Significance
The GATA-1 transcription factor has been extensively characterized and shown to play crucial roles in the development of erythroid cells, magakaryocytes, eosinophils, and mast cells. However, the role of GATA-1 in basophils remained unidentified. We demonstrate that knockdown of Gata1 gene expression in basophils results in impaired cytokine production upon allergen-mediated activation. Moreover, ΔdblGATA mice carrying the mutated Gata1 promoter have reduced numbers of basophils and their progenitors and show impaired responses in basophil-mediated protective immunity against parasitic infections. Thus, GATA-1 plays an important role in both generation and activation of basophils, and ΔdblGATA mice display numerical and functional aberrancy in basophils, in addition to the well-known eosinophil deficiency.
Abstract
Developmental processes of hematopoietic cells are orchestrated by transcriptional networks. GATA-1, the founding member of the GATA family of transcription factors, has been demonstrated to play crucial roles in the differentiation of erythroid cells, magakaryocytes, eosinophils, and mast cells. However, the role of GATA-1 in basophils remains elusive. Here we show that basophils abundantly express Gata1 mRNAs, and that siRNA-mediated knockdown of Gata1 resulted in impaired production of IL-4 by basophils in response to the stimulation with IgE plus antigens. ΔdblGATA mice that carry the mutated Gata1 promoter and are widely used for functional analysis of eosinophils owing to their selective loss of eosinophils showed a decreased number of basophils with reduced expression of Gata1 mRNAs. The number of basophil progenitors in bone marrow was reduced in these mice, and the generation of basophils from their bone marrow cells in culture with IL-3 or thymic stromal lymphopoietin was impaired. ΔdblGATA basophils responded poorly ex vivo to stimulation with IgE plus antigens compared with wild-type basophils as assessed by degranulation and production of IL-4 and IL-6. Moreover, ΔdblGATA mice showed impaired responses in basophil-mediated protective immunity against intestinal helminth infection. Thus, ΔdblGATA mice showed numerical and functional aberrancy in basophils in addition to the known deficiency of eosinophils. Our findings demonstrate that GATA-1 plays a key role in the generation and function of basophils and underscore the need for careful distinction of the cell lineage responsible for each phenotype observed in ΔdblGATA mice.
The differentiation of hematopoietic cells from pluripotent progenitors is regulated by the coordinated action of transcription factors (1). GATA proteins comprise a family of transcription factors that have highly conserved zinc finger DNA binding domains (2, 3). GATA-1, GATA-2, and GATA-3 among six members play major roles in the hematopoietic and immune systems (4). Each GATA factor shows a tissue- and cell-restricted pattern of expression. GATA-1 is expressed in erythroid cells, megakaryocytes, mast cells, and eosinophils among hematopoietic lineages (5–9), and in Sertoli cells of the testis (10). The critical role for GATA-1 in erythropoiesis has been clearly illustrated by establishing GATA-1−null mice that die during embryogenesis due to severe anemia (11). A series of engineered mice carrying genetic modifications in the promoter region of the Gata1 gene have been established (12–15), and some of these are not embryonic lethal, despite displaying anemia. Studies using these mutant mice revealed that GATA-1 also plays important roles in the development of platelets, mast cells, and eosinophils (12–18). ΔdblGATA mice were generated by deleting a high-affinity double GATA site in the Gata1 promoter region (14). The double GATA site is also present in the regulatory regions of eosinophil-specific genes (19). Of note, ΔdblGATA mice show selective loss of the eosinophil lineage, with only mild anemia but no apparent anomaly in platelets and mast cells, and therefore are widely used as eosinophil-deficient mice for the analysis of eosinophil function in vivo (19).
Basophils are the least common granulocytes, and represent less than 1% of peripheral blood leukocytes (20). In addition, they share certain features with tissue-resident mast cells, including the presence of basophilic granules in the cytoplasm, the surface expression of the high-affinity Fc receptor for IgE (FcεRI), and the release of chemical mediators, such as histamine, after stimulation. Accordingly, basophils have often erroneously been considered as minor and redundant relatives of mast cells or blood-circulating precursors of tissue-resident mast cells, and have long been neglected in immunological studies (21). However, recent development of analytical tools for basophil function in vivo, including basophil-deficient mice, has identified pivotal and nonredundant roles for basophils in a variety of immune responses, such as allergic reactions, protective immunity against parasitic infections, and regulation of innate and acquired immunity (22–28). Nevertheless, the origin and developmental pathway of basophils, including transcription factors regulating their differentiation, still remain ill-defined compared with those of other hematopoietic cells. Although a developmental relationship between basophils and eosinophils has been suggested in humans (29), a bipotent progenitor of basophils and mast cells, in addition to a unipotential basophil progenitor, has been identified in mice (30, 31), suggesting a closer lineage relationship of basophils with mast cells in mice. The ordered expression of two transcription factors, GATA-2 and C/EBPα, appears to regulate the commitment of progenitor cells into the basophil lineage (30, 32). Of note, mice deficient in the distal promoter-derived Runt-related transcription factor 1 (P1-Runx1) show severe basophilopenia with no apparent anomaly of mast cell, neutrophils, or eosinophils (33), indicating the requirement of P1-Runx1 for the later stage of basophil development. Recent studies demonstrated that basophils express GATA-1 (30, 32, 34), but its functional significance remains to be determined.
