Significance
Mosquitoes use their acute sense of smell to locate hosts, oviposition sites, and repellents. Here, we investigated by next generation sequencing the key molecular components of the olfactory system of the southern house mosquito—a vector of West Nile virus. We studied differential expression of genes in antennae—the main olfactory organ—and nonolfactory tissues. Additionally, we prospected for unknown genes with transcripts enriched in antennae. Our approach, which was validated by quantitative real-time polymerase chain reaction, cloning, and deorphanization, led to the identification of a large repertoire of putative olfactory genes. This study paved the way for a better understanding of the sense of smell of this mosquito species and led to a naturally occurring mosquito repellent.
Keywords: RNA-Seq, citronellal polymorphism isoforms
Abstract
The southern house mosquito, Culex quinquefasciatus, has one of the most acute and eclectic olfactory systems of all mosquito species hitherto studied. Here, we used Illumina sequencing to identify olfactory genes expressed predominantly in antenna, mosquito’s main olfactory organ. Less than 50% of the trimmed reads generated by high-quality libraries aligned to a transcript, but approximately 70% of them aligned to the genome. Differential expression analysis, which was validated by quantitative real-time PCR on a subset of genes, showed that approximately half of the 48 odorant-binding protein genes were enriched in antennae, with the other half being predominantly expressed in legs. Similar patterns were observed with chemosensory proteins, “plus-C” odorant-binding proteins, and sensory neuron membrane proteins. Transcripts for as many as 43 ionotropic receptors were enriched in female antennae, thus making the ionotropic receptor family the largest of antennae-rich olfactory genes, second only to odorant receptor (OR) genes. As many as 177 OR genes have been identified, including 36 unique transcripts. The unique OR genes differed from previously annotated ORs in internal sequences, splice variants, and extended N or C terminus. One of the previously unknown transcripts was validated by cloning and functional expression. When challenged with a large panel of physiologically relevant compounds, CquiOR95b responded in a dose-dependent manner to ethyl 2-phenylacteate, which was demonstrated to repel Culex mosquitoes, and secondarily to citronellal, a known insect repellent. This transcriptome study led to identification of key molecular components and a repellent for the southern house mosquito.
Insects cause direct and indirect harm to public health. They may be vegetarian and harmful to our food supply as well as vectors of pathogens that inflict tremendous suffering and human losses. Culex mosquitoes are vectors for human diseases, including filariasis and various types of encephalitis, throughout the world (1). In the United States, mosquitoes within the Culex pipiens complex are major vectors of West Nile Virus (2). The southern house mosquito, Culex pipiens quinquefasciatus (Cx. quinquefasciatus), is a significant bridge vector in urbanized centers in the Western United States, particularly southern California, due to its opportunistic feeding on avian and mammalian hosts, including humans (2, 3). In general, female mosquitoes at some physiological stages in their adult life need to find (i) hosts (for a blood meal), (ii) suitable sites for oviposition, or (iii) plants to acquire energy for flights. Thus, for their fitness in the environment, they need an acute and eclectic olfactory system. Olfaction is orchestrated at various levels starting with reception of semiochemicals at the peripheral sensory system (mainly antennae), processing of signals at the antennal lobes, integration of olfactory and other sensory modalities in the higher processing centers of the brain and, ultimately, translation of olfactory signals into behavior. Thus, the cornerstone of a sophisticated olfactory system is the ability of the insect’s peripheral system to selectively detect odorants (4) that guide their navigations toward suitable vertebrate or plant hosts and oviposition sites. The advent of insect genome sequences triggered an exponential growth in our knowledge of the molecular basis of insect olfaction. We now know that the major peripheral olfactory proteins involved in the reception of odorants in insects are the odorant-binding proteins (OBPs), chemosensory proteins (CSPs), odorant-degrading enzymes (ODEs), odorant receptors (ORs), ionotropic receptors (IRs), and sensory neuron membrane proteins (SNMPs) (4). The southern house mosquito may possess one of the most, if not the most, acute olfactory system in mosquitoes (3) and has the largest repertoire of putative odorant receptors (ORs) of all dipteran species whose genomes have been hitherto sequenced (5–7). Given the diversity of olfactory genes involved in reception of semiochemicals, our understanding of the molecular basis of olfaction heavily depends on genomic data. Unfortunately, Cx. quinquefasciatus and related Culex pipiens complex mosquitoes have a very high degree of polymorphism and the highest density of single nucleotide polymorphism (SNP) of all mosquito species studied thus far (8). Thus, genetic differences between the Johannesburg strain (the source of genome sequences) and our California strain has retarded progress in our attempts to isolate, clone, and deorphanize ORs on the basis of genomic data (7, 9), particularly given the number of potential pseudogenes and genes unlikely to be expressed in female antennae. Here, we used next generation sequencing (Illumina) on olfactory tissues (antennae) and nonolfactory tissues (legs) of adult female mosquitoes to identify and determine differential expression of key molecular components making the acute olfactory system of the southern house mosquito. We identified a large number of putative ORs, including one that led to the identification of ethyl 2-phenylacetate as a mosquito repellent.
Results and Discussion
RNA Sequencing and Gene Mapping.
