from structural studies and isolated mutagenesis efforts into a cohesive model that explains how specific residues in this class II viral fusion protein enable virus infectivity.
Dengue virus (DENV) infection of target cells proceeds through a membrane-fusion step, catalyzed by a viral fusion protein. The DENV genome, like that of other flaviviruses, encodes a single polyprotein that is processed into three structural proteins—capsid (C), precursor membrane (prM), and envelope (E)—and seven nonstructural proteins that are required for viral replication. The capsid protein and the viral RNA genome form a nucleocapsid that buds at the endoplasmic reticulum (ER) in association with 180 copies each of prM and E, as well as host-derived lipids, to form the immature virion (1). This noninfectious virion passes through the cell’s secretory pathway, maturing in the Golgi where most of the prM proteins are cleaved into pr and M proteins by the host protease furin (2). The mature virions bud from the cell via exocytosis and are then available to infect new cells.
DENV infects a cell by binding of E protein to one or more receptors on the cell surface, followed by clathrin-mediated endocytosis and transport to endosomes (3). The three distinct ectodomains (DI, DII, and DIII) of E protein lie over a helical stem region tethered to the virus membrane by a transmembrane anchor. In the acidic endosome, protonation of E protein histidine residues is thought to mediate conformational changes that trigger the dissociation of the E dimer into monomers and subsequent rearrangements among DI, DII, and DIII (4, 5). These conformational changes enable formation of the fusogenic E trimer and insertion of E protein into the endosomal membrane by means of a fusion loop (6, 7). Fusion between the viral and endosomal membranes is then driven by a portion of the E stem region lifting off the viral membrane and “zippering” up the trimer by interacting with a groove formed by the DII domains of neighboring E proteins (8, 9). This zippering interaction pulls the viral and endosomal membranes together, culminating in their fusion and the release of the DENV genome into the cytoplasm.
A number of crystal and cryo-electron microscopy (cryo-EM) structures of the DENV E protein describe distinct stages of DENV maturation and infectivity. Initially, the immature virus is covered by 60 spikes, each composed of E trimers with associated prM proteins (10). As the virion passes through the cell’s secretory pathway, prM is cleaved and the acid pH results in a rearrangement of E to the immature dimer structure, in which E maintains interactions with pr and M (11). The association of E and pr is lost upon encountering the neutral pH of the extracellular environment, forming the mature virus dimer, which has been visualized by several crystal structures of E dimers (crystallized as soluble ectodomains) (12, 13) and also by cryo-EM structures of mature virions (which show the entire E protein) (14, 15). Following host cell infection, dissociation of the E dimer, triggered by low pH in the target cell endosome, leads to the formation of a fusogenic E trimer structure, crystallized as soluble ectodomains (16, 17), including a structure that shows some of the stem-trimer zippering interactions that drive membrane fusion (9). These E structures differ not only by oligomeric status, but also display changes in relative conformations and angles among the three E protein domains. Similar E protein structures at various stages of virus development and infection have also been solved for related flaviviruses, such as tick-borne encephalitis virus, yellow fever virus, West Nile virus, Japanese encephalitis virus, and St. Louis encephalitis virus (8, 18–24).
Although the many structures of DENV E protein provide static images of the protein at various stages of infectivity, the key functional residues that mediate the dynamic fusion process remain to be determined. Here we report a cohesive model for the mechanism by which DENV E protein mediates its structural changes to effect membrane fusion. Using a comprehensive library of functional point mutations covering all 390 residues of the E protein ectodomain, we identified key residues that are required for infectivity but not for E protein expression, folding, virion assembly, or budding. A first set of residues was critical for fusion-loop functionality, either directly mediating membrane fusion (e.g., residues W101 and F108) or controlling fusion-loop exposure pre- and post-triggering (e.g., H149). A second set of residues was critical for hinge functionality, mediating the conformational changes that occur between domains I and II after pH triggering (e.g., R186 and I276) and between domains I and III (e.g., S296). Finally, a third set of residues was critical for E protein–M protein “latch” functionality, constraining E protein close to the membrane in the mature virion before releasing it in the endosome to enable formation of the fusogenic trimer (e.g., M258 and H259). Our results complement the existing crystallography studies of DENV E protein to form a hypothesis-driven functional model that describes how specific residues in this class II viral fusion protein (i) mediate fusion-loop functionality before, during, and after triggering; (ii) enable hinge movements between E domain interfaces during triggered structural transformations; and (iii) drive membrane fusion through trigger interactions with M protein and late-stage zipper contacts with E stem.
Results
Identification of E protein Residues Critical for DENV Infectivity.