In the present study, we explored the possible involvement of GATA-1 in the ontogeny and function of murine basophils. Knockdown of Gata1 gene expression in basophils resulted in impaired production of IL-4 in vitro in response to stimulation with IgE plus antigens. Moreover, ΔdblGATA mice had basophilopenia with reduced expression of Gata1 and poor IL-4 production in basophils, and showed impaired responses in basophil-mediated protective immunity against intestinal helminth infection. Thus, GATA-1 plays an important role in both generation and activation of basophils, and ΔdblGATA mice display aberrancy in basophils, in addition to eosinophil deficiency.
Results
Knockdown of Gata1 in Basophils Impairs IL-4 Production in Response to Stimulation with IgE Plus Antigens.
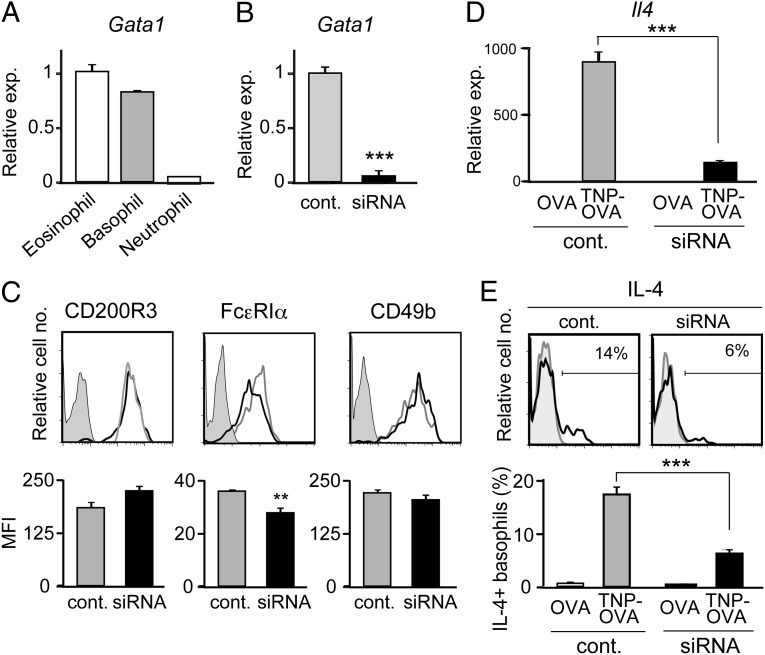
We first compared the level of Gata1 mRNA expression in three distinct types of murine granulocytes. Basophils expressed Gata1 mRNAs at the level that was as high as ∼80% of that in eosinophils, whereas neutrophils showed little or no expression (Fig. 1A). To examine the functional significance of Gata1 expression in basophils, Gata1-specific siRNAs were introduced into IL-3−cultured bone marrow-derived basophils to repress Gata1 expression (Fig. 1B). Gata1-knockdown basophils reproducibly showed a slightly lower level of surface FcεRIα expression compared with control basophils, whereas surface CD200R3 and CD49b expression was comparable between them (Fig. 1C). When stimulated with IgE plus antigens, Gata1-knockdown basophils produced significantly lower amounts of IL-4 than did control siRNA-treated basophils at both mRNA and protein levels (Fig. 1 D and E). Theses results suggested that GATA-1 in basophils contribute to the regulation of FcεRI expression, and IL-4 production triggered by FcεRI cross-linking.
Fig. 1.
Knockdown of Gata1 in basophils impairs their IL-4 production. (A) Eosinophils, basophils, and neutrophils were separately isolated from the bone marrow of BALB/c mice, and subjected to RT-PCR analysis. The relative expression of Gata1 is shown (mean ± SEM, n = 3 each); the level of expression in eosinophils is set as 1. (B−E) Gata1-specific or control siRNAs were introduced into IL-3−cultured basophils generated from BALB/c bone marrow cells. Two days later, siRNA-treated basophils were subjected to RT-PCR analysis for Gata1 expression (B, mean ± SEM, n = 3 each), and flow cytometric analysis for indicated surface markers (C). In C, representative staining profiles are shown (Upper); gray, black, and shaded histograms indicate those of control and Gata1 siRNA-treated basophils, and control staining with isotype-matched antibodies, respectively. The mean fluorescence intensity (MFI) of each surface marker is shown (Lower) (mean ± SEM, n = 3 each). In D and E, basophils treated with Gata1-specific or control siRNAs were stimulated with TNP-specific IgE plus TNP-OVA or control OVA for 3 h (D) or 6 h (E), and subjected to RT-PCR analysis for Il4 expression (D, mean ± SEM, n = 3 each) or flow cytometric analysis for IL-4 production (E). In E, representative staining profiles are shown (Upper); black and gray histograms indicate those of TNP-OVA and OVA-treated basophils, respectively. The percentage of IL-4–producing cells among basophils is shown (Lower) (mean ± SEM, n = 3 each). Data in A−E are representative of at least three independent experiments. **P < 0.01; ***P < 0.001.