Our preparations led to high-quality RNA samples and libraries. In electrophoresis analysis, the fragments from the antenna and leg libraries showed bell-shaped distributions, with peaks of 336 and 341 bp, respectively. Ideally, the fragments are in the 300- to 350-bp range. To avoid possible lane-to-lane variation, RNA sequencing (RNA-Seq) libraries constructed with mRNA derived from female antennae and legs were barcoded (antennae, GAGTGG; legs, ACTGAT) and run on the same lane of an Illumina HiSeq sequencer (University of California Davis Genome Center). First, the paired-end reads (2 × 100 bp) were processed by three software packages, Quick Read Quality Control (10), Syche (11), and Sickle (12), to assess overall quality by sample and to trim 3′-end adapter contamination and low-quality sequences. Our sequencing led to more than 200 million clean reads from female antennae (Table S1), possibly the highest number of reads thus far obtained from an insect olfactory tissue. Next, the short RNA-Seq reads were aligned to the genome of Cx. quinquefasciatus Johannesburg strain (VectorBase; www.vectorbase.org/organisms/culex-quinquefasciatus) by using Burrows–Wheeler alignment tool (13). Surprisingly, less than 50% of the reads from each sample aligned to a transcript (Table S1), thus indicating that the genome annotation is far from been completed. Simultaneously, the reads were aligned to the genomic supercontigs by using TopHat/Cufflinks software package (14). The quality of the mRNA samples that generated the libraries is inferred by the low percentage (<0.02%) of transcripts aligned to mitochondrial DNA (Table S1). However, more than 130 million reads (approximately 70%) were aligned to the genome (Table S1). These findings strongly suggest that a significant number of transcripts are yet to be annotated. Thus, our transcriptome data were used not only to measure antennae/leg differential gene expression, but also to identify hitherto unknown transcripts, particularly OR genes, including putative isoforms. The tool cuffdiff (from the TopHat/Cufflinks software package; ref. 14) was used to generate a table of antennae/legs differential gene expression, whereas cuffmerge and cuffcompare tools were used to find unique transcripts and splice variants (15). This approach increased the number of transcripts from 23,049 (16) to 42,720. It is always prudent to further interrogate putative transcripts (see example below for unique putative OR genes) given that noncoding and aberrant transcripts could be caveats of any bioinformatics algorithms/tools.
Overall Comparison of Expression Profiles in Olfactory and Nonolfactory Tissues.
We examined the differential gene expression data obtained by Cuffdiff to identify olfactory genes enriched in female antennae. Typically, olfactory genes are enriched in antennae compared with nonolfactory tissues, but a large number of them have no basal expression in nonolfactory tissues (e.g., legs). Therefore, ranking olfactory genes on the basis of an antennae-to-leg ratio (commonly calculated with log2FoldChange) may be misleading because no or very low transcript levels in legs leads to infinite or very high rate even if the levels of transcripts in antennae are very low. We then reanalyzed the data by using DESeq package (17), which moderate the fold-change estimates with variance stabilizing transformation. It is worth mentioning that DESeq normalizes the data, thus the transcript counts are given in mean normalized counts (17), whereas transcripts analyzed by Cufflinks are given in fragments per kilobase of transcript per million mapped fragments (FPKM) (14). With DESeq, we were able to rank differential expression in a more biologically meaningful way.
Examination of the top 100 transcripts, including genes yet to be annotated, showed that more ORs (30 genes) than OBPs (20 genes) were enriched in antennae (Fig. S1), although CquiOBP7 (moderated logtwofold changes, mod_lfc, 9.89) was on the top of the list and the most enriched OR, CquiOR125 (mod_lfc, 8.78), was the 25th in the list. Interestingly, more than one-fourth of the transcripts are annotated as “conserved hypothetical proteins” (14%) or other putative genes (others, 15%) unlikely related to olfaction: e.g., juvenile hormone-inducible protein, 3-oxoacyl-[acyl-carrier-protein] reductase, and δ-9-desaturase. Two main surprises were the numbers of P450s (6 genes) and IRs (10 genes), with CYP9J33 (mod_lfc, 9.5) and IR41m (mod_lfc, 8.18) being the most enriched genes in each of the two groups. Next, we examined closely the differential expression of OBPs, CSPs, “plus-C” OBPs, SNMPs, IRs, and ORs.
Odorant-Binding Proteins Highly Expressed in Antennae and Legs.
Analysis of differential expression of olfactory proteins became more demanding given the changes in nomenclature and identification numbers in VectorBase (latest release, June 2013), which took place after we have already mapped all RNA-Seq reads (see above). However, this problem was not encountered with OBPs because VectorBase essentially kept our suggested nomenclature (18), with only two new ID numbers added (Dataset S1). Two putative salivary OBPs (19), which were previously named CquiOBP45 (AAR18456) and CquiOBP50 (AAR18408) (18), have been purged from OBP sequences as were putative CquiOBP47, 48, 49, and 50. Transcripts for all currently annotated OBPs (48 genes) were found, except for CquiOBP41 (CPIJ007935). CquiOR22 and CquiOR15 were detected at low, but nearly equal, levels in antennae and legs, thus their modified log folds were nearly zero (Dataset S1). Interestingly, expression profile of Cx. quinquefasciatus OBPs was bimodal, with the number of transcripts preferentially expressed in antennae almost equal to the number of transcript enriched in legs (Fig. S2). Nineteen OBPs (7, 3, 5, 2, 11, 1, 53, 8, 14, 4, 12, 6, 13, 9, 17, 18, 29, 52, and 51) had mod_lfc above the commonly accepted threshold of 2 (a standard borrowed from microarray analysis), whereas 5 OBPs (10, 43, 16, 46, 23) were more expressed in antennae than legs, but with mod-lfc lower than 2 (Dataset S1). By contrast 21 OBPs were more expressed in legs than in antennae (Fig. S2). Exclusive expression in olfactory tissues is not essential for olfactory function. The odorant receptor coreceptor (Orco) (20), for example, is undoubtedly functional and sine qua non for OR activity (21). CquiOrco transcripts were found in legs (see below), but Orco transcript levels were 113× higher in antennae (mod_lfc of 6.8). Thus, basal expression of a gene in nonolfactory tissue does not negate olfactory function. However, it is unlikely that proteins predominantly expressed in nonolfactory tissues (as suggested by their transcript levels in legs) play a role in odorant reception in antennae. These proteins highly expressed in nonolfactory tissues may be carriers of ligands other than odorants (22). Because proteins are tentatively classified as OBPs primarily on the basis of their well-conserved cysteine patterns (22–24), the OBP family includes olfactory and nonolfactory proteins. Our transcriptome analysis showed that half of the OBP repertoire is predominantly expressed in legs (Fig. 1) and, thus, unlikely to be involved specifically in olfaction. We suggest that the above-described OBPs (24 genes) predominantly expressed in antennae, particularly 19 of them with mod_lfc > 2, are involved in odorant reception in the female antennae of Cx. quinquefasciatus.