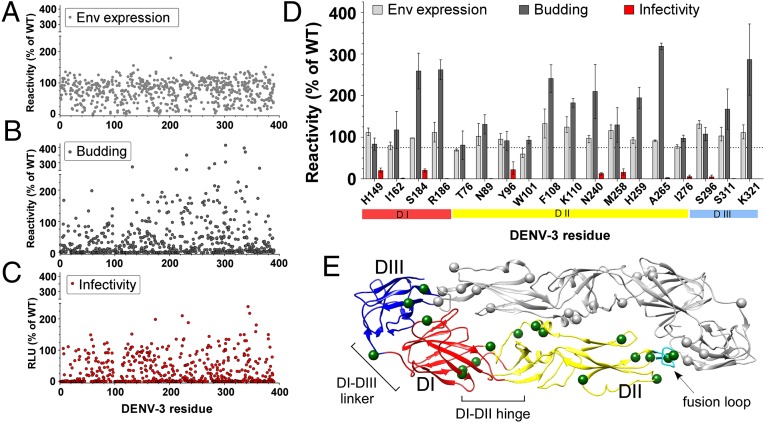
To identify E protein critical residues required for DENV infectivity, we created a comprehensive “shotgun mutagenesis” mutation library (25) of DENV-3 prM/E variants, with at least one mutation at each of the 390 residues of the E protein ectodomain. The library contained a total of 1,433 mutant clones representing multiple substitutions at each amino acid position (an average of 2.3 per residue). The entire mutation library was transfected into human HEK-293T cells in a 384-well array format (one clone per well) and evaluated in parallel for expression, budding, and infectivity. Expression was measured by immunofluorescent antibody staining and detection by flow cytometry (Fig. 1A). Viral budding was measured by ELISA capture of released virus particles (Fig. 1B). Finally, infection of target cells was measured by luciferase expression by replication-incompetent dengue reporter viruses (26) (Fig. 1C).
Fig. 1.
Identification of E protein residues critical for DENV infectivity. Each clone in the DENV-3 mutation library was tested for (A) DENV E protein expression levels by detecting cellularly expressed prM/E protein with a mixture of diverse E protein mAbs, (B) viral particle budding levels by capturing and then detecting DENV virions released by producer cells, and (C) infectivity levels by detecting luciferase expression levels in target cells infected with DENV RVPs made with each mutated prM/E protein. RVPs are antigenically identical to live virus and have been used in numerous studies of DENV function (26, 35, 36). RVPs carry a reporter gene in place of structural genes and so are capable of only a single round of infection. (D) Clones with wild-type levels (≥75%) of expression and budding, but low (≤25%) infectivity were selected for confirmation in repeat assays. These screens identified 18 residues as critical for infectivity. Shown are the values and ranges for E protein expression levels (n = 3), viral budding (n = 2), and viral infectivity (n = 2), all as a percentage of wild-type activities. (E) E protein residues critical for infectivity are mapped in green on the DENV-3 protein E-dimer structure [Protein Data Bank (PDB) ID: 1UZG]. Domain I is red, domain II yellow, domain III blue, and the fusion loop cyan. All structure numbering in this article follows that of the DENV-3 dimer crystal structure (PDB ID: 1UZG) unless stated otherwise.
The results of these assays were compared, and clones were selected that demonstrated no signs of structural impairment (i.e., wild-type levels of total expression, reactivity with multiple mAbs, and assembly and release of intact DENV virions), yet were deficient for infection (≤25% infectivity, Fig. 1D). To further confirm that these mutant proteins were not misfolded, each was tested for individual reactivity with eight diverse DENV E protein–specific mAbs that recognize seven distinct epitopes on the protein. Each mutant E protein demonstrated reactivity (>50% wild type) with all eight mAbs in nearly every case (Table S1).
The majority of the identified critical residues (11/18) mapped to DII, including three residues directly within the fusion loop. An additional four residues were located in DI and another three in DIII (Fig. 1E). Based on the selection criteria used, these residues represent critical amino acids that are individually required for E protein functionality during infection but that do not disrupt its expression, folding, viral assembly, or budding, thus localizing the functional importance of these residues to late stages of infectivity.
Network of Residues Proximal to the Fusion Loop Determines Its Exposure and Mediates pH Triggering.
The available structures of DENV E protein represent distinct conformations of the protein, including the immature trimer of prM/E heterodimers, the mature dimer, and the fusogenic trimer, but the key residues responsible for mediating the conformational changes between these structures have not previously been determined. To understand the contribution of each identified critical residue to E protein functionality, the position of that residue was evaluated within seven E protein structures. Individual interatomic contact points (within 4.2 Å) made by the critical residues in each structure were evaluated (Table S2), revealing interactions between residues that are necessary for membrane fusion and infection. The change in these interaction networks observed between the different structures (Table S2, highlighted contact distances) offers insight into how each infectivity critical residue may be involved in mediating the functionality of E protein.
