Significance
For decades, researchers have been trying to unravel one of the key questions in cell biology regarding keratin intermediate filament function in protecting epithelial cells against mechanical stress. For many different reasons, however, this fundamental hypothesis was still unproven. Here we answer this pivotal question by the use of keratin KO cells lacking complete keratin gene clusters to result in total loss of keratin filaments. This lack significantly softens cells, reduces cell viscosity, and elevates plastic cell deformation on force application. Reexpression of single keratin genes facilitates biomechanical complementation of complete cluster loss. Our manuscript therefore makes a very strong case for the crucial contribution of keratins to cell mechanics, with far-reaching implications for epithelial pathophysiology.
Abstract
Keratins are major components of the epithelial cytoskeleton and are believed to play a vital role for mechanical integrity at the cellular and tissue level. Keratinocytes as the main cell type of the epidermis express a differentiation-specific set of type I and type II keratins forming a stable network and are major contributors of keratinocyte mechanical properties. However, owing to compensatory keratin expression, the overall contribution of keratins to cell mechanics was difficult to examine in vivo on deletion of single keratin genes. To overcome this problem, we used keratinocytes lacking all keratins. The mechanical properties of these cells were analyzed by atomic force microscopy (AFM) and magnetic tweezers experiments. We found a strong and highly significant softening of keratin-deficient keratinocytes when analyzed by AFM on the cell body and above the nucleus. Magnetic tweezers experiments fully confirmed these results showing, in addition, high viscous contributions to magnetic bead displacement in keratin-lacking cells. Keratin loss neither affected actin or microtubule networks nor their overall protein concentration. Furthermore, depolymerization of actin preserves cell softening in the absence of keratin. On reexpression of the sole basal epidermal keratin pair K5/14, the keratin filament network was reestablished, and mechanical properties were restored almost to WT levels in both experimental setups. The data presented here demonstrate the importance of keratin filaments for mechanical resilience of keratinocytes and indicate that expression of a single keratin pair is sufficient for almost complete reconstitution of their mechanical properties.
Formation of a barrier capable of protecting tissue from external damage, chemical factors, and pathogens while resisting mechanical stress, external pressure, or shear force is one of the main functions of epithelial tissues. Keratinocytes represent the major cell type of mammalian epidermis and are mainly responsible for barrier functionality (1, 2). On the molecular level, mechanical cell properties mainly depend on cytoskeletal fibrous structures (3), namely actin filaments, microtubules, and intermediate filaments (IFs). Although the contribution of actin filaments and microtubules to the resilience of many cell types is widely accepted (4), it has been hypothesized for many years that the resilience of epithelia against various types of deformation depends largely on keratins (5–7). These form a stable network spanning from the cell periphery to the nucleus. Peripheral filaments dynamically enlarge into thicker filaments and progressively intermingle with the preexisting network by continuously moving centripetally until, in the center of the cell, a dense network of keratin filaments encircles the nucleus (8, 9).
Keratins are encoded by a large multigene family of more than 50 genes that are specifically expressed depending on distinct developmental pathways and physiological requirements (10, 11). Based on their amino acid sequence, type I keratins display an overall acidic character and differ considerably from the more basic type II keratins (12). Filament assembly requires both keratin types because of the obligatory heterodimer composition of keratin IFs (13). Given that most epithelia express 4–10 different keratin subunits (14), the total and isotype-specific contribution of the overall keratin network to mechanical properties of epithelia remains highly challenging to analyze in vivo. An additional difficulty in experimentation is the absence of drugs to specifically disrupt the keratin IF system.
Most results on the mechanical properties and functions of IFs are based on biomimetic systems and disease models. One of the hallmarks of IFs is their low bending stiffness. In conjunction with nonlinear strain stiffening observed in networks at large deformations, these features supposedly enable IFs to serve as a mechanical buffer system protecting cells from environmental stress (15–17). High tensional loads have already been observed for keratins in vivo (18). Strain stiffening goes along with a predominantly elastic behavior in biomimetic systems (19). A similar elastic response was also found in particle-tracking microrheology experiments on epithelial cells (20). Furthermore, on the level of transgenic mice and patients with heritable fragility of the epidermis, a clear correlation between molecular integrity of keratin filaments (K5 and K14) and mechanical toughness of epithelia was shown (21, 22).
Generation of keratin KO mouse strains by deleting either the type I or type II keratin gene cluster (23) results in complete loss of keratin filaments in epidermal cells and provides the unique opportunity to accomplish accurate and exact biomechanical analyses of keratin-free keratinocytes for the very first time. Until now, these cell systems have been characterized for various cell biological and biochemical functions (24, 25). Of note, increased cell motility and altered desmosomal and hemidesmosomal adhesion were identified in these studies.
In the current work, we analyze the keratin type I and type II mutant keratinocyte cell lines and compare them to WT controls established in parallel and rescue cell lines reexpressing keratins 5 and 14. Mechanical properties are examined in living cells from outside by atomic force microscopy (AFM) (26, 27) and from inside by magnetic tweezers (28) (Fig. S1). The data provide compelling evidence for the hypothesis that keratins determine the mechanical stability and integrity of epithelial cells by direct effects on various mechanical parameters such as elasticity, viscosity, spring constant, and relaxation time.
Results
Loss of Keratin Filaments Leads to Significant Cell Softening.
Keratin filaments are believed to strengthen epithelial cells against various types of mechanical stress, mainly indentation, local deformation, and strain. To investigate this hypothesis, we characterized the mechanical properties of WT and keratin-lacking cell lines by complementary approaches, i.e., by AFM and magnetic tweezers experiments. To consolidate our data beyond doubt, both keratin type I−/− mutant cells (KtyI−/−) and keratin type II mutant cells (KtyII−/−), as well as corresponding rescue cell lines, were examined in parallel.