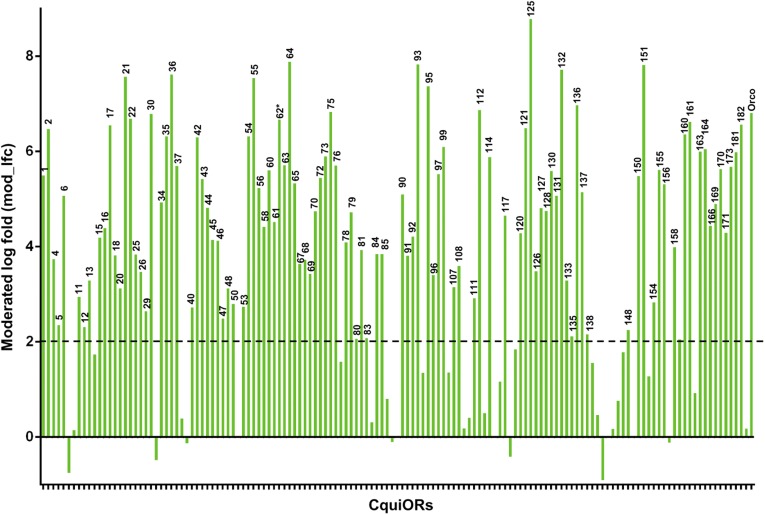
Fig. 1.
Differential expression of currently annotated odorant receptors (141 CquiOR genes). One hundred and seven CquiOR genes were significantly expressed in antennae, whereas 8 genes were enriched in legs, and transcripts for 3 genes were not detected.
One way to validate transcriptome data is to perform quantitative real-time PCR (qPCR) and compare differential expression by these two independent methods. All OBP transcripts that we have previously demonstrated to be enriched in Cx. quinquefasciatus female antennae (figure 6A in ref. 18) were confirmed by RNA-Seq to be enriched in antennae. Additionally, RNA-Seq data for one OBP gene we previously found to be more expressed in legs than antennae, CquiOBP19 (18), mirrored the qPCR data. Given the practical limitation of number of transcripts that can be analyzed by qPCR, as opposed to RNA-Seq, we previously missed a few OBPs enriched in antennae (mod_lfc > 2): e.g., CquiOBP53 (CPIJ010789), CquiOBP17 (CPIJ012716), CquiOBP18 (CPIJ012717), CquiOBP52 (CPIJ010788), and CquiOBP51 (CPIJ010787). Thus, these comparisons of qPCR and RNA-Seq performed in the same laboratory with the same strain of Cx. quinquefasciatus not only validate the transcriptome analysis, but also reinforces the power of next generation sequencing in quantifying the entire repertoire of genes, an “across-the-border-,” “large-scale qPCR.”
Expression Profiles of CSPs and Other OPs.
Previously, we have identified a family of 27 CSP genes in the genome of Cx. quinquefasciatus (18). We have now identified transcripts for all these genes, except for CquiCSP3 (Dataset S2). Five CSP genes previously shown to be ubiquitously expressed, namely CquiCSP2, 5, 22, 23, and 24, are enriched in legs (Fig. S3), thus fortifying our conclusion that they might not be involved specifically in olfaction. However, five CSPs emerged as putative olfactory genes. Specifically, CquiCSP4, 12, 13, 14, and 17 are enriched in antennae (mod_lfc > 2), whereas CquiCSP15, 16, and 18 are more abundant in antennae, but their mod_lfc are moderate. Interestingly, the CSP genes enriched in antennae are Culex-specific genes and members of a Culicinae expansion group (18), expect for CquiCSP4. The latter is a member of the CSP3 group with orthologs in Anopheles gambiae and Aedes aegypti (18). Demonstration of their role in Culex olfaction is an interesting topic for future research.
Transcripts for all previously identified plus-C OBPs (18) have been detected by RNA-Seq. As previously shown by qPCR, two of them, CquiOBP+C1 and CquiOBP+C2, were highly enriched in antennae (Fig. S3). Two other genes, CquiOBP+C3 and CquiOBP+C4, showed significantly higher expression in antennae (mod_lfc > 2), whereas two more genes, CquiOBP+C5 and CquiOBP+C6, were moderately (mod_lfc < 2) more abundant in antennae (Fig. S3). The plus-C OBPs more expressed in antennae than in legs belong to phylogenetic group A (18), which share significant amino acid identity across different species.