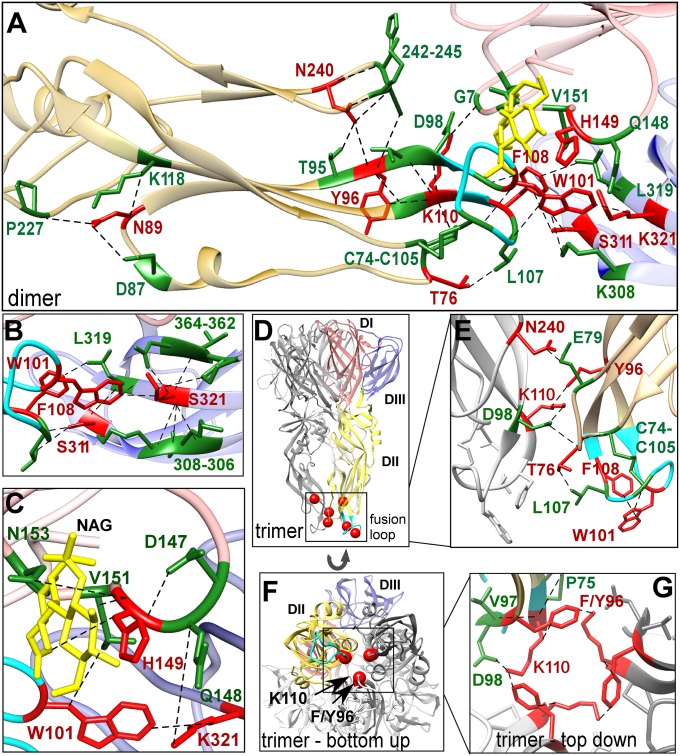
Consistent with their known role in mediating membrane fusion, fusion-loop residues W101, F108, and K110, which are 100% conserved among 33 different flaviviruses, were identified as critical for infectivity (Table S1). In the fusion loop (residues 99–110), W101 and F108 contact each other at the base of the fusion loop to form an “aromatic anchor” in the fusion trimer (16, 17) and are essential for host-virus membrane fusion (27).
In the mature E dimer, the fusion loop lies at the center of a network of intra- and inter-monomer contacts (within 4.2 Å) involving seven additional critical residues—T76, N89, Y96, H149, N240, S311, and K321—on all three domains (Fig. 2A, critical residues shown in red). In the mature structure of E, this network appears to shield and constrain the fusion loop to prevent its premature exposure (Fig. 2A, fusion loop shown in cyan). For example, critical residues S311 and K321 make intradomain and cross-monomer contacts (Fig. 2B, contact residues shown in green), including direct contacts with W101 and F108 that stabilize the position of the fusion loop relative to DIII. Similarly, critical residues T76, N89, Y96, and N240 form contacts that hold in parallel the proximal structures leading into and out of the fusion loop (Fig. 2A).
Fig. 2.
An interaction network of fusion loop-related critical residues prevents premature exposure of the loop in the mature virion and stabilizes the fusion loop in the fusion trimer. (A) Fusion loop-related critical residues T76, N89, Y96, W101, F108, K110, H149, N240, S311, and K321 (in red) and their contacts (in green) are mapped onto the DENV-3 E protein dimer (PDB ID: 1UZG) and participate in an extensive fusion loop stabilization network that prevents premature triggering. The glycan modification of N153 is shown in yellow, and the fusion loop is shown in cyan. Contacts shown in figures represent the closest interactions between two residues. (B) S311 and K321 contact the fusion loop of the opposing monomer and tether it to DIII, in cooperation with four other critical residues (T76, W101, F108, and H149) to prevent premature fusion-loop exposure. (C) H149 contacts neighboring residues 147–151, N153, and the N153-linked glycan. H149 shares contact of V151 with fusion-loop critical residue W101 of the opposing monomer and shares contact of Q148 with DIII critical residue K321. W101 and K321 also contact one another, strengthening H149’s fusion-loop tethering network. (D and E) In the fusogenic trimer (PDB ID: 1OK8), T76 and N240 establish new contacts with an opposing monomer (in gray), whereas W101 and F108 form the “aromatic anchor” of the fusion loop, which will ultimately insert in the endosome membrane. T76 establishes cross-monomer contact with D98, shared with fusion-loop critical residue K110. N240 mediates the change from E dimer to trimer by releasing contact with T95, Y96, A243, and K244 and then establishing a cross-monomer contact with E79 during fusion trimer formation. (F) Bottom-up view of the fusion trimer with Y96 and K110 in red on all three monomers (PDB ID: 1OK8). (G) Y96 and K110 intra- and intermonomer contacts stabilize the fusion trimer (top–down cross-section view).
Interestingly, in the transition from mature E dimer to fusogenic trimer, nearly all of these critical residues lose dimer contacts and then form new contacts in the fusogenic trimer structure (Fig. 2D). For example, in the fusogenic trimer, critical residues T76, Y96, and N240 all form new contacts across the monomer interfaces of the trimer (Fig. 2E), with Y96 forming intra- and intermonomer contacts with K110 to stabilize the trimeric fusion-loop structure (Fig. 2 F and G). These critical residues thus appear to tether the fusion loop in the mature dimer structure, whereas in the fusogenic trimer these same residues form new contacts, together identifying critical interatomic connections that help explain how E protein both prevents premature triggering and stabilizes the post-triggered trimer structure.
The connectivity of residues surrounding the fusion loop suggests that critical residue H149 may function as a “switch” that controls the formation and dissolution of a network of interactions around the fusion loop. In the mature E protein dimer, H149 is part of a network that tethers DI residues 142–157 (a loop containing a glycan) over the fusion loop (Fig. 2C; glycan on N153 highlighted in yellow). Protonation of H149 at low pH is predicted to disrupt these interactions and spring the glycan loop from a rigid interdomain conformation to a disordered strand, as suggested by the absence of this loop from E protein in the immature and fusogenic trimer structures (Table S2). The N153-linked glycan is largely conserved among flaviviruses and has been implicated in DENV infectivity in mammalian cells (28). The glycan loop also contains a second, absolutely conserved histidine, H144, which has been proposed to play a role in the destabilization of the DI–DIII interface during triggering (4, 5), but which did not express or bud sufficiently in our study or other studies (4) to enable conclusive analysis. H315 may also contribute to the triggered dissociation of the E dimer because it shares contacts at G7 and Q314 with critical residue K110 and has been previously proposed to have a role in pH sensing in tick borne encephalitis virus (4, 5). Thus, the critical nature of H149 suggests that its protonation, and possibly that of H144 and H315, could form an underlying switch that determines whether the fusion loop is shielded in its mature monomer form or exposed in its fusogenic trimer form.