In a first series of experiments, WT and KtyI−/− cell lines were characterized for their resilience against external force by AFM indentation. Immunocytochemistry stainings revealed well-developed keratin networks in WT cells with thin keratin filaments in the cell periphery and more prominent keratin bundles within the cell body surrounding the nucleus as a stable cage (Fig. 1). In contrast, keratin filaments were completely absent in KtyI−/− cells. AFM indentation experiments were performed above the nucleus and the cell body (Fig. 1 D–F and Fig. S1A). Subsequently, cell elasticities E were calculated for increasing indentation depths (Fig. S2 and SI Text, AFM Apparent Modulus Plots). Plateau regions indicating ideal indentation depths for cell elasticity analysis were identified in an indentation range of 300–600 nm above the nucleus and estimated around 200 nm for the cell body. These indentation depths were always less than 10% of the respective overall cell thickness [7.0 ± 1.4 (SD) µm for WT (n = 6) and 5.0 ± 0.8 µm for KtyI−/− cells (n = 4) on the nucleus and ∼4 µm for both cell types on the cell body]. In all AFM experiments, at least three force-distance curves were recorded for each cell position. The total number of analyzed cells was >100 for WT and KtyI−/−. Superposition of the resulting raw data curves that are exemplarily given for one position in Fig. 2A indicated reliable reproducibility. The data showed that cells responded elastically with no permanent plastic deformation or active response to probing. Calculating the applied forces necessary to indent 100–600 nm directly from the raw data revealed significant differences between WT and KtyI−/− already for indentation depths of 200 nm for nuclear and cell body regions (Fig. 2 B and C). The differences became even more pronounced at higher indentation depths.
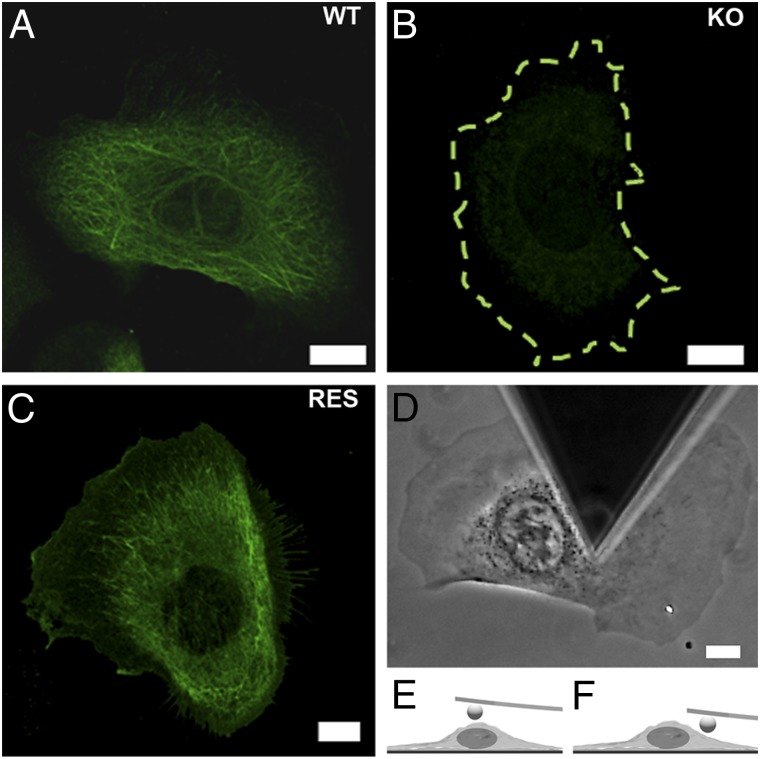
Fig. 1.
Keratin network characterization and AFM analysis regions. (A–C) Immunfluorescence micrographs of the keratin network in WT, KtyI−/− KO, and K14 rescue (RES) keratinocytes. The KtyI−/− cell outline is given by a dashed line. (Scale bars, 10 µm.) (D) Top view phase contrast image of an AFM tip-less cantilever aligned with the cell body of a living adherent keratinocyte. (Scale bar, 10 µm.) (E) Side view diagram of a cantilever positioned above the nucleus and (F) the cell body.
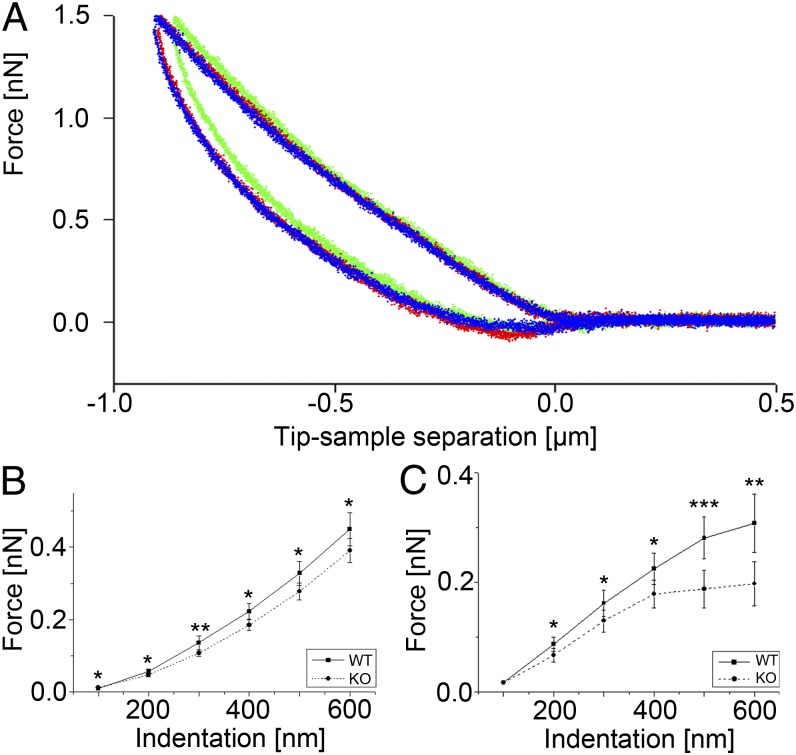
Fig. 2.