ΔdblGATA Mice Show Basophilopenia with Reduced Expression of Gata1 and Surface FcεRI in Basophils.
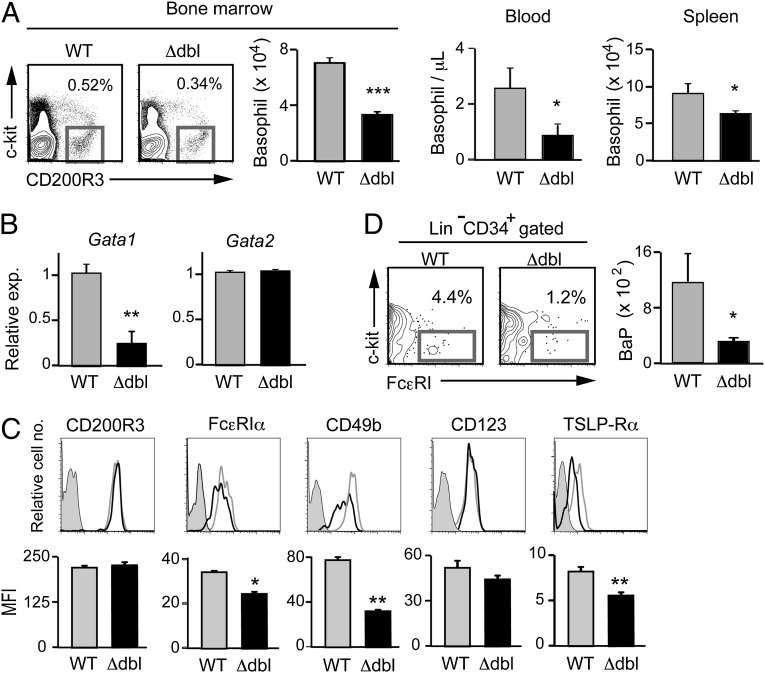
The results obtained from the Gata1 knockdown experiments prompted us to examine ΔdblGATA mice for possible aberrancy of their basophils. In contrast to the nearly complete loss of eosinophils, basophils were detectable in ΔdblGATA BALB/c mice. However, the number of basophils was significantly reduced in these mice, compared with that in wild-type mice, particularly in the bone marrow and peripheral blood, to approximately a half and one-third of normal, respectively (Fig. 2A). Similar basophilopenia was observed in ΔdblGATA C57BL/6 mice compared with control littermates (Fig. S1). Of note, the Gata1 mRNA expression in ΔdblGATA basophils was not null but reduced to approximately a quarter of that in wild-type basophils, whereas the level of Gata2 mRNAs was comparable between them (Fig. 2B). Flow cytometric analysis revealed that the expression of cell surface CD200R3 was equivalent between them, whereas the expression of FcεRIα and CD49b was slightly but significantly reduced in ΔdblGATA basophils (Fig. 2C). Considering the fact that the Gata1 gene is expressed in basophil progenitors (30), the discrepancy between the reduced and unaltered expression of CD49b on ΔdblGATA basophils (Fig. 2C) and siRNA-treated basophils (Fig. 1C), respectively, could be attributed to the effect of reduced versus normal GATA-1 expression on the process of basophil development.
Fig. 2.
ΔdblGATA BALB/c mice show basophilopenia with a reduced number of basophil progenitors. (A) Numbers of basophils, defined as FSClowSSClowCD200R3+c-kit− cells, in the bone marrow, peripheral blood, and spleen of wild-type (WT) and ΔdblGATA (Δdbl) BALB/c mice (mean ± SEM, n = 3 each). (B) Basophils isolated from the bone marrow of WT and Δdbl mice were subjected to RT-PCR analysis. The relative expression of Gata1 and Gata2 mRNAs is shown (mean ± SEM, n = 4 each); the level of expression in WT basophils is set as 1. (C) Cell surface phenotype of bone marrow basophils isolated from WT and Δdbl mice. Representative staining profiles are shown (Upper); gray, black, and shaded histograms indicate those of WT and Δdbl basophils and control staining with isotype-matched antibodies, respectively. MFI of each surface marker is shown (Lower) (mean ± SEM, n = 4 each). (D) Numbers of basophil progenitors (BaP) in the bone marrow of WT and Δdbl mice (mean ± SEM, n = 4 each). Data in A−D are representative of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
ΔdblGATA Mice Have a Reduced Number of Basophil Progenitors and Show Poor Generation of Basophils.