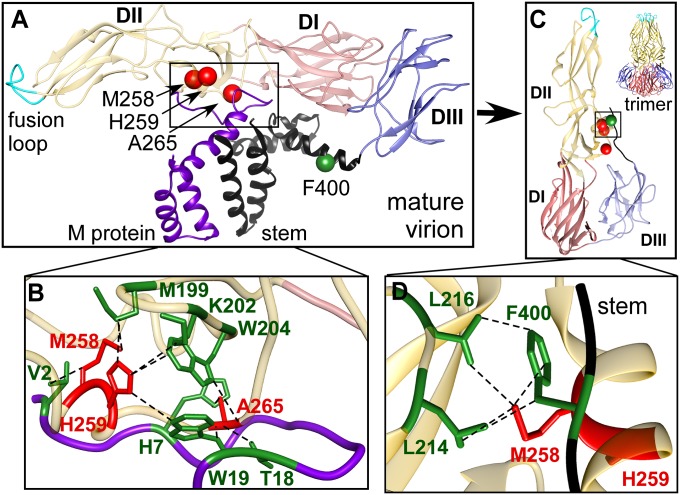
Latch Between E Protein and M Protein Controls Triggering.
Critical residues M258, H259, and A265 lie on the underside of E (Fig. 3A, red residues) and have been proposed to form an E protein–M protein “latch” with the highly conserved M protein residues V2, H7, and W19, which holds the E protein and M protein together in the mature structure (Fig. 3B) and mediates their triggered dissociation in the endosome (14, 15). Upon low pH exposure, protonation of H259, as well as the strictly conserved M protein-H7 contacted by A265 and H207 (14, 15), disrupts the M258, H259, A265 “latch” interactions with M and promotes dissociation of the E dimer, consistent with the role of K202 (contacted by H259) in influencing the pH threshold of fusion (12). As was the case with residues in the fusion-loop network, many of these latch residues lose their contacts in the dimer and then form new contacts in the activated trimer structure that are crucial in later stages of membrane fusion (Fig. 3C). Critical residue M258 in particular appears to enable part of the stem region (residues 393–401) to zipper between adjacent E monomer components of the fusion trimer by contacting the highly conserved stem residue F400 (highlighted in green, Fig. 3D) (9). Together, the identification of these residues as critical for infectivity, and the nature of their interatomic connections, help explain how mature E protein is held in its prefusion dimer conformation by M protein and then subsequently locks into the post-triggered fusogenic trimer conformation and drives membrane fusion by enabling zippering interactions with stem residues.
Fig. 3.
DII critical residues M258, H259, and A265 prevent premature E protein triggering in the mature virion. (A) M258 and H259 (poorly exposed on the mature virion) and A265 (exposed on the underside of E) interact with M protein (purple) (PDB ID: 3J27). (B) Critical residues M258, H259, and A265 (red) make multiple contacts with highly conserved M protein residues (V2, H7, and W19) that prevent premature triggering of E protein (contacts shown in green). (C and D) In the fusion trimer (PDB ID: 4GT0), M258 is partially exposed at the interface between monomers. Stem residue F400 packs against a hydrophobic pocket composed of L214, L216, and M258 (stem shown in black).
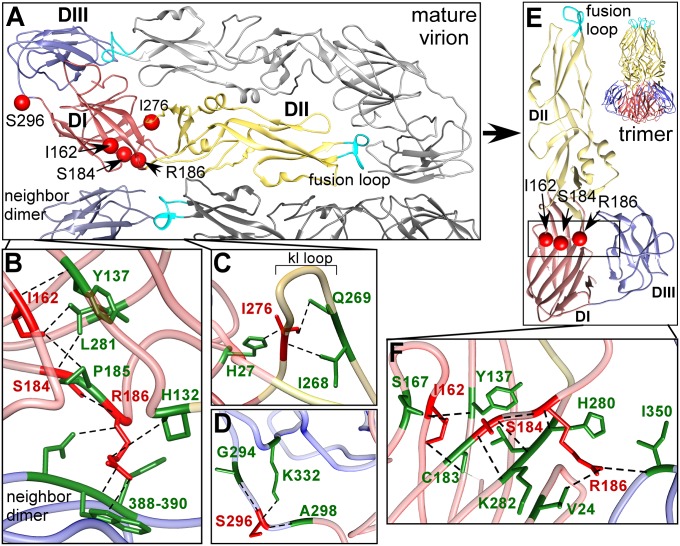
Hinge Residues at the Domain Interfaces Mediate Conformational Changes Required for Fusion Trimer Formation.