AFM force-distance curves and force-indentation plots. (A) Plot showing the superposition of three AFM force-distance curves (green, red, blue) recorded successively at 5-s intervals on the same position above the nucleus of a WT keratinocyte. (B) Plot showing the average forces needed to reach a certain indentation depth above the nucleus (n = 109–125, depending on indentation depth) and (C) cell body (n = 54–117). Significant differences between WT and KtyI−/− cells (KO) are indicated (*P = 0.05, **P = 0.01, ***P = 0.001); error bars are SEM.
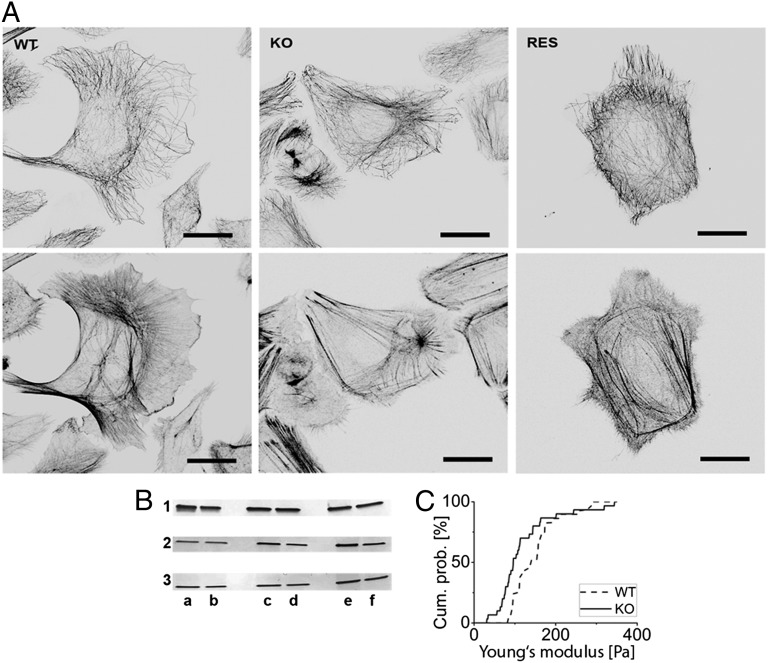
To accurately calculate cellular elasticities, force-distance curves were analyzed by the classical Hertzian spherical indenter model (Eq. 1) as implemented by the AFM manufacturer (Fig. 3A). For all areas analyzed, the resulting Young’s moduli indicated a significantly higher (Fig. 3 B and C) stiffness of WT cells compared with KtyI−/− cells [nucleus: EWT = 459 ± 31 Pa (SEM) (n = 125) and EKtyI−/− = 343 ± 18 Pa (n = 109); significance level, 0.0030; cell body: EWT = 752 ± 100 Pa (n = 117) and EKtyI−/− = 412 ± 75 Pa (n = 113); significance level, 0.0002]. Force maps from whole WT and KtyI−/− cells also demonstrated an overall softened cell phenotype on keratin loss, most prominently above the nucleus (Fig. 4 A–D).
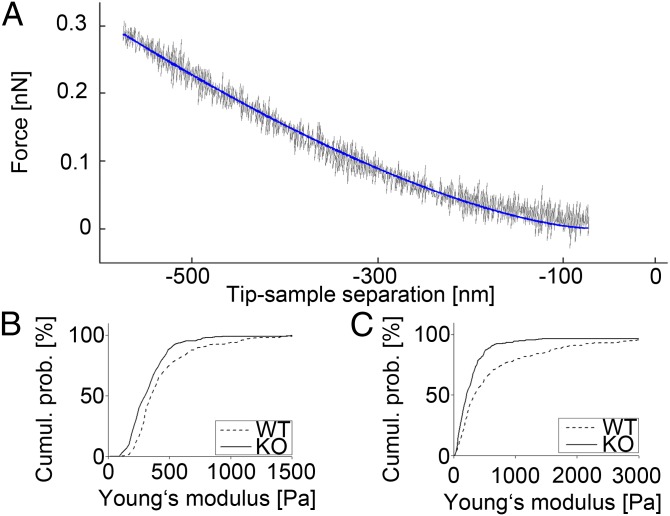
Fig. 3.
(A) AFM force distance curve (gray) and data fit to Hertz model (blue). (B) Cumulative histogram of Young's modulus E obtained from indentation experiments performed on the nucleus of WT (n = 110) and KtyI−/− (KO; n = 125) cells. (C) Same plots showing data measured on the cell body (nWT = 117, nKO = 113).
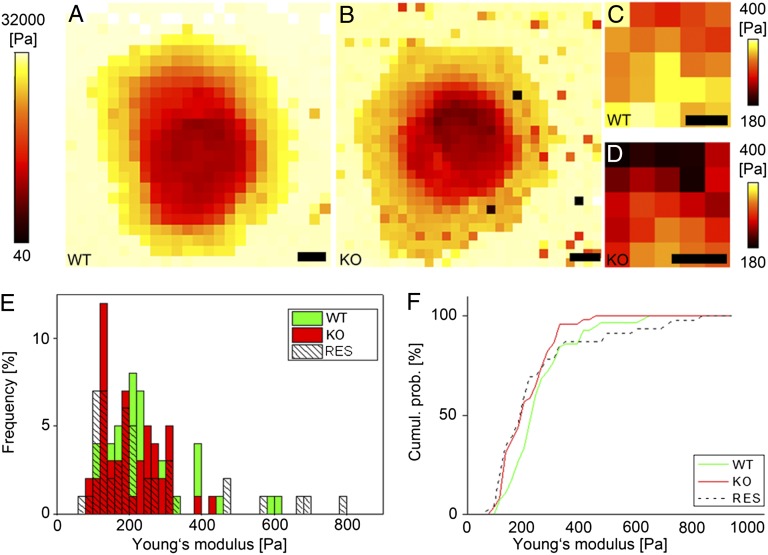
Fig. 4.
Stiffness maps and rescue measurements. Stiffness maps of a live WT (A) and KtyI−/− (KO) (B) cell displaying the Young's moduli computed at every position. (Scale bar, 10 µm.) (C and D) Zoom-ins of the nuclear region of A and B, respectively. (Scale bars, 3 µm.) (E) Histogram showing the distribution of Young's moduli obtained from indentation experiments performed on the nucleus of WT (n = 58), KO (n = 48), and RES (n = 44) cells and (F) corresponding cumulative histograms.