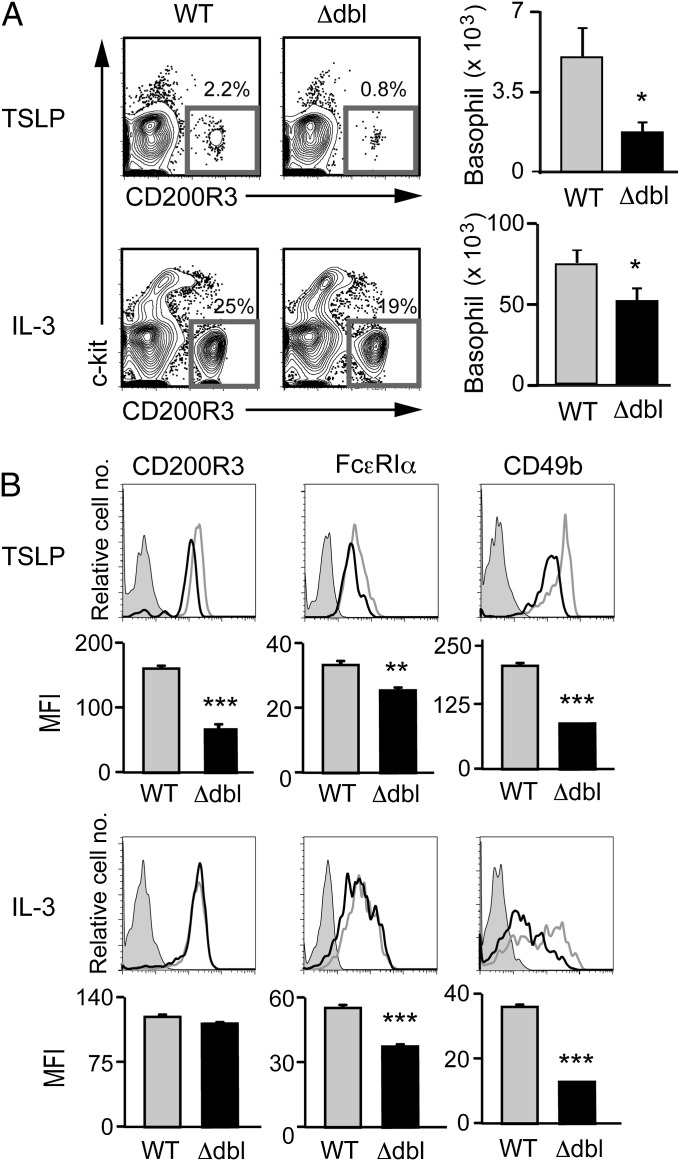
Basophilopenia in the bone marrow suggested the possible impairment of basophil hematopoiesis in ΔdblGATA mice. Indeed, the number of CD34+c-kit−FcεRI+ basophil progenitors (30) in the bone marrow of ΔdblGATA mice was only a quarter of that in wild-type mice (Fig. 2D). A recent study demonstrated that thymic stromal lymphopoietin (TSLP) and IL-3 independently promote basophil hematopoiesis (35). We found that the surface expression of TSLP-Rα on bone marrow basophils was significantly lower in ΔdblGATA mice than in wild-type mice, whereas IL-3R (CD123) expression was not significantly different between them (Fig. 2C). This prompted us to compare the generation of basophils from bone marrow cells isolated from wild-type and ΔdblGATA mice, when cultured ex vivo with TSLP or IL-3. ΔdblGATA bone marrow cells generated only one-third of CD200R3+c-kit− basophils in a 5-d culture with TSLP, compared with wild-type cells (Fig. 3A, Upper). The IL-3−elicited generation of basophils was also impaired in ΔdblGATA bone marrow cells, albeit to a lesser extent than the TSLP-elicited ones (Fig. 3A, Lower). Thus, ΔdblGATA mice showed poorer generation of basophils both in vivo and ex vivo than wild-type mice. In accordance with a previous report (35), the levels of surface marker expression differed between IL-3– and TSLP-elicited basophils even from wild-type mice (Fig. 3B). Notably, as observed in primary basophils, both IL-3– and TSLP-elicited basophils generated from bone marrow cells of ΔdblGATA mice displayed reduced expression of FcεRIα and CD49b, compared with those from wild-type mice (Fig. 3B).
Fig. 3.
ΔdblGATA bone marrow cells show impaired generation of basophils when cultured ex vivo with TSLP or IL-3. Bone marrow cells isolated from WT or Δdbl BALB/c mice were cultured ex vivo with TSLP or IL-3 for 5 d. (A) Basophils were identified as FSClowSSClowCD200R3+c-kit− cells as shown at Left, and the number of basophils in each group is summarized at Right (mean ± SEM, n = 4 each). (B) The cell surface phenotype of bone marrow-derived basophils. Representative staining profiles are shown (TSLP and IL-3 rows); gray, black, and shaded histograms indicate those of WT and Δdbl basophils and control staining with isotype-matched antibodies, respectively. MFI of each surface marker is shown (MFI rows) (mean ± SEM, n = 4 each). Data shown are representative of at least two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
ΔdblGATA Basophils Show Impaired Degranulation and Cytokine Production ex Vivo.