Five critical residues were identified that could facilitate the repositioning of the E domains, relative to each other, which are required for formation of the fusion trimer. Critical residues I162, S184, R186, and I276 are located on the four-strand DI–DII hinge (Fig. 4A) that mediates the post-pH–triggered reorganization of these domains. Critical residue R186 in DI appears to play a particularly important role, contacting residues H132, N388, W389, and Y390 in the neighboring dimer DIII of the mature virion (Fig. 4B), but losing these contacts upon fusogenic trimer formation (Fig. 4E) and making new contacts with residues V24, E26, S167, and T168 on DI and I350 on DIII (Fig. 4F). Critical residue I276 maintains the DI–DII hinge region “kl loop” structure through interactions with H27 (and other residues) that are lost in the fusogenic trimer structure (Fig. 4C). Consistent with the role of I276, the kl loop structure is known to be crucial for initiating the pH-mediated conformational change, and mutations within it can affect the pH threshold for fusion, resulting in diminished virus fusion and replication in mammalian cells (12, 29). At the other hinge region, between DI and DIII, critical residue S296 appears to play a similar critical role. On the mature virion, S296 contacts adjacent G294 and A298 on the DI–DIII linker and K332 on a DIII β-strand (Fig. 4D), but its interaction with K332 weakens following dimer dissociation, thus potentially enabling repositioning of DIII relative to DI. Together, the connections of these critical residues help explain the changes in orientation between domains DI and DII and between domains DI and DIII, which allow the E protein to form a functional fusion trimer.
Fig. 4.
Critical resides required for hinge movement of DI–DII and DI–DIII. (A) Critical residues (red) are mapped on the mature E protein structure (PDB ID: 3J27). (B) Critical residues I162, S184, and R186 form the DI–DII hinge. I162 and S184 make stabilizing contacts in the DI core whereas R186 establishes contact with the neighboring dimer. (C) I276 supports the kl loop of the four-strand DI–DII hinge region through cross-loop contact with I268 and Q269. (D) S296 makes contact with residues 294 and 298 on the DI–DIII linker and with conserved residue K332 on a DIII loop. (E) Location of critical residues I162, S184, and R186 on the fusion trimer structure (PDB ID: 1OK8). (F) In the E fusion trimer, R186 establishes new DIII contacts and cross-monomer DI contacts (cross-monomer contacts not shown).
Discussion
Model for DENV E protein Functionality.
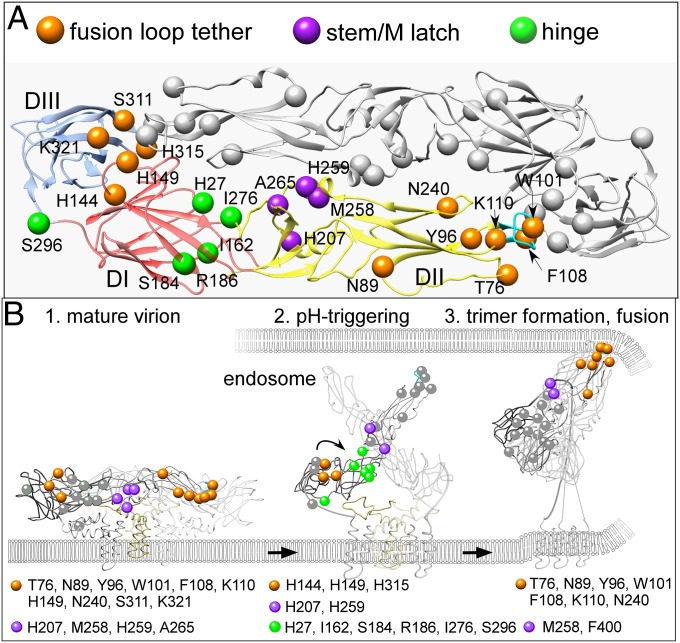
The location and interatomic contacts of each identified critical residue allow them to be grouped into distinct functional roles crucial for infection: fusion-loop tether and trigger, hinge mobility, and stem/M latch functionality (Fig. 5A). These critical residues, complemented by previous structural and mutagenesis studies, enabled us to form a hypothesis-driven functional model of how DENV E protein mediates infectivity.
Fig. 5.
Model for the functions of critical E protein residues in infectivity. (A) Residues critical for DENV-3 infectivity shown on the DENV-3 dimer, grouped into distinct roles crucial for infection: fusion loop tether (orange), stem/M latch (purple), and hinge (green). (B) A model for critical residues in mediating the structural changes that initiate membrane fusion. (1) On the mature virion, critical residues important for latch functionality maintain DII/M protein interactions, whereas fusion-loop support residues tether and position the fusion loop to retain E in dimer conformation until uptake by cells and transport to the endosome. (2) In the low pH of the endosome, histidine residues are protonated to disrupt E–M interactions and fusion-loop support residue networks to dissociate the E dimer. This is followed by changes in the relative positions of DI, DII, and DIII, facilitated by hinge critical residues. (3) Fusion-loop residues insert in the endosomal membrane as E monomers form the fusion trimer. Viral and endosomal membranes are pulled together by E stem zippering between the interstices of DI, facilitated by stem/latch critical residues.