Using an alternative approach for force-distance curve analysis by accurately determining the indenter contact points and by describing curves via a standard power law function, the presented results were fully confirmed with an even better fit accuracy (SI Text, Alternative Force-Distance Curve Analysis and Figs. S3–S6).
Identical AFM experiments on keratin KtyII−/− mutant cells lacking the complete set of type II keratins resulted in very similar data. Mutant cells were significantly softer (∼40%) than the corresponding WT cells (n ≥ 10 cells each).
Reexpression of Keratin 14 Rescues Keratin Network and Cell Elasticity.
To prove that the absence of keratins was responsible for altered cell stiffness, rescue cells stably expressing keratin 14-YFP (yellow fluorescent protein) were generated (K14), leading to IF formation by pairing with the endogenously produced K5. Cells were analyzed by the above-described AFM protocol. Proper keratin network formation was assessed by fluorescence microscopy (Fig. 1C) before indentation in each of the analyzed cells (n = 46). Elastic moduli were retrieved by fitting the resulting force distance curves with the Hertz model. To minimize errors, WT and KtyI−/− cells (n = 57 and n = 51, respectively) were measured in parallel using the same cantilever (Fig. 4 E and F). We found that the average cell elasticity was significantly increased on reexpression of K14. Interestingly, Young's moduli of rescue cells showed a biphasic distribution, with ∼50% of cells presenting reduced elasticity as in KtyI−/− cells and the other ∼50% presenting WT or even more elevated stiffness.
Keratin Stiffens Cells Against Intracellular Deformation and Elevates Cell Elasticity.
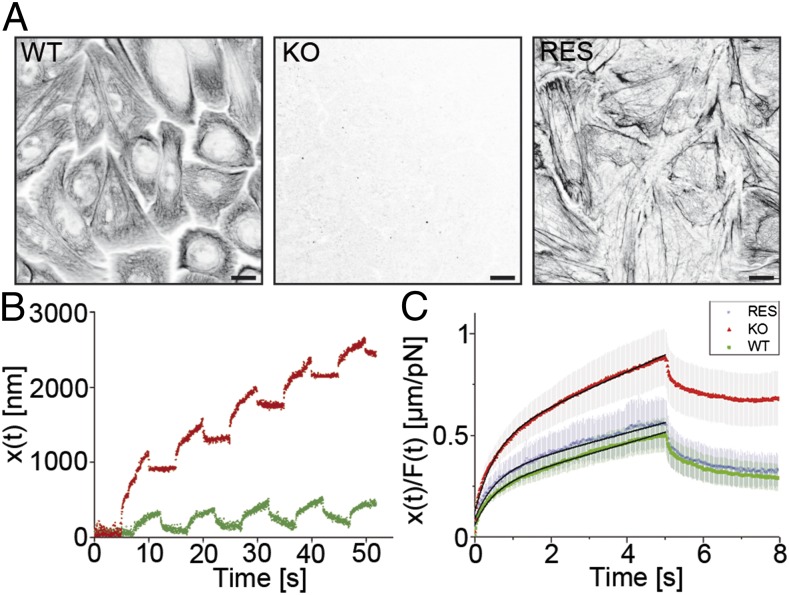
Although AFM analyses are based on the application of extracellular indentation forces, we sought for an alternative method to probe for intracellular force measurements of adherent cells. To this end, we adapted magnetic tweezers (for a scheme, see Fig. S1B) to examine intracellular deformation by local straining in keratin-free cells. In this set of experiments, KtyII−/− cells, corresponding WT cells and K5-YFP expressing rescue cells, were used. The KtyII−/− cells much like the KtyI−/− cells were completely devoid of keratin filaments, and reexpression of a single keratin, in this instance K5, was sufficient for restitution of an extended keratin filament network (Fig. 5A). Cyclic force pulses applied to superparamagnetic beads resulted in viscoelastic response curves exemplified by Fig. 5B for WT and KtyII−/−. Major bead displacements, however, were noted in KtyII−/− cells that resulted in incremental bead movement toward the magnetic tweezers tip with repeated pulses, indicating a lower resistance of the keratin-free cytoplasm. The response curves were then rescaled, normalized by the amplitude of the applied magnetic force, and averaged. The resulting curves revealed a significantly different viscoelastic behavior of WT vs. KtyII−/− cells. Stable reincorporation of GFP-keratin 5 (K5) in KtyII−/− cells and subsequent direct fluorescence microscopic preselection for cells with a well-developed keratin network almost fully restored viscoelastic behavior to WT levels (Fig. 5C).
Fig. 5.
Keratin network localization and magnetic tweezers analyses. (A) Immunfluorescence micrographs of the keratin network of WT, KO (KtyII−/−), and K5 rescue (Res) cells. (Scale bars, 10 µm.) (B) Example of raw magnetic tweezers data: displacement of the superparamagnetic bead incorporated in the cytoplasm of a WT and a KtyII−/− cell (KO) following the application of 5-s force pulses exerted at 10-s intervals. (C) Average overall rescaled peaks recorded on the three phenotypes as a function of time (nWT = 40 for 9 cells; nKO = 39 for 10 cells; nRES = 22 for 6 cells). In black, fits of Eq. 3. Error bars are SEM.
Extension portions of these processed response curves recorded on each cell phenotype were fitted via Eq. 2. After an initial quick elastic response [initial jump of amplitude 1/(k0 + k1) in the creep compliance], the system enters a viscous regime (with a characteristic transition time equal to τ) until a flow regime dominated by η0 (viscous friction coefficient) is established.
Parameters obtained from the fits (Table 1) clearly indicate a lowering of the cytoplasmic viscosity by more than 40% in the absence of a keratin network. This reduction is comparable to the reduced cell elasticity found above by AFM experiments and is accompanied by an overall softening of KtyII−/− cells in comparison with WT and rescue cells. The relaxation times remain comparable.