There were no surprises regarding SNMPs. Earlier RT-PCR data (18) demonstrating that CquiSNMP2 was expressed in antennae and legs (as well as maxillary palps and proboscis) were substantiated by differential expression (Fig. S3), showing that SNMP2 transcripts were enriched in legs. By contrast, transcripts of SNMP1 paralogs (SNMP1a, SNMP1b, and SNMP1c) (18, 25) were all highly enriched in antennae, although the precise role(s) of SNMPs in mosquito olfaction is (are) yet to be elucidated.
Most IR Transcripts Are Enriched in Antennae.
We have identified as many as 59 IR transcripts, with 43 of them being more expressed in antennae than in legs, including 34 with statistically significant (mod_lfc > 2) differential expression. As previously pointed out, 10 of these IR transcripts are among the top 100 transcripts enriched in female antennae (see above). Thus, IR genes form the second largest family of putative olfactory proteins in Cx. quinquefasciatus, second only to OR genes family (see below). Strictly speaking, these transcripts are for ionotropic glutamate receptors (iGluRs), with IRs being a related family found initially in Drosophila melanogaster (26) and later in various insect species, including Cx. quinquefasciatus (27). They belong to five phylogenetic groups, namely, NMDA iGluRs, non-NMDA iGluRs, IR25a/IR8a, divergent IRs, and antennal IRs. The Culex “antennal IRs” classification (27) was based entirely on orthology, i.e., it has been demonstrated that the Drosophila orthologs are antennae specific (28), but hitherto there was no experimental evidence to demonstrate that the Culex orthologs are transcribed specifically or predominantly in antennae. Our transcriptome data showed that the largest majority of the antennal IRs is enriched in antennae (Fig. S4). Of notice, transcript levels for all 15 members of the Culex IR75 subfamily were significantly higher in antennae than in legs, with only one of them, CquiIR75i.1, having mod_lfc smaller than 2. By contrast, CquiIR75g.1 is one of the most abundant transcripts in antennae (mod_lfc, 8.03). Three antennal IR genes (CquiIR92f, 40a, and 68a) are enriched in antennae, but below the statistical threshold (mod_lfc = 2), whereas CquiIR41p is the only exception as its transcripts were predominantly detected in legs than in antennae (mod_lfc, −2.23) (Dataset S3). Recently, an antennal IR, DmelIR76b, was unambiguously demonstrated to be a salt detector in Drosophila (29). Their orthologs in Culex, CquiIR76b.1 and CquiIR76b.2, were significantly enriched (mod_lfc > 2) in female antennae.
IR8a and IR25a are well-conserved receptors in insects (27). The Culex ortholog of DmelIR8a, CquiIR8a, was readily identified in our libraries, but despite several attempts, we did not find transcripts for CquiIR25a. Additionally, we blasted CquiIR25a amino acid and nucleotide sequences (27) against Culex genome in VectorBase and best hits were low (<40%), for small fragments of the proposed DNA sequence (27), and these hits led to other CquiIRs, particularly CquiIR8a. Attempts to retrieve CquiIR25a from our libraries via Blast2Go were also unsuccessful. Additionally, we blasted locally the proposed DNA sequence for CquiIR25a (27) against a local database created with BioEdit and derived from 42,720 RNA-Seq transcript sequences (see above) from antennae and/or legs, but found no transcripts for CquiIR25a (27). We therefore concluded that CquiIR25a might not be expressed in Culex female antennae and legs. By contrast, the other member of the family, CquiIR8a, was highly enriched in antennae (mod_lfc, 7.3). Heterologous expression in Xenopus laevis oocytes clearly demonstrated that only when coexpressed with DmelIR8a, the receptor DmelIR75a responded to propionic acid, whereas DmelIR84a⋅DmelIR8a was activated by phenylacetaldehyde (28). Mutations in DmelIR8a as well as DmelIR25a eliminated responses in IR-expressing OR neurons in D. melanogaster (28), thus both DmelIR8a and DmelIR25a are considered to be coreceptors, although evidence in heterologous system is still lacking for the latter.
The Largest Majority of OR Repertoire Was Significantly Expressed in Female Antennae.
RNA-Seq data further supported our previous suggestion (7) to purge the following 29 sequences from the annotated ORs in Cx. quinquefasciatus genome (5): CquiOR3, 9, 19, 31, 33, 41, 49, 59, 66, 74, 94, 101, 102, 103, 104, 124, 129, 134, 147, 152, 159, 167, 168, 172, 174, 176, 177, 178, and 180. These sequences are short (e.g., OR3, 945 nucleotides) and very short transcripts (e.g., OR9, 288 nucleotides). Consequently, their encoded proteins were predicted by OCTOPUS (11) to have fewer than 7 transmembrane segments (e.g., OR59, 4 TMP; OR152, 3 TMP; OR19, 2 TMP; OR174, 1 TMP), or lack a transmembrane domain (e.g., OR31). Transcripts of these genes were enriched in antennae compared with legs (e.g., OR3, mod_fold, 5.37; OR101, 3.54; OR152, 2.8; OR134, 2.57), others showed high levels of transcripts in legs and antennae (e.g., OR102, OR103, OR159, OR167, and OR174), and some had low levels (e.g., OR9) or no significant transcript levels. Transcripts for CquiOR41 were either short (encoded protein with 329-aa residues and 3 TMP) or too long (520 aa, 6 TMP with a predicted atypical C terminus of 195-aa residues). By contrast, our transcriptome data prompted us to reconsider some of genes recently omitted from Culex OR repertoire (7). Thus, CquiOR8, 15, and 35 should remain as putative OR genes. Their genes encode proteins with 483-, 379-, and 385-aa residues, respectively, with predicted seven transmembrane domains, and their transcripts are identical to those annotated in VectorBase. CquiOR35 and CquiOR15 are highly enriched in female antennae (mold_fold 6.3 and 4.2, respectively) (Dataset S4), whereas CquiOR8 showed low transcript levels in antennae (0.014 FPKM), which could be due to basal expression of a gene enriched in another olfactory tissue (e.g., maxillary palps) or in larval stage. Other genes retained for further phylogenetic analysis are as follows: CquiOR76, 105, 133, 135, 138, 139, 144, 160, and 170, with transcripts typically identical to those annotated in VectorBase and which encode six predicted transmembrane proteins. Thus, further evidence is necessary to rule out their role as ORs. Some of these genes were highly enriched in antennae: The moderated log folds for CquiOR76, 133, 160, and 170 were 5.7, 3.3, 6.4, and 5.6, respectively. Transcript levels for CquiOR105, 135, and 139 were higher in antennae than in legs (mod_fold 1.4, 2.1, and 1.5, respectively), whereas CquiOR144 and CquiOR138 showed low transcript levels in antennae (0.013 and 0.09 FPKM, respectively).