On the mature virion (Fig. 5B, 1), seven residues (T76, N89, Y96, H149, N240, S311, and K321), distributed among all three domains, form a network of interactions with and around the fusion loop (via critical residues W101, F108, and K110) that appears to function to prevent its premature exposure. H149 is involved in a network of interactions that tether a loop, containing an N-linked glycan, over the fusion loop. Our model suggests that H149, together with H144 and H315, functions as a switch that triggers the fusion loop from a shielded conformation on the mature dimer to a fully exposed monomer available to interact with the endosomal membrane and to form a fusogenic trimer. Further behind the fusion loop, three residues in DII (M258, H259, and A265) form “latch” contacts with M protein residues that help to prevent premature triggering of E protein.
In our model, when a virus is internalized by a cell and transported to the endosome (Fig. 5B, 2), protonation of H207 and H259 on E protein and H7 on M protein disrupts E/M contacts. The concurrent protonation of H144, H149, and H315 results in complete dissociation of the E dimer. The ensuing interdomain rearrangements are facilitated by critical hinge residues essential for fusion trimer formation. I162 and S184 stabilize the β-strand core of DI and coordinate R186 contacts with a neighboring dimer, whereas I276 modulates the DI–DII angle during the dimer-to-trimer conformational change, assisted by protonation of H27. Similarly, S296 in the DI–DIII linker enables DIII to fold down against DI during trimer formation to enable stem zippering.
After dissociation of the E dimer, the resulting monomers form fusogenic trimers whose fusion loops insert in the endosome membrane to initiate membrane fusion (Fig. 5B, 3). Critical residues W101 and F108 form the fusion loop’s “aromatic anchor,” essential for membrane insertion and complete fusion, whereas Y96 makes intermonomer contacts with K110 to stabilize the three fusion loops in a trimeric configuration. DII critical residues T76, Y96, and N240, proximal to the fusion loop, may provide structural support to the trimer’s fusion-loop cluster. To promote membrane fusion, a portion of the E stem region zips up along DII in our model mediated by M258 contact with F400, pulling together the viral and endosomal membranes.
Contribution of Other Residues.
Cumulatively, our results and those of others suggest that at least 6 of E protein’s 13 histidine residues (H27, H144, H149, H207, H259, and H315) mediate the pH sensing function of DENV E protein for fusogenic triggering. It is likely that these histidines act in concert to promote triggering (30). Additional histidines may also participate in DENV pH sensing, but in our studies their mutation disrupted E protein expression, folding, and/or budding and did not make direct contact with any of the critical residues that we identified and so could not be included in our model. H242 mediates the interaction with prM residue D63 and so is thought to play a role in maturation but not in fusion (10, 31).
Mutation of several other residues of interest eliminated both budding and infectivity, including important connecting residues identified in our analysis (e.g., F400), all of the 12 Cys residues in DENV E protein, and both of the glycosylation sites of E protein (N67 and N153 and their associated glycosylation recognition sites at T69 and T155), emphasizing the importance of these structures to E protein folding and budding earlier in the viral life cycle. Other important residues likely also participate in E protein infectivity, but could not be definitively distinguished here based on the criteria used to select critical mutants that are not misfolded.
The functional model described here combines our own data with information from seven solved structures and numerous individual mutagenesis studies into a cohesive model that describes how specific E protein residues and their interactions mediate DENV infectivity. Although our final model of how each residue functions is hypothesis-driven, critical residues were identified using stringent criteria: their mutation did not significantly disrupt expression, folding, viral assembly, or budding; many of our critical residues form networks around E protein structures known to be important during infectivity; and the importance and functionality of many of these residues is confirmed by individual mutagenesis and structural studies of DENV or related flaviviruses by other investigators.
Our studies do not experimentally distinguish between different steps of late-stage E protein functionality, such as receptor binding, pH dependence, triggering, zippering, and fusion, but infer the dynamic role of specific residues from their change in interactions between defined fusogenic structures. We also cannot specifically exclude the possibility that mutations have affected capsid, cleavage or replicon genome incorporation (i.e., forming noninfectious virus particles), but the residues of E protein that we identified are not suspected of influencing these interactions and we have thus far detected no such defects. Although some of the residues identified by our analyses were previously shown to be important for infectivity, our work independently validates and extends these earlier results and incorporates them into a cohesive model to explain the residues’ interdependent functionality. Indeed, it would have been surprising and disconcerting if we had not identified known crucial residues. We expect that our model of DENV infectivity can be further refined in future studies, including using additional serotypes of DENV.
Related flaviviruses, including West Nile virus, Japanese encephalitis virus, and tick-borne encephalitis virus, share the same overall structural features as DENV E protein, suggesting that the functional model derived here for DENV may apply to other similar class II viral fusion proteins. The identification of functionally critical residues throughout DENV E protein also provides attractive candidate residues and structures for antibody and/or drug-mediated inhibition of the virus.
Materials and Methods
Construction of DENV-3 Chimera Plasmid.
To generate a DENV construct capable of high-level structural polyprotein CprM/E expression and viral production, we created a chimera construct encoding residues 1–100 of the capsid protein from DENV-2 (S16803) and codon-optimized residues 101–760 from DENV-3 (CH53489). These residues encode the ER anchor for capsid (residues 101–114) and prM/E (residues 115–760). The wild-type DENV-2 capsid gene fragment was cloned by PCR, and the codon-optimized prM/E genes from DENV-3 were cloned in-frame in pTRex-pBR322, a modified version of expression plasmid pTRex-DEST-30 (Invitrogen).