Table 1.
Viscoelastic cell parameters
| Parameter | KO | RES | WT |
| keff = k0 + k1 (pN/µm) | 8 | 11.8 | 12.3 |
| SD | 1.1 | 3.2 | 1.8 |
| η (pN s/µm) | 10.8 | 18.5 | 18.3 |
| SD | 1.3 | 4.2 | 3.0 |
| τ (s) | 0.4 | 0.43 | 0.48 |
| SD | 0.11 | 0.19 | 0.17 |
Viscoelastic parameters obtained from fitting Eq. 2 to the averaged normalized magnetic tweezers data of WT, K5 rescue (RES), and KtyII−/− mutant cells (KO). keff, effective spring constant; η, viscous friction coefficient; τ, characteristic relaxation time.
Keratin Loss Does Not Affect Actin and Microtubule Networks.
To verify that reduced cell resilience and viscosity in the absence of keratins were not significantly influenced by altered expression of actin and tubulin, Western blot analysis was performed (24). Overall protein levels for all cell lines analyzed here indicated no major changes in protein concentration of actin and tubulin (Fig. 6B). In agreement with previous findings, staining for actin and tubulin demonstrated that the overall actin and microtubule organization was unaffected by the absence of keratins (Fig. 6A). To further exclude a putative influence of actin on altered mechanical properties of KO cells, actin filaments were disassembled by latrunculin A in WT and KtyI−/− cells. Subsequent AFM measurements on those cells identified a continuous, basically unchanged difference in elasticity and therefore significantly (significance level, 0.003) stiffer cell properties for WT cells compared with KtyI−/− cells, strongly supporting the dominant role of the keratin network in determining cell stiffness (Fig. 6C).
Fig. 6.
Independency of biomechanical deficiencies on actin and microtubules. (A) Immunofluorescent staining of microtubules (Upper) and actin (Lower) in WT, KtyI−/− (KO), and rescue cells. (B) Crude protein extract analysis by Western blotting. Protein concentrations were equalized using GAPDH as constitutively expressed marker (1) and subsequently analyzed for tubulin (2) and actin (3) expression. (a and b) Corresponding WTs. (c) KtyI−/−. (d) KtyII−/−. (e) K14 rescue. (f) K5 rescue. (C) Elasticity measurements above the nucleus of WT and KtyI−/− cells by AFM after depolymerization of actin filaments by latrunculin A. n = 30 cells each. (Scale bars, 20 µm.)
Discussion
The elasticity of cells and their resistance to external mechanical stress have been intensively analyzed for decades because of their crucial contribution to many cell functions such as differentiation (29) or cancer progression (30). Mechanical stability is especially relevant for epidermal keratinocytes, which are subjected to intense deformation in all directions. Although one of the main characteristics of epithelial cytoskeleton is the predominance of keratin filaments spanning the entire cytoplasm and surrounding the nucleus as a dense network (8), mechanical analyses have largely focused on actin filaments. On the other hand, it has been proposed that IFs serve as a major mechanical buffer system protecting cells from environmental stress (5). Seminal publications have tried to shed light on this hypothesis by use of biomimetic model systems on artificial IF networks (15), extracted keratin networks (20, 31), and analyses on whole cells or mouse models (32, 33). In essence, many of these analyses identified a very low bending stiffness for keratin filaments and a minor effect of keratin on cell elasticity. At the same time, however, rearrangements of the keratin network in various cancer cell lines were hypothesized to induce significant changes in cell elasticity (5).
Using two independent keratinocyte cell lines, both lacking the entire keratin cytoskeleton, and matched rescue cell lines reexpressing the basal cell–specific keratin pair K5/14, we conclusively demonstrate that elasticity, effective spring constant, and cell viscosity are significantly affected by keratins, leading to softened, less viscous cells in their absence. The use of two independent keratin-free cell models, the previously established keratin type II−/− mutant cells and the newly isolated keratin type I−/− keratinocyte mutant cells, in combination with matched WT controls and rescue cell lines largely excludes cell line–specific artifacts and considerably strengthens the tenacity of our findings. Both mutant cell lines lacked keratin filaments completely, showed very similar cell morphologies, and presented very similar biomechanical deficiencies as reflected by almost identical reductions in Young’s moduli that were identified in our AFM analyses. Furthermore, all alterations in mechanical cell properties were shown to be fully dependent on keratin loss. Here, neither actin nor MT protein levels nor corresponding cytoskeletal structures were affected. These results are well in line with results on protein levels found before for KtyII−/− cells (23, 24). Additionally, pharmacological actin network disruption reduced the overall cell elasticity as one would expect on removal of cortical actin but did not influence the clear biomechanical differences between WT and KtyI−/− cells. These data therefore clearly show that our AFM measurements were sufficiently deep to deform not only cortical actin but also the keratin network beneath.
Keratin material properties additionally explain the even more drastic keratin mutant phenotypes observed in magnetic tweezers straining experiments. Although AFM-induced indentations work primarily against the comparatively low IF bending stiffness of only 4–12 × 10−27 Nm2 (15), microrheological magnetic tweezers experiments locally stretch the keratin network to such an extent that they work against increasing stiffness at high strain. Keratin filament extensions of more than 2.5-fold have been observed in vitro (34), and stretched keratins have been imaged in cultured keratinocytes that were elongated by 133% (18) without filament rupture. The absence of plastic deformations on application of successive magnetic tweezers pulses in WT cells in the current study provides further compelling evidence for the effectiveness of keratin networks as mechanical buffer system. Consequently, the absence of this buffer system in keratin-deficient cells resulted in reduced viscosity, lowered effective spring constants, and plastic cell deformation. These observations are in line with reports on other IF systems. For example, vimentin-deficient fibroblasts presented a reduced stiffness on mechanical stress application by extracellular magnetic twisting cytometry (35). In another study, Brown et al. (36) noted that the rigidity of T lymphocytes depends primarily on intact vimentin filaments as determined by flow cytometry. On the other hand, AFM measurements on rat fibroblasts with a disrupted vimentin cytoskeleton identified localized stiffening (37).