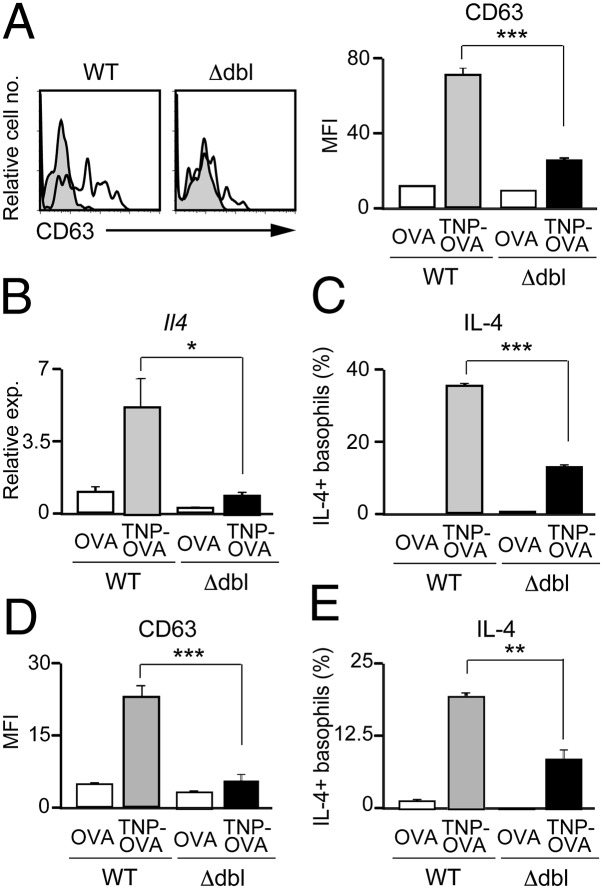
We next examined the functional consequence of reduced Gata1 expression in ΔdblGATA basophils. When stimulated with IgE plus antigens, IL-3−elicited ΔdblGATA basophils showed poorer responses in up-regulation of surface CD63 expression (Fig. 4A) and release of β-hexosaminidase (Fig. S2A) than wild-type counterparts, indicating impaired degranulation of ΔdblGATA basophils. Moreover, they produced significantly lower amounts of IL-4 and IL-6 at both mRNA and protein levels than did wild-type basophils (Fig. 4 B and C and Fig. S3 A and B). When stimulated with phorbol ester and ionomycin, IL-4 production was comparable between IL-3−elicited wild-type and ΔdblGATA basophils (Fig. S2B), suggesting that the machinery necessary for IL-4 production remained intact in ΔdblGATA basophils. Importantly, impaired degranulation and cytokine production was also detected in primary basophils freshly isolated from ΔdblGATA mice (Fig. 4 D and E and Figs. S3 C and D and S4).
Fig. 4.
ΔdblGATA basophils show defects in degranulation and cytokine production. (A−C) IL-3−elicited WT or Δdbl basophils were stimulated with anti-TNP IgE plus TNP-OVA or control OVA for 20 min (A), 3 h (B), or 6 h (C), and subjected to flow cytometric analysis for surface CD63 expression (A), RT-PCR analysis for Il4 expression (B), or flow cytometric analysis for IL-4 production (C). In A, open and shaded histograms indicate CD63 expression when stimulated with TNP-OVA and control OVA, respectively. (D and E) Primary basophils isolated from the bone marrow and spleen of WT or Δdbl mice were stimulated with anti-TNP IgE plus TNP-OVA or control OVA for 20 min (D) or 6 h (E), and subjected to flow cytometric analysis for surface CD63 expression (D) or flow cytometric analysis for IL-4 production (E). Data shown are the mean ± SEM (n = 3 or 4 each), and representative of three independent experiments, and displayed as in Fig. 1. *P < 0.05; **P < 0.01; ***P < 0.001.
ΔdblGATA Mice Show Impaired Acquired Protection Against Helminth Infection Due to Their Basophil Anomaly.
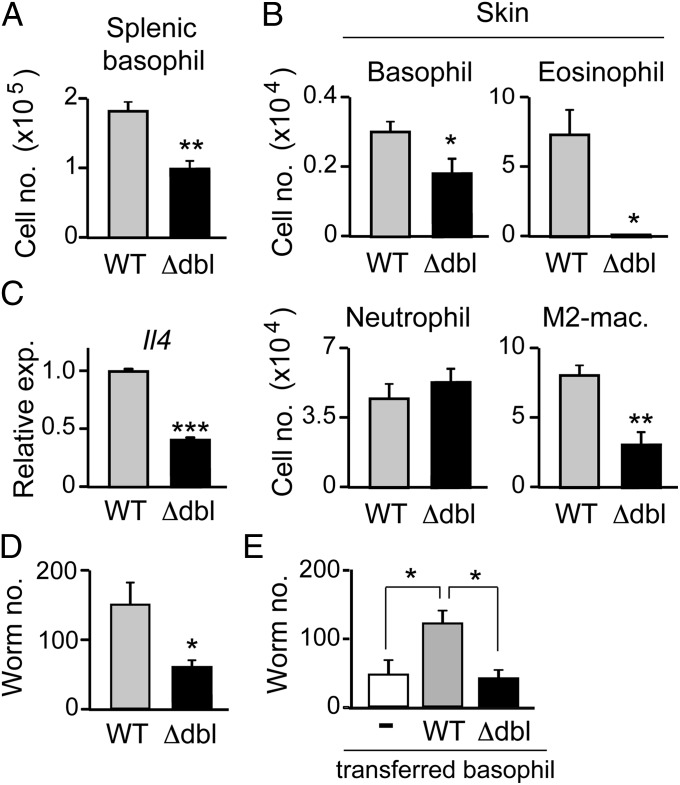
We then investigated whether the numerical and functional aberrancy of basophils in ΔdblGATA mice indeed has any significant impact on in vivo immune responses. We have recently demonstrated that basophils but not eosinophils play a key role in the protection against reinfection of an intestinal helminth Nippostrongylus brasiliensis by means of restraining infectious larvae from migration out of their entry point in the skin toward the lung and intestine (36). IgE-armed basophils recruited to the skin lesions of the second larval infection are activated in response to N. brasiliensis antigens to secrete IL-4, which in turn acts on monocytes/macrophages to promote their differentiation into M2-type macrophages in the skin. Arginase 1 produced by M2-type macrophages is involved in the larval trapping in the skin. Depletion of either basophils or M2-type macrophages abolishes the larval trapping in the skin, but it has no significant impact on eosinophil accumulation in the infected skin (36). As shown in Fig. 5A, the number of basophils in the spleen of N. brasiliensis-infected ΔdblGATA mice was approximately half of that of N. brasiliensis-infected wild-type mice, although helminth-elicited basophilia was observed in both mice (compare Figs. 2A, Right, and 5A). This was also the case in the number of basophils infiltrating the skin of larva inoculation site during the second N. brasiliensis infection (Fig. 5B). Moreover, the amount of Il4 mRNAs per cell in skin-infiltrating basophils isolated from ΔdblGATA mice was less than half of that isolated for wild-type mice (Fig. 5C). In accordance with the reduced number and IL-4 production of basophils, the number of PD-L2+ M2-type macrophages generated in the skin lesion of ΔdblGATA mice was approximately one-third of that of wild-type mice (Fig. 5B). Importantly, the larval trapping in the skin during the second N. brasiliensis infection was significantly reduced in ΔdblGATA mice (Fig. 5D), and adoptive transfer of basophils isolated from wild-type but not ΔdblGATA mice normalized the larval trapping in ΔdblGATA mice (Fig. 5E). These results clearly demonstrated that ΔdblGATA mice have previously-unrecognized basophil anomaly that can affect immune responses in vivo.