Construction of DENV-3 prM/E Mutation Library.
A shotgun mutagenesis mutation library of prM/E was created as previously described (25). Briefly, the parental plasmid expressing DENV-3 CH53489 CprM/E polyprotein was used as a template to make a library of random mutations across prM/E (residues 115–760, excluding capsid), created using PCR-based mutagenesis (Diversify PCR Random Mutagenesis Kit, Clontech). Each mutant clone was sequence-verified. A complete mutation library was assembled by selection of at least two mutant clones per residue, preferably representing a conserved and nonconserved residue at each position. A total of 917 DENV-3 prM/E variants contained single mutations, and the remaining 516 clones contained mutations at two or more positions.
Immunofluorescence Assay.
Mutation libraries and controls were expressed in human HEK-293T cells. Twenty-two hours post-transfection cells were washed and fixed in 4% (wt/vol) paraformaldehyde, permeabilized with 0.1% (wt/vol) saponin, incubated with a mixture of diverse mAbs recognizing the DENV Envelope protein (4.8A, D11C, 4.2C-13.7F, 9.4F-8.10E) (a gift from John Schieffelin, Tulane University, New Orleans) (32), followed by AlexaFluor 488-conjugated secondary antibody. Microplates were measured using the Intellicyt HTFC screening system. Antibody reactivities against each mutant prM/E protein clone were calculated relative to wild-type protein reactivity by subtracting the signal from mock-transfected controls and normalizing to the signal from wild-type prM/E-transfected controls.
Critical clones were also tested individually against eight different E protein–specific mAbs [D11C and 4.2C-13.7F, a gift from John Schieffelin (32), and 1C17, 2D7, 5J7, 1B13, 1D7, and 1M12, a gift from James Crowe (Vanderbilt University, Nashville, TN) (33, 34)], representing seven different epitopes, to confirm that the expressed mutant E proteins are not misfolded. Two of the mAbs bind to the fusion loop and so do not react with some of the mutated residues in the fusion loop (32, 33).
DENV Reporter Virus Particle Production.
The BHK-DRRZ cell line, which stably propagates a DENV replicon, was produced by transduction of BHK21 cells with DENV reporter virus particles (RVPs) that encapsidate a replicon encoding both Renilla luciferase and the ShBle gene (conferring resistance to zeocin). The DENV replicon was described previously (35), and DENV RVPs have been extensively characterized for antigenic equivalence to live virus (26, 35). Following infection, selection of infected cells was conferred by supplementing complete DMEM with 300 µg/mL zeocin. For RVP production, BHK-DRRZ cells were added to mutation array microtiter plates in DMEM complete medium with Hepes added to 25 mM, pH 8.0. Plates were incubated at 37 °C at 5% (vol/vol) CO2 to allow for transfection and RVP production initiation. RVP supernatant from each well was then transferred to a 384-well white flat-bottom microtiter plate and stored at −80 °C for at least 4 h to eliminate any viable BHK-DRRZ cells that were present before use for RVP Budding ELISA or RVP Infectivity Assays.
DENV RVP Budding ELISA.
A 384-well white flat-bottom microtiter plate was coated with mouse prM mAb 2H2 (American Type Culture Collection #HB-114) (5 µg/mL) in 0.1 M NaHCO3, pH 8.6, and incubated overnight at 4 °C. The plate was blocked with 3% (wt/vol) BSA in PBS for 2 h at room temperature. A 384-well RVP production plate containing frozen RVPs was thawed, and 6–10 µL of supernatant was transferred to the blocked ELISA plate and incubated overnight at 4 °C to allow capture of RVPs. A human monoclonal antibody mixture (2.11B-18.12D, 3.1E-61.7F, 2.9G, 4.4F; final 1 µg/mL) (a gift from John Schieffelin) in blocking buffer was allowed to incubate on the plate for 1 h at room temperature, followed by addition of rabbit anti-human HRP-conjugated secondary antibody (1:5,000) in blocking buffer for 1 h at room temperature. Reactivity was detected using SuperSignal West Femto Chemiluminescent Substrate (Pierce). Serial dilutions of both capture and detection antibodies were used to optimize detection conditions (Z′ > 0.3). All luminescence values were background-subtracted and normalized to the average luciferase signal generated from wild-type RVPs.
DENV RVP Infectivity Assay.
RVPs produced from the mutation array were used for infectivity assays, as described previously (26), except that a Renilla luciferase reporter replaced GFP. Twenty microliters of frozen RVPs were used to infect BHK DC-SIGN cells at a density of 10,000 cells per well in DMEM with Hepes added to 10 mM, pH 8.0, in a 384-well microplate format. The plate was then incubated at 37 °C at 5% CO2. After infection, the genomic replicon delivered by the RVPs replicates and expresses luciferase. Seventy-two hours postinfection, cells were washed with PBS and lysed for 20 min at room temperature (1X Lysis Buffer, Promega). Luciferase substrate was diluted 1:100 in assay buffer (Promega), and luminescence was detected on a Wallac or Envision luminometer. All luminescence values were background-subtracted and normalized to the average signal from wild-type RVP infection.