Different type I and type II keratins are simultaneously expressed in resting keratinocytes. The most prominent keratins of basal epidermal cells are the type II K5 and the type I keratins K14 and K15 (14). K5 deletion results in pronounced cytolysis and neonatal lethality (38), whereas K14 depletion leads to epidermolysis bullosa simplex-like disease where mechanical trauma exacerbates blistering (39). Assuming a similar role of K14 and K15, we decided to test for rescue of overall type I KO by reexpression of K14 only. At the same time, K5 rescues were analyzed here to induce formation of the same keratin network by expression of K5 as sole type II keratin. Both rescue clones formed a well-detectable keratin network (Figs. 1 and 5). To our surprise, reexpression of K5 rescued basically all biomechanical phenotypes of KtyII−/− cells to WT levels when analyzed by magnetic tweezers. Similar results were found for KtyI−/− cells reexpressing K14 recuperating elasticity levels similar to the WT when analyzed by AFM indentation. A slightly enhanced spread in cell elasticity found for this rescue may be caused by different K14 expression levels.
In summary, our data clearly prove the vital character of keratin filaments for biomechanical cell properties as a buffer system to withstand mechanical stresses with far-reaching implications for epithelial pathophysiology.
Materials and Methods
Cell Culture.
Keratinocyte cell line KtyII−/− lacking type II keratins and control WT cell lines were generated and described previously (23, 24). Generation of the type II rescue cell line KII KO K5-GFP reexpressing GFP-tagged keratin 5 was described in previously (25).
For generation of the cell line KtyI−/− lacking the complete set of type I keratins, homozygous C57B16 mice harboring two floxed alleles of the type I keratin cluster were crossed with 129S1hprt Cre mice. The resulting heterozygous mice were crossed to get 25% type I keratin cluster-deficient mice. Keratinocytes were isolated from the epidermis of E18.5 WT and KtyI−/− embryos and cultivated under conditions that permit spontaneous immortalization as described in Turksen (40).
To rescue KtyI−/− cells, human keratin 14-YFP cDNA was cloned into lentiviral pLVX-puro vector using XhoI and NotI restriction sites. Lentivirus for transduction was produced using the Lenti-X expression system (Clontech) according to the manufacturers' protocol. Supernatant containing lentivirus was harvested, and titers were checked with Lenti-X GoStix (Clontech). KtyI−/− cells were incubated with lentivirus for 24 h in the presence of 8 µg/mL polybrene (Sigma-Aldrich). Subsequently, cells were selected with puromycin (8 µg/mL; PAA), and clones stably expressing hK14-YFP were generated by limiting dilution cloning. Cultivation of keratinocytes was performed as described previously (25). Cells were grown at 5% (vol/vol) CO2 and the appropriate temperature. Rescue cells were incubated in medium containing 8 µg/mL puromycin for selection purposes. For some experiments, KtyI−/− and WT cells were incubated with medium containing 0.5 µM latrunculin A for 60 min at 37 °C. Directly before analysis, medium was replaced, and AFM measurements were performed within the next 60 min. The effectiveness of actin depolymerization was verified by immunocytochemistry on identically incubated cells.
Western Blotting.
SDS/PAGE on crude protein extracts of all cell lines analyzed in this work was performed as described previously (41). Separation of total protein extracts was performed by standard procedures (4–20% SDS-PAGE). Using GAPDH as constitutively expressed marker (antibody G8795; Sigma-Aldrich), overall protein levels of actin and tubulin were determined using mAB8457 (CST) and MAB1864 (Millipore) as antibodies.
Immunocytochemistry.
For immunocytochemical experiments, 2.5 × 104 cells were grown on collagen I–coated glass coverslips for 48 h. Labelings were performed as described previously (25) using fluorescently labeled phalloidin (Invitrogen) for actin and antibodies against tubulin (see above) or keratin (rabbit anti-pan keratin; ProGen Biotechnik). Images were recorded either on an ApoTome.2 (Zeiss) using AxioVision LE software (Zeiss) or acquired using a laser scanning confocal microscope (LSM 710; Zeiss), both equipped with a Plan-Apochromat 63×/1.4 oil immersion objective.
AFM Measurements.
AFM imaging and force spectroscopy were performed using a JPK Nanowizard Life Science version instrument (JPK) equipped with a fluid chamber (Biocell; JPK) for live cell analysis and an inverted optical microscope (Axiovert 200; Zeiss) for sample observation. AFM imaging of fixed and live cells was performed as described in SI Text, AFM Measurements. During indentation experiments, each analyzed position was recorded by at least three force curves and a cantilever speed of v = 1.5 µm/s. Force set points of 1.48 and 0.5 nN were used for the nucleus and cell body, respectively.
AFM Data Analysis.
The approaching parts of the recorded force-distance curves were analyzed using the JPK DP software (Hertz fit, sphere model) to obtain values of Young’s modulus E (Fig. 2A). With this option, the software uses the fit function described by Sneddon (42)
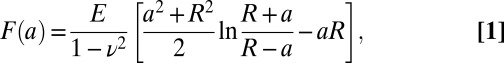 |
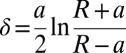 |
Here, 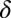 is the indentation depth, a is the contact radius of the indenter, R is the measured silica bead radius,
is the indentation depth, a is the contact radius of the indenter, R is the measured silica bead radius, 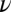 is the sample's Poisson ratio (set to 0.5 assuming cell’s incompressibility), and E is the cell’s Young’s modulus. In the fits, Young's modulus E and the contact point were used as fit parameters. For estimating the optimal indentation depth, so-called “apparent modulus” plots were used based on ref. 43 (Fig. S2 and SI Text, AFM Apparent Modulus Plots).
is the sample's Poisson ratio (set to 0.5 assuming cell’s incompressibility), and E is the cell’s Young’s modulus. In the fits, Young's modulus E and the contact point were used as fit parameters. For estimating the optimal indentation depth, so-called “apparent modulus” plots were used based on ref. 43 (Fig. S2 and SI Text, AFM Apparent Modulus Plots).