On the basis of transcriptome data and given the position in the genome, we renamed CquiOR27 into CquiOR182, CquiOR28 into CquiOR183, CquiOR100 into CquiOR158, and CquiOR179 into CquiOR173. Thus, CquiOR27, 28, 158, and 179 have been omitted. Lastly, we omitted seven postulated OR sequences, namely CquiOR57, 71, 88, 122, 123, 165, and 175, because their transcripts encodes for short proteins (CquiOR57: 179 amino acid residues; OR71: 289; OR88: 305; OR123: 156; and OR175: 201) or were not found (CquiOR122). Transcripts levels for CquiOR165 were very high (mod_fold 4.4), thus suggesting they may have other antennae-specific function(s). RNA-Seq data explained our failure to clone CquiOR87, CquiOR110 (7), and CquiOR38 (9) given that their transcripts are very low in antennae (0.06, 0.05, and 0.08 FPKM, respectively), thus indicating they are not adult antennal OR genes. More importantly, this transcriptome analysis provides a road map for cloning and deorphanization OR genes enriched in female antennae.
Transcripts for 107 OR genes, including CquiOrco, were significantly enriched (mod_lfc > 2) in antennae (Fig. 1), whereas only 8 OR genes (CquiOR8, 32, 39, 87, 115, 118, 142, and 157) showed higher transcripts levels in legs than in antennae. Twenty-three genes (CquiOR182, 119, 146, 14, 77, 139, 105, 94, 153, 116, 183, 162, 86, 145, 113, 141, 110, 38, 83 109, 183, 144, and 10) were more expressed in antennae but below the threshold (0 < mod_lfc < 2), and transcript for three genes (CquiOR89, 143, and 149) were not detected. Thus, differential expression was compared for a total of 141 OR genes (Dataset S4). The top OR genes most enriched in antennae were CquiOR125 (mod_lfc, 8.79), CquiOR64 (7.89), CquiOR93 (7.84), CquiOR151 (7.82), and CquiOR132 (7.72). Among deorphanized Culex ORs, CquiOR121 (previously named CquiOR2) (6), CquiOR21 (formerly known as CquiOR10) (30), CquiOR1, CquiOR44, CquiOR73, CquiOR161 (7), CquiOR37, and CquiOR99 (9), CquiOR21 (mod_lfc, 7.58), was the most enriched in antennae. It is a common practice to validate RNA-Seq differential analysis with quantitative analysis of a subset of genes by qPCR. Thus, we compared by qPCR expression levels of the top five OR genes (plus CquiOR21) in female antennae and legs (Fig. S5). As it was the case with OBPs (see above), qPCR data mirrored the quantification by RNA-Seq, with small variations in ranking. One of the obvious advantages of RNA-Seq is that it goes beyond quantification of target genes and leads to identification of novel genes and/or isoforms (see below).
Identification of Unique Putative OR Genes and Isoforms.