Supplementary Material
Acknowledgments
We thank Joe Rucker and Eli Berdougo for helpful analysis and discussion in this project; Trevor Barnes, David Tucker, Chida Sulli, Rachel Fong, Jason Goodman, Yana Thaker, and Morganne Phillips for help with cloning, mutagenesis, and assay preparation; and Drs. John Schieffelin and James Crowe for providing valuable antibodies. This work was supported by National Institutes of Health Grant AI062100 and National Institute of Allergy and Infectious Diseases Contract HHSN272200900055C.
Footnotes
Conflict of interest statement: E.A.C., K.M.K., K.M., J.M.P., C.P., E.D., and B.J.D. are all current employees of Integral Molecular. B.J.D. is a shareholder of Integral Molecular.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310962110/-/DCSupplemental.
References
- 1.Welsch S, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5(4):365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71(11):8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Schaar HM, et al. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 2008;4(12):e1000244. doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fritz R, Stiasny K, Heinz FX. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J Cell Biol. 2008;183(2):353–361. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez-San Martín C, Liu CY, Kielian M. Dealing with low pH: Entry and exit of alphaviruses and flaviviruses. Trends Microbiol. 2009;17(11):514–521. doi: 10.1016/j.tim.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaufmann B, et al. Capturing a flavivirus pre-fusion intermediate. PLoS Pathog. 2009;5(11):e1000672. doi: 10.1371/journal.ppat.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stiasny K, Brandler S, Kössl C, Heinz FX. Probing the flavivirus membrane fusion mechanism by using monoclonal antibodies. J Virol. 2007;81(20):11526–11531. doi: 10.1128/JVI.01041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bressanelli S, et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23(4):728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein DE, Choi JL, Harrison SC. Structure of a dengue virus envelope protein late-stage fusion intermediate. J Virol. 2013;87(4):2287–2293. doi: 10.1128/JVI.02957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, et al. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science. 2008;319(5871):1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 11.Yu IM, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319(5871):1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 12.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA. 2003;100(12):6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005;79(2):1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat Struct Mol Biol. 2013;20(1):105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostyuchenko VA, Zhang Q, Tan JL, Ng TS, Lok SM. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J Virol. 2013;87(13):7700–7707. doi: 10.1128/JVI.00197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 17.Nayak V, et al. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J Virol. 2009;83(9):4338–4344. doi: 10.1128/JVI.02574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luca VC, AbiMansour J, Nelson CA, Fremont DH. Crystal structure of the Japanese encephalitis virus envelope protein. J Virol. 2012;86(4):2337–2346. doi: 10.1128/JVI.06072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luca VC, Nelson CA, Fremont DH. Structure of the St. Louis encephalitis virus postfusion envelope trimer. J Virol. 2013;87(2):818–828. doi: 10.1128/JVI.01950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH. Crystal structure of the West Nile virus envelope glycoprotein. J Virol. 2006;80(23):11467–11474. doi: 10.1128/JVI.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375(6529):291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG. Structure of immature West Nile virus. J Virol. 2007;81(11):6141–6145. doi: 10.1128/JVI.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12(9):1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Structures of immature flavivirus particles. EMBO J. 2003;22(11):2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paes C, et al. Atomic-level mapping of antibody epitopes on a GPCR. J Am Chem Soc. 2009;131(20):6952–6954. doi: 10.1021/ja900186n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattia K, et al. Dengue reporter virus particles for measuring neutralizing antibodies against each of the four dengue serotypes. PLoS ONE. 2011;6(11):e27252. doi: 10.1371/journal.pone.0027252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang CY, et al. The dengue virus type 2 envelope protein fusion peptide is essential for membrane fusion. Virology. 2010;396(2):305–315. doi: 10.1016/j.virol.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 28.Mondotte JA, Lozach PY, Amara A, Gamarnik AV. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J Virol. 2007;81(13):7136–7148. doi: 10.1128/JVI.00116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butrapet S, et al. Amino acid changes within the E protein hinge region that affect dengue virus type 2 infectivity and fusion. Virology. 2011;413(1):118–127. doi: 10.1016/j.virol.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 30.Nelson S, Poddar S, Lin TY, Pierson TC. Protonation of individual histidine residues is not required for the pH-dependent entry of West Nile virus: Evaluation of the “histidine switch” hypothesis. J Virol. 2009;83(23):12631–12635. doi: 10.1128/JVI.01072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng A, Umashankar M, Kielian M. In vitro and in vivo studies identify important features of dengue virus pr-E protein interactions. PLoS Pathog. 2010;6(10):e1001157. doi: 10.1371/journal.ppat.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costin JM, et al. Mechanistic study of broadly neutralizing human monoclonal antibodies against dengue virus that target the fusion loop. J Virol. 2013;87(1):52–66. doi: 10.1128/JVI.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith SA, et al. Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol. 2012;86(5):2665–2675. doi: 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Alwis R, et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci USA. 2012;109(19):7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansarah-Sobrinho C, Nelson S, Jost CA, Whitehead SS, Pierson TC. Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology. 2008;381(1):67–74. doi: 10.1016/j.virol.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Austin SK, et al. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog. 2012;8(10):e1002930. doi: 10.1371/journal.ppat.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.