Fig. 5.
ΔdblGATA mice show impaired acquired protection against intestinal helminths due to basophil anomaly. (A−D) WT and Δdbl BALB/c mice were infected twice with N. brasiliensis larvae, and on day 2 of the second infection, their spleen and skin of larva inoculation site were isolated, and subjected to numerical counts of basophils in the spleen (A), indicated cell types accumulating in the skin (B, Upper and Lower), and skin-trapped worms (D). M2-Mac, M2-type macrophage. In C, basophils were sorted from the skin preparation and subjected to RT-PCR analysis for Il4 expression. (E) WT and Δdbl mice were infected once with N. brasiliensis larvae, and basophils were sorted from their bone marrow cells on day 18 of infection. These N. brasiliensis-sensitized basophils isolated from WT or Δdbl mice were intraperitoneally transferred into Δdbl mice (4 × 104 cells per mouse) that had been infected with larvae 18 d before. On the day of cell transfer, the recipient mice were subjected to the second infection, and 2 d later, the skin of the larva inoculation site was isolated, and subjected to a numerical count of larvae. In parallel, twice-infected Δdbl mice without basophil transfer (−) were analyzed as control. Data shown are the mean ± SEM (n = 3 or 4 each), and representative of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
Basophils express GATA-1 and GATA-2 but not GATA-3 among hematopoietic GATA transcription factors (34). GATA-2 has been demonstrated to play a key role in basophil hematopoiesis (30, 32), and positively regulate ST2 (IL-33 receptor) expression in basophils (37). In contrast, the role of GATA-1 in basophils remained to be identified despite the fact that GATA-1 has been extensively characterized in terms of its functions in the development of erythroid cells, megakaryocytes, eosinophils, and mast cells (11–18). We illustrated in the present study that GATA-1 plays an important role in both generation and function of basophils, and that ΔdblGATA mice show numerical and functional aberrancy in basophils, in addition to the known eosinophil deficiency.
Although several cell types, including Th2 cells and basophils, can produce IL-4, each cell type uses distinct cis-regulatory element for the Il4 gene expression (38). Basophils, unlike Th2 cells, do not express Th2 transcription factors GATA3, RBPJκ, or c-Maf (34). Instead, basophils express GATA-1, GATA-2, and C/EBPα in contrast to Th2 cells. C/EBPα has been shown to directly regulate Il4 gene transcription in basophils, in cooperation with NFAT (34). Although GATA-1 and GATA-2 have been reported to bind to the intronic enhancer of the Il4 gene in mast cells (39), basophils used the HS4 element rather than the intronic enhancer as a lineage-specific Il4 enhancer (38). Thus, the role of GATA-1 and GATA-2 in the Il4 gene regulation in basophils remained uncertain. In the present study, we illustrated the key role for GATA-1 in IL-4 production by basophils. Gata1 knockdown resulted in significantly reduced IL-4 expression at both mRNA and protein levels by basophils when stimulated with IgE plus antigens. In accordance with this, ΔdblGATA basophils expressing reduced amounts of Gata1 mRNAs also showed impaired production of IL-4. In both Gata1-knockdown basophils and ΔdblGATA basophils, the surface FcεRI expression was slightly but significantly reduced, compared with that in normal counterparts, as observed in mast cells from GATA-1low and GATA-KD mice that are genetically engineered to have mutations in the promoter region of the Gata1 gene, and hence show reduced expression of GATA-1 (18, 40). However, the extent of MAPK (ERK1/2 and p38) phosphorylation in response to FcεRI-mediated stimulation was comparable between mutant and wild-type basophils (Fig. S5). Therefore, impaired IL-4 production in mutant basophils may not be attributed to the reduced FcεRI expression, and is likely due to a reduced amount of GATA-1 that regulates Il4 gene expression.