Because infinite sample thickness is assumed in the Hertz model, we limited the indentation to at most 5–10% of the sample thickness. Accordingly, curves recorded when indenting on the nuclei were fit for a range of 0 to 500 nm after the contact point. For the cell body, comparably thinner, this distance was taken to be 200 nm.
Distributions of resulting elasticities E were analyzed for differences with the parameter-free two-sided Mann-Whitney-Wilcoxon test.
Magnetic Tweezers Setup and Measurements.
Construction and calibration of the self-made magnetic tweezers and measurement procedures are described in detail in Fig. S7 and in SI Text, Magnetic Tweezers Measurements. In short, the instrument is based on a self-etched, water-cooled magnetic core of an electromagnet attached to a micromanipulator. M-270 Dynabeads (Invitrogen) were incorporated as superparamagnetic beads using a PDS-1000/He particle delivery system (BioRad).
Magnetic Tweezers Data Analysis.
Single peaks were normalized by the magnetic force applied at every time frame [calculated from the calibration curve F = F(d), d being the bead-tip distance; Fig. S8 and SI Text, Magnetic Tweezers Measurements] and averaged for every cell type. Following Bausch et al. (28), the extensional parts of these averaged creep-response curves (i.e., distance divided by force) were fitted by a four-element mechanical equivalent circuit composed of a dashpot in series with a Zener body
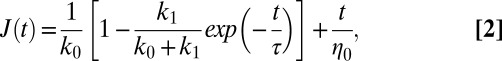 |
where 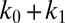 describes the effective spring constant of the system,
describes the effective spring constant of the system, 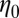 indicates the viscous friction coefficient of the bead, and
indicates the viscous friction coefficient of the bead, and 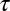 is the relaxation time characteristic of the transition from an elastic to a viscous regime. Uncertainties of fit results were determined as SDs from 500 times fitting synthetic data created by adding to the fit result normally distributed random numbers with scatter taken from the experiment.
is the relaxation time characteristic of the transition from an elastic to a viscous regime. Uncertainties of fit results were determined as SDs from 500 times fitting synthetic data created by adding to the fit result normally distributed random numbers with scatter taken from the experiment.
Supplementary Material
Acknowledgments
We thank Florian Kumpfe (JPK) for technical advice and constructive discussions. This work was supported by the START Program of the Faculty of Medicine, Rheinisch-Westfaelische Technische Hochschule Aachen, and by the German Research Council (LE566/18-1, WI1731/6-1, WI1731-8-1, MA1316-9/3, MA1316-15, INST 268/230-1, Translational Centre for Regenerative Medicine, and Leipzig No. 0315883).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313491110/-/DCSupplemental.
References
- 1.Fuchs E. Keratins and the skin. Annu Rev Cell Dev Biol. 1995;11:123–153. doi: 10.1146/annurev.cb.11.110195.001011. [DOI] [PubMed] [Google Scholar]
- 2.Sun TT, Shih C, Green H. Keratin cytoskeletons in epithelial cells of internal organs. Proc Natl Acad Sci USA. 1979;76(6):2813–2817. doi: 10.1073/pnas.76.6.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasza KE, et al. The cell as a material. Curr Opin Cell Biol. 2007;19(1):101–107. doi: 10.1016/j.ceb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Stricker J, Falzone T, Gardel ML. Mechanics of the F-actin cytoskeleton. J Biomech. 2010;43(1):9–14. doi: 10.1016/j.jbiomech.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beil M, et al. Sphingosylphosphorylcholine regulates keratin network architecture and visco-elastic properties of human cancer cells. Nat Cell Biol. 2003;5(9):803–811. doi: 10.1038/ncb1037. [DOI] [PubMed] [Google Scholar]
- 6. Kreplak L, Fudge D (2007) Biomechanical properties of intermediate filaments: From tissues to single filaments and back. BioEssays 29(1):26–35. [DOI] [PubMed]
- 7.Lulevich V, Yang HY, Isseroff RR, Liu GY. Single cell mechanics of keratinocyte cells. Ultramicroscopy. 2010;110(12):1435–1442. doi: 10.1016/j.ultramic.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Windoffer R, Beil M, Magin TM, Leube RE. Cytoskeleton in motion: The dynamics of keratin intermediate filaments in epithelia. J Cell Biol. 2011;194(5):669–678. doi: 10.1083/jcb.201008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Windoffer R, Wöll S, Strnad P, Leube RE. Identification of novel principles of keratin filament network turnover in living cells. Mol Biol Cell. 2004;15(5):2436–2448. doi: 10.1091/mbc.E03-09-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bragulla HH, Homberger DG. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J Anat. 2009;214(4):516–559. doi: 10.1111/j.1469-7580.2009.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hesse M, Zimek A, Weber K, Magin TM. Comprehensive analysis of keratin gene clusters in humans and rodents. Eur J Cell Biol. 2004;83(1):19–26. doi: 10.1078/0171-9335-00354. [DOI] [PubMed] [Google Scholar]
- 12.Steinert PM, Jones JC, Goldman RD. Intermediate filaments. J Cell Biol. 1984;99(1 Pt 2):22s–27s. doi: 10.1083/jcb.99.1.22s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichner R, Sun TT, Aebi U. The role of keratin subfamilies and keratin pairs in the formation of human epidermal intermediate filaments. J Cell Biol. 1986;102(5):1767–1777. doi: 10.1083/jcb.102.5.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magin TM, Vijayaraj P, Leube RE. Structural and regulatory functions of keratins. Exp Cell Res. 2007;313(10):2021–2032. doi: 10.1016/j.yexcr.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Janmey PA, Euteneuer U, Traub P, Schliwa M. Viscoelastic properties of vimentin compared with other filamentous biopolymer networks. J Cell Biol. 1991;113(1):155–160. doi: 10.1083/jcb.113.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin YC, et al. Origins of elasticity in intermediate filament networks. Phys Rev Lett. 2010;104(5):058101. doi: 10.1103/PhysRevLett.104.058101. [DOI] [PubMed] [Google Scholar]