Lastly, we analyzed the transcriptome to prospect for unique OR genes and/or isoforms. First, we analyzed all transcripts (strictly speaking transfrags, i.e., assembled transcripts fragments, TCONS) related to previously annotated OR genes. If the transcripts and database sequences were identical or had minor differences, the VectorBase sequence was adopted. For example, two transcripts were identified for CquiOR1, one (TCONS_00029650) identical to the sequence in the database and another one (TCONS_00029651) that encoded a protein with an additional residue: Val-323. Likewise, there were two transcripts for CquiOR2: one identical to the sequence in VectorBase and the other encoding a protein with an additional residue, Ser-324. Because a pool of 2,000 antennae was used, RNA-Seq analysis had enough sensibility to detect small variations in genome such as different alleles of the same gene in this laboratory population. Other ORs had only one transcript per gene (e.g: CquiOR4, 5). Synonymous substitutions were not considered. If the transcripts differed significantly from those in the VectorBase, they were further analyzed to identify putative ORs with sequences suggesting splice variants, having two or more amino acid differences, or having a longer N or C terminus. First, the transcript (identified by a TCONS number) was translated (Expasy Translate) and then the topology of the encoded protein was predicted by OCTOPUS. Simultaneously, unique transcripts were analyzed by Blast2Go (12). For this approach, a fasta file was created by removing all cufflinks transcripts identical, potential isoforms, and nonexact overlap to leave a file only with unique transcripts (5,833 entries) found in regions with no VectorBase genes. Separately, Blast2Go was performed on the complete cufflink by selecting only sequences with a length between 900 and 1,250 nt. These Blast2Go analyses suggest a large number of potentially unique receptors (71 among 1,024 annotated genes) and approximately 7.5% of OR genes in the specified nucleotide range (Fig. S6). After these blasts, mapping and annotation, the transfrags identified as potentially unique OR genes, were manually analyzed as described above. For example, TCONS_00023454 and TCONS_00023455 encoded proteins that shared amino acid sequences (85–92% identity) with CquiOR22 (TCONS_00023453). Thus, they were named CquiOR22b and CquiOR22c, and the VectorBase sequence was renamed CquiOR22a considering that they may be products of the same gene. As an example of OR identified anew, TCONS_00000073 was identified as related to odorant receptor 47b by Blast2Go. After determining that this transcript had higher expression levels in antennae (13.6 FPKM) than in legs (0.06 FPKM), it was analyzed by the above-described procedure. Because the protein encoded by this transcript shared amino acid sequence with AaegOR33 (58% identity), it was named CquiOR33a so as to avoid confusion with CquiOR33, a previously purged OR sequence (see above). Using this multiple approaches, we identified 36 unique OR transcripts, namely: CquiOR21b, 22b, 33a, 36b, 55b, 58b, 58c, 65b, 67b, 68b, 71a, 73b, 79b, 81b, 90b, 90c, 95b, 96b, 102b, 102c, 104b, 108b, 114b, 117b, 119b, 125b, 129b, 131b, 153a, 154b, 159b, 160b, 168b, 169b, 173b, and 182b (Dataset S5 and Table S2).
As analyses of the Culex genome are refined, the numbers of putative ORs are being updated. Our initial estimate of 158 OR genes (6) and the report of 180 putative OR genes, along with the comprehensive genome analysis (5), were updated to 130 putative OR genes (7). Our current studies with the discovery of unique genes and isoforms suggests a repertoire of 177 Cx.. quiquefasciatus OR genes, including CquiOrco. Of notice, a pair of genes, CquiOR114 (CPIJ013945) and CquiOR117 (CPIJ013953), which seem to have derived from gene duplication (31) because they encode ORs with identical amino acid sequences (Table S3), but different nucleotide sequences, and are located in different supercontigs, were counted separately.
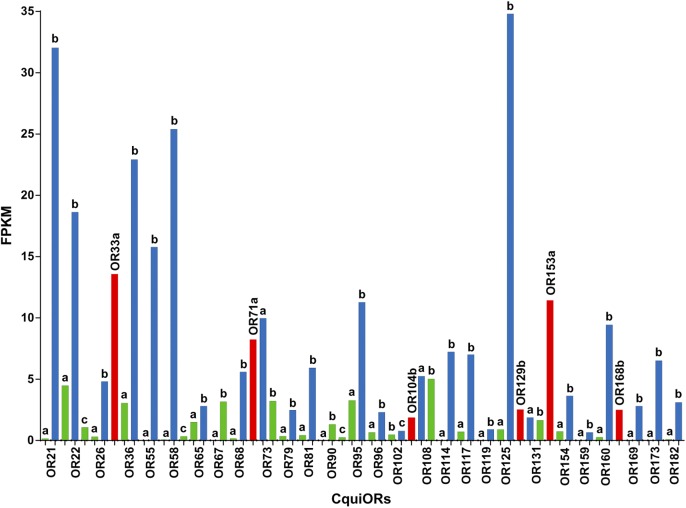
Most of the newly identified ORs showed high transcript levels in antennae (Fig. 2), very low (<0.09 FPKM), low (<0.12 FPKM), or no transcripts in legs, except for 58b and 68b (3.17 and 0.28 FPKM, respectively). Interestingly, transcript levels for the unique isoforms were typically higher than those for the previously annotated sequences (e.g: CquiOR125b, 34.8 FPKM; CquiOR125a, 0.89 FPKM), except for CquiOR108 and CquiOR131, with nearly the same levels of unique and previous isoforms, and CquiOR73 with lower transcript levels for the unique isoform (CquiOR73b, 3.21 FPKM; CquiOR73a, 9.97 FPKM). Some of the unique isoforms and previously annotated ORs have identical sequences, but differed in having longer C terminus (e.g., CquiOR95a and CquiOR95b: 383 and 398 aa residues, respectively). Given the importance of C terminus in receptor function (32), we further scrutinized their sequences by making certain that the nucleotides encoding the C terminus amino acid sequences were in the same supercontigs and downstream of the previously annotated sequences. For example, nucleotide sequence encoding for the C terminus of CquiOR95b is in supercont3.314:398962:401483:1, downstream (400,934-400,981) of the predicted stop codon (400,881-3 in exon CPIJ802591:4) in the annotated sequence, and followed by a stop codon. It is beyond the scope of this research to determine whether these differences are intrinsic errors of large-scale genome annotations or variations from different strains of Culex mosquitoes. Of notice, the genome was sequenced from the Johannesburg strain of Cx. quinquefasciatus, whereas our transcriptome data were derived from a Californian strain. To validate our RNA-Seq–based prospect of unique OR genes and isoforms, we cloned CquiOR95 (isoform b with an extended C terminus) and investigated whether the receptor protein was functional.
Fig. 2.
Transcript levels (FPKM) for unique CquiORs. Bars in blue are for unique isoforms of currently annotated OR genes (green) and those in red represent unique CquiOR genes. Largely, the transcript levels in legs (Dataset S5) were low and omitted for clarity.