GATA-1 has been shown to be involved in the differentiation of mast cells. Morphologically abnormal alcian blue+ mast cells and numerous mast cell precursors are detected in connective tissues and peritoneal lavage of GATA-1low and GATA-KD mice (17, 18). Moreover, in vitro generation of c-kit+FcεRI+ mast cells from bone marrow cells is reduced in these mice (18). Thus, GATA-1 is required for generation and maturation of connective tissue mast cells. Although a close developmental relationship between mast cells and basophils has been suggested in mice (30, 31), the functional significance of GATA-1 expression in basophils in terms of their development remained to be clarified. In the present study, we found that ΔdblGATA mice had a decreased number of basophils with reduced expression of Gata1 mRNAs. ΔdblGATA bone marrow cells generated fewer basophils ex vivo in culture with IL-3 or TSLP than did wild-type cells, in accordance with a decreased number of basophil progenitors in the bone marrow of ΔdblGATA mice, suggesting that GATA-1 is involved in the development of basophils. This reduced basophilopoiesis does not seem to be linked to eosinophil deficiency, because two other mouse strains with eosinophil deficiency have a normal number of basophils (41). Because GATA-1 regulates GATA-2 expression (9), and GATA-2 is involved in the regulation of basophil development (30, 32), GATA-1 may contribute to the basophil development through GATA-2 regulation. However, GATA-2 expression in ΔdblGATA basophils remains unaltered despite significant reduction in GATA-1 expression, suggesting that GATA-1 may regulate the expression of a gene(s) involved in basophil differentiation, either independently of GATA-2 or by competing or cooperating with GATA-2. Of note, in ΔdblGATA mice, basophilopoiesis is not completely arrested as in the case of erythropoiesis. This could be attributed to the reduced but not null expression of GATA-1 in ΔdblGATA basophils, even though we cannot formally exclude the possibility that GATA-2 expressed in ΔdblGATA basophils may compensate some of GATA-1 functions. A previous study on ΔdblGATA mice demonstrated that in vitro generation of bone marrow-derived mast cells in culture with IL-3 and stem cell factor seems intact, assessed by toluidine blue staining and flow cytometric analysis of c-kit and FcεRI expression (14). Therefore, the ΔdblGATA mutation appears to impair the generation of eosinophils and basophils but not mast cells.
ΔdblGATA mice have been widely used for functional analysis of eosinophils because they show almost complete loss of the eosinophil lineage, with only mild anemia but no apparent anomaly in platelets and mast cells (14). We illustrated in the present study that ΔdblGATA mice showed impaired acquired protection against hilminth infection due to basophil anomaly. Considering recent advances in our understanding of functional significance of basophils in various immune responses (22–28), this finding raises concern about the possibility that certain functions of basophils might have been erroneously interpreted as those of eosinophils in studies using ΔdblGATA mice as eosinophil-deficient mice. Two distinct eosinophil-deficient mouse strains, ΔdblGATA and PHIL, were analyzed to clarify the role of eosinophils in pathogenesis of asthma, resulting in some conflicting results (42, 43). This discrepancy reportedly stems from the different genetic background of these mice (44), but may also in part come from the difference in quantity and quality of basophils between these two strains. Thus, the present study underscores the need for careful distinction of the cell lineage responsible for each phenotype observed in ΔdblGATA mice, by means of parallel analyses with other eosinophil-deficient models and basophil-deficient mice.
Materials and Methods
Mice.
Wild-type and ΔdblGATA BALB/c mice (14) were purchased from CLEA Japan and Jackson Laboratory, respectively. ΔdblGATA BALB/c mice were backcrossed to C57BL/6 mice for five generations to obtain ΔdblGATA C57BL/6 mice. All animal studies were approved by the Institutional Animal Care and Use Committee of Tokyo Medical and Dental University.
Antibodies.