- 17.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435(7039):191–194. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 18.Fudge D, et al. The intermediate filament network in cultured human keratinocytes is remarkably extensible and resilient. PLoS ONE. 2008;3(6):e2327. doi: 10.1371/journal.pone.0002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada S, Wirtz D, Coulombe PA. The mechanical properties of simple epithelial keratins 8 and 18: Discriminating between interfacial and bulk elasticities. J Struct Biol. 2003;143(1):45–55. doi: 10.1016/s1047-8477(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 20.Sivaramakrishnan S, DeGiulio JV, Lorand L, Goldman RD, Ridge KM. Micromechanical properties of keratin intermediate filament networks. Proc Natl Acad Sci USA. 2008;105(3):889–894. doi: 10.1073/pnas.0710728105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs E, Cleveland DW. A structural scaffolding of intermediate filaments in health and disease. Science. 1998;279(5350):514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- 22.Simpson CL, Patel DM, Green KJ. Deconstructing the skin: Cytoarchitectural determinants of epidermal morphogenesis. Nat Rev Mol Cell Biol. 2011;12(9):565–580. doi: 10.1038/nrm3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vijayaraj P, et al. Keratins regulate protein biosynthesis through localization of GLUT1 and -3 upstream of AMP kinase and Raptor. J Cell Biol. 2009;187(2):175–184. doi: 10.1083/jcb.200906094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kröger C, et al. Keratins control intercellular adhesion involving PKC-α-mediated desmoplakin phosphorylation. J Cell Biol. 2013;201(5):681–692. doi: 10.1083/jcb.201208162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seltmann K, et al. Keratins mediate localization of hemidesmosomes and repress cell motility. J Invest Dermatol. 2013;133(1):181–190. doi: 10.1038/jid.2012.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azeloglu EU, Costa KD. Atomic force microscopy in mechanobiology: Measuring microelastic heterogeneity of living cells. Methods Mol Biol. 2011;736:303–329. doi: 10.1007/978-1-61779-105-5_19. [DOI] [PubMed] [Google Scholar]
- 27.Radmacher M, Fritz M, Kacher CM, Cleveland JP, Hansma PK. Measuring the viscoelastic properties of human platelets with the atomic force microscope. Biophys J. 1996;70(1):556–567. doi: 10.1016/S0006-3495(96)79602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bausch AR, Ziemann F, Boulbitch AA, Jacobson K, Sackmann E. Local measurements of viscoelastic parameters of adherent cell surfaces by magnetic bead microrheometry. Biophys J. 1998;75(4):2038–2049. doi: 10.1016/S0006-3495(98)77646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 30.Suresh S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007;3(4):413–438. doi: 10.1016/j.actbio.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter N, Busch T, Seufferlein T, Spatz JP. Elastic moduli of living epithelial pancreatic cancer cells and their skeletonized keratin intermediate filament network. Biointerphases. 2011;6(2):79–85. doi: 10.1116/1.3601755. [DOI] [PubMed] [Google Scholar]
- 32.Cao T, Longley MA, Wang XJ, Roop DR. An inducible mouse model for epidermolysis bullosa simplex: Implications for gene therapy. J Cell Biol. 2001;152(3):651–656. doi: 10.1083/jcb.152.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hesse M, Franz T, Tamai Y, Taketo MM, Magin TM. Targeted deletion of keratins 18 and 19 leads to trophoblast fragility and early embryonic lethality. EMBO J. 2000;19(19):5060–5070. doi: 10.1093/emboj/19.19.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreplak L, Bär H, Leterrier JF, Herrmann H, Aebi U. Exploring the mechanical behavior of single intermediate filaments. J Mol Biol. 2005;354(3):569–577. doi: 10.1016/j.jmb.2005.09.092. [DOI] [PubMed] [Google Scholar]
- 35.Wang N, Stamenović D. Contribution of intermediate filaments to cell stiffness, stiffening, and growth. Am J Physiol Cell Physiol. 2000;279(1):C188–C194. doi: 10.1152/ajpcell.2000.279.1.C188. [DOI] [PubMed] [Google Scholar]
- 36.Brown MJ, Hallam JA, Colucci-Guyon E, Shaw S. Rigidity of circulating lymphocytes is primarily conferred by vimentin intermediate filaments. J Immunol. 2001;166(11):6640–6646. doi: 10.4049/jimmunol.166.11.6640. [DOI] [PubMed] [Google Scholar]
- 37.Plodinec M, et al. The nanomechanical properties of rat fibroblasts are modulated by interfering with the vimentin intermediate filament system. J Struct Biol. 2011;174(3):476–484. doi: 10.1016/j.jsb.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Peters B, Kirfel J, Büssow H, Vidal M, Magin TM. Complete cytolysis and neonatal lethality in keratin 5 knockout mice reveal its fundamental role in skin integrity and in epidermolysis bullosa simplex. Mol Biol Cell. 2001;12(6):1775–1789. doi: 10.1091/mbc.12.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan Y, et al. A human keratin 14 “knockout”: The absence of K14 leads to severe epidermolysis bullosa simplex and a function for an intermediate filament protein. Genes Dev. 1994;8(21):2574–2587. doi: 10.1101/gad.8.21.2574. [DOI] [PubMed] [Google Scholar]
- 40. Turksen K (2005) Epidermal Cells: Methods and Protocols (Springer, New York), p 480.
- 41.Waschbüsch D, et al. Presenilin 1 affects focal adhesion site formation and cell force generation via c-Src transcriptional and posttranslational regulation. J Biol Chem. 2009;284(15):10138–10149. doi: 10.1074/jbc.M806825200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sneddon IN. The relation between load and penetration in the axisymmetric Boussinesq problem for a punch of arbitrary profile. Int J Eng Sci. 1965;3(1):47–57. [Google Scholar]
- 43.Rosenbluth MJ, Lam WA, Fletcher DA. Force microscopy of nonadherent cells: a comparison of leukemia cell deformability. Biophys J. 2006;90(8):2994–3003. doi: 10.1529/biophysj.105.067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.