Deorphanization of Unique OR, CquiOR95b.
CquiOR95b⋅CquiOrco-expressing oocytes were challenged with a large panel of physiologically relevant odorants, including repellents, oviposition attractants, host-derived kairomones, and other compounds involved in the chemical ecology of the southern house mosquito. Of 82 compounds tested, only two compounds elicited significant current amplitudes, namely citronellal and ethyl 2-phenylacetate (Fig. S7A). They responded to both compounds in a dose-dependent fashion (EC50, 16 µM, ethyl 2-phenyl acetate; 276 µM, citronellal) (Fig. S7B), thus demonstrating that the receptor was functional and interestingly narrowly tuned to two compounds of unrelated chemical structures, one of which, citronellal, is a known insect repellent (33). Next, we tested the effect of ethyl 2-phenylacteate on mosquito behavior.
Repellency Assay.
Given that CquiOR95b did not respond to other known insect repellents, N,N-diethyl-3-methylbenzamide (DEET) and p-menthane-3,8-diol, we initially envisioned that ethyl 2-phneylacteate might be a plant attractant (kairomone). However, our attempts to measure attractancy by the sugar-feeding assay (34) demonstrated that mosquitoes were actually repelled by ethyl 2-phenylacetate. Then, we systematically tested repellency by the surface landing assay (34) by using DEET as a positive control. Although not as strong as the gold standard of insect repellents, DEET, ethyl 2-phenylacteate is clearly a repellent (Fig. 3). Significantly more mosquitoes landed on the side of the chamber treated with solvent than in the side treated with ethyl 2-phenylacetate. Thus, the “chemical curtain” created by ethyl 2-phenylacteate loaded on the filter paper surrounding the warm surface prevented mosquitoes from landing. As previously demonstrated with DEET (34), mosquitoes approached both sides of the chamber, but landed on the solvent (control) side and avoided ethyl 2-phenylacetate.
Fig. 3.
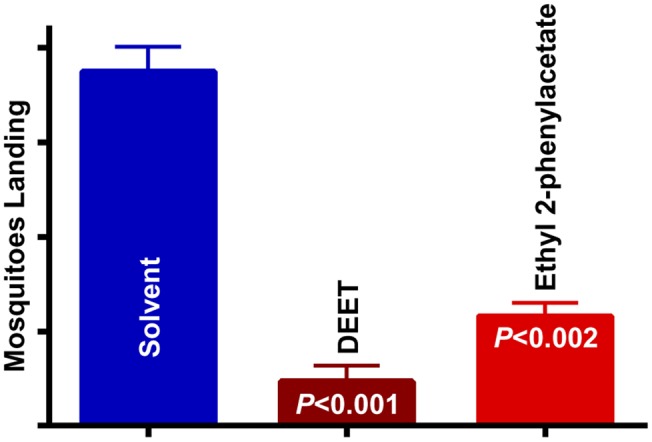
DEET-induced and ethyl 2-phenylacetate-induced repellency of mosquitoes responding to physical stimuli. Combined results of 2 two-choice assays comparing solvent with DEET, and solvent with the best ligand for CquiOR95b, ethyl 2-phenylacetate (n = 5–6).
Conclusions
RNA-Seq–based differential expression analysis of olfactory genes from antennae and legs of adult female Cx. quinquefasciatus showed that a large number of putative OBP, plus-C OBP, and CSP genes are enriched in nonolfactory tissues, thus reducing the number of candidates for future interrogation on their role(s) in mosquito olfaction. Our analysis unraveled a significant number of IR genes most likely involved in olfaction given their predominant expression in antennae. The largest majority of OR genes were predominant in antennae. These differential expression data were validated with previous and current qPCR analysis of a subset of OBP and OR genes. Prospecting for unique OR transcript led to the identification of 36 OR genes. Our libraries generated from a large pool of mosquitoes allowed the detection of unique isoforms of currently annotated OR genes. Additionally, we identified unique putative OR genes differing in internal sequences (indels) or having predicted proteins with extended N and/or C terminus. For validation of the prospecting approach, we cloned a gene encoding an extended C terminus OR, CquiOR95b, and demonstrated that this receptor was functional when expressed in Xenopus oocytes. This reverse chemical ecology approach led to the discovery of a unique mosquito repellent, ethyl 2-phenylacetate.
Materials and Methods
A laboratory colony of Cx. quinquefasciatus originated from adult mosquitoes collected in Merced, CA, in the 1950s (34) was used in this study. Antennae and legs dissected from blood-fed adult females were used for RNA-Seq as well as to generate templates for validation by qPCR and gene cloning. Deorphanization of a unique OR gene was performed with the Xenopus oocyte expression system (9, 35). Oocytes expressing CquiOR95b along with CquiOrco were challenged with large panel of physiologically relevant odorants. Activity of the best ligand was tested with the surface-landing assays for mosquito repellents (34). Greater detail is provided in SI Materials and Methods. Sequences were deposited on National Center for Biotechnology Information’s Sequence Reads Archive (accession no. SRP030034).
Supplementary Material
Acknowledgments
We thank Dr. Monica Britton (University of California Davis Genome and Biological Sciences Facility) for her invaluable contributions to initial bioinformatics analysis and guidance throughout this project; Dr. Anthony Cornel for providing mosquitoes that allowed us to duplicate his colony at the Davis campus; and members of the laboratory, particularly Dr. Fen Zhu and Mr. Kevin Cloonan, for their help with antennae collection and RNA extraction. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award R01AI095514. C.U.-V. and C.S.B.d.S. were supported by the National Council of Scientific and Technological Development (Brazil) for International Post-Doctoral Scholarship from the Science Without Borders Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession nos. SRP030034, SRS483777, and SRS483778).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316059110/-/DCSupplemental.
References
- 1.Nasci RS, Miller BR. 1996. Culicine mosquitoes and the agents they transmit. The Biology of Disease Vectors, eds Beaty BJ, Marquardt WC (Univ Press of Colorado, Niwot, CO)
- 2.Andreadis TG. The contribution of Culex pipiens complex mosquitoes to transmission and persistence of West Nile virus in North America. J Am Mosq Control Assoc. 2012;28(4) Suppl:137–151. doi: 10.2987/8756-971X-28.4s.137. [DOI] [PubMed] [Google Scholar]
- 3.Syed Z, Leal WS. Acute olfactory response of Culex mosquitoes to a human- and bird-derived attractant. Proc Natl Acad Sci USA. 2009;106(44):18803–18808. doi: 10.1073/pnas.0906932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leal WS. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 5.Arensburger P, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330(6000):86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pelletier J, Hughes DT, Luetje CW, Leal WS. An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PLoS ONE. 2010;5(4):e10090. doi: 10.1371/journal.pone.0010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu P, et al. Silent, generic and plant kairomone sensitive odorant receptors from the Southern house mosquito. J Insect Physiol. 2013;59(9):961–966. doi: 10.1016/j.jinsphys.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, et al. High degree of single nucleotide polymorphisms in California Culex pipiens (Diptera: Culicidae) sensu lato. J Med Entomol. 2012;49(2):299–306. doi: 10.1603/me11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu F, Xu P, Barbosa RM, Choo YM, Leal WS. RNAi-based demonstration SRP030034 of direct link between specific odorant receptors and mosquito oviposition behavior. Insect Biochem Mol Biol. 2013;43(10):916–923. doi: 10.1016/j.ibmb.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buffalo V. 2011. Quick read quality control. (Bioconductor, Seattle) Available at www.bioconductor.org/packages/release/bioc/html/qrqc.html. Accessed October 17, 2013.
- 11.Buffalo V. 2011. Scythe—A very simple adapter trimmer. (GitHub, San Francisco) Available at https://github.com/ucdavis-bioinformatics/scythe. Accessed October 17, 2013.
- 12.Joshi N. 2011. Sickle—A windowed adaptive trimming tool for FASTQ files using quality (GitHub, San Francisco) Available at https://github.com/najoshi/sickle. Accessed October 17, 2013.
- 13.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown SM, Goecks J, Taylor J. RNA sequencing with NGS. In: Brown SM, editor. Next-Generation DNA Sequencing Informatics. Plainview, NY: Cold Spring Harbor Lab Press; 2013. pp. 171–186. [Google Scholar]
- 16.VectorBase 2013. VectorBase. Available at www.vectorbase.org. Accessed October 17, 2013.
- 17.Andres S, Huber W. 2013. Differential expression of RNA-Seq data at the gene level—The DESeq package. Available at www.bioconductor.org/packages/devel/bioc/vignettes/DESeq/inst/doc/DESeq.pdf. Accessed October 17, 2013.
- 18.Pelletier J, Leal WS. Characterization of olfactory genes in the antennae of the Southern house mosquito, Culex quinquefasciatus. J Insect Physiol. 2011;57(7):915–929. doi: 10.1016/j.jinsphys.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Ribeiro JM, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004;34(6):543–563. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Larsson MC, et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43(5):703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 21.DeGennaro M, et al. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature. 2013;498(7455):487–491. doi: 10.1038/nature12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bigiani A, Mucignat-Caretta C, Montani G, Tirindelli R. Pheromone reception in mammals. Rev Physiol Biochem Pharmacol. 2005;154:1–35. doi: 10.1007/s10254-004-0038-0. [DOI] [PubMed] [Google Scholar]
- 23.Breer H, Krieger J, Raming K. A novel class of binding proteins in the antennae of the silkworm Antheraea pernyi. Insect Biochem. 1990;20:735–740. [Google Scholar]
- 24.Vogt RG. Odorant binding protein homologues of the malaria mosquito Anopheles gambiae; possible orthologues of the OS-E and OS-F OBPs OF Drosophila melanogaster. J Chem Ecol. 2002;28(11):2371–2376. doi: 10.1023/a:1021009311977. [DOI] [PubMed] [Google Scholar]
- 25.Vogt RG, et al. The insect SNMP gene family. Insect Biochem Mol Biol. 2009;39(7):448–456. doi: 10.1016/j.ibmb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136(1):149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croset V, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6(8):e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abuin L, et al. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69(1):44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340(6138):1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes DT, Pelletier J, Luetje CW, Leal WS. Odorant receptor from the southern house mosquito narrowly tuned to the oviposition attractant skatole. J Chem Ecol. 2010;36(8):797–800. doi: 10.1007/s10886-010-9828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci USA. 2003;100(Suppl 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4(2):e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J-K, et al. Evaluation of repellency effect of two natural aroma mosquito repellent compounds, citronella and citronellal. Entomol Res. 2005;35(2):117–120. [Google Scholar]
- 34.Syed Z, Leal WS. Mosquitoes smell and avoid the insect repellent DEET. Proc Natl Acad Sci USA. 2008;105(36):13598–13603. doi: 10.1073/pnas.0805312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu P, Leal WS. Probing insect odorant receptors with their cognate ligands: insights into structural features. Biochem Biophys Res Commun. 2013;435(3):477–482. doi: 10.1016/j.bbrc.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.