Biotinylated anti-CD49b (DX5), PE-conjugated anti-CD200R3 (Ba13) and anti-CD63 (NVG-2), PE-Cy7-conjugated anti−c-kit (2B8), APC-conjugated anti-CD200R3 (Ba13), IL-4 (11B11) and CD34 (HM34), and FITC-conjugated CD4 (RM4-5), CD8α (53-6.7), CD11b (M1/70), B220 (RA3-6B2), Gr-1 (RB6-8C5), and anti-FcεRIα (MAR-1) were purchased from BioLegend. Biotinylated anti-CD123 (5B11), PE-conjugated IL-6 (MP5-20F3), APC-conjugated anti-CD49b (HMα2), FITC-conjugated CD11c (HL3), Ax488-conjugated anti-phosphorylated ERK1/2 (20A), Ax647-conjugated anti-phosphorylated p38 MAPK (36/p38), and streptavidin were from BD Pharmingen. PE-conjugated anti−TSLP-Rα was from R&D Systems.
Quantitative RT-PCR.
Total mRNAs from cells were isolated by Reliaprep RNA cell miniprep system (Promega). The first-strand cDNAs were generated with reverse transcription using oligo-dT, random primers (Life Technologies Corporation), and ReverTra Ace-α (Toyobo). Quantitative PCR of the cDNA was performed on Applied Biosystems StepOnePlus Real-Time PCR system using a Fast SYBR Green Master Mix (Life Technologies Corporation) and the following primer sets: for Gata1, forward 5′-CACTCCCCAGTCTTTCAGGTGTA-3′ and reverse 5′-GGTGAGCCCCCAGGAATT-3′; for Gata2, forward 5′-CACCTGTTGTGCAAATTGTCAGA-3′ and reverse 5′-GGATCCCTTCCTTCTTCATGGT-3′; for Il4, forward 5′-ACTTGAGAGAGATCATCGGCA-3′ and reverse 5′-AGCTCCATGAGAACACTAGAGTT-3′; for Il6, forward 5′-CTGCAAGAGACTTCCATCCAG-3′ and reverse 5′-AGTGGTATAGACAGGTCTGTTGG-3′; and for Hprt, forward 5′-GGCCAGACTTTGTTGGATTTG-3′ and reverse 5′-CGCTCATCTTAGGCTTTGTATTTG-3′.
Gene expression was analyzed using Hprt as an endogenous control in each sample.
Flow Cytometric Analysis and Cell Preparation.
For flow cytometric analysis, cells were preincubated with anti-CD16/32 mAb and normal rat serum on ice for 15 min before incubation with the indicated combination of Abs to prevent the nonspecific binding of irrelevant Abs. Stained cells were analyzed using FACSCanto II (BD Biosciences). Basophils were isolated from bone marrow cells using biotinylated anti-CD49b antibody and streptavidin-conjugated magnetic particles (BD Pharmingen), followed by sorting FSClowSSClowCD200R3+c-kit− cells with FACSAria II (BD Bioscience). Eosinophils (FSClowSSChighSiglec-F+) and neutrophils (Gr-1high) were directly sorted from bone marrow cells. Basophil progenitors were identified as Lin (CD4, CD8, B220, CD11b, Gr-1, CD11c)−CD34+c-kit−FcεRI+ cells in the bone marrow (30). Bone marrow-derived basophils were generated by culturing bone marrow cells ex vivo with 1 ng/mL of IL-3 or 1 μg/mL of TSLP for 5 d.
Knockdown of Gata1.
Gata1-specific siRNA (Silencer Predesigned siRNA, no. 66474, Life technologies Corporation) and control siRNA (Silencer Negative Control #1 siRNA) were introduced into IL-3−cultured bone marrow-derived basophils by using Neon Transfection System (Life Technologies Corporation).
Stimulation of Basophils.
Basophils were enriched from freshly isolated bone marrow cells or IL-3−cultured bone marrow cells by using biotinylated anti-CD49b antibody and streptavidin-conjugated magnetic particles (BD Pharmingen). For RT-PCR analysis, FSClowSSClowc-kit−CD45lowCD49b+ basophils were further sorted from them. Basophil-enriched CD49b+ cells or purified basophils were first sensitized with 1 μg/mL of hapten 2,4,6-trinitrophenol (TNP)-specific IgE, followed by incubation with 100 ng/mL of TNP-conjugated ovalbumin (OVA) or control OVA for 20 min for flow cytometric analysis of the CD63 up-regulation, for 30 min for β-hexosaminidase release assay (Sigma-Aldrich), for 3 h for RT-PCR analysis, and for 6 h in the presence of momesin for cytokine production. For detection of cytokine production, stimulated cells were subjected to fixation and permeabilization using Cytofix/Cytoperm buffer (BD Pharmingen), followed by intracellular staining with antibodies specific to IL-4 or IL-6. In some experiments, basophils were stimulated for 6 h with phorbol 12-myristate 13-acetate (PMA) (0.1 μg/mL, Sigma-Aldrich) plus ionomycin (0.5 μg/mL, Sigma-Aldrich).
Helminth Infection.
Mice were first injected s.c. with 500 third-stage larvae (L3) of N. brasiliensis in the back, and 18 d later, they were injected intradermally with 500 L3 in the flank. Two days after the second larva inoculation, the skin of the inoculation site was isolated, and subjected to numerical counts of skin-trapped larvae and skin-infiltrating cells as described previously (36).
Statistical Analysis.
Statistical analysis was performed using unpaired Student t test. A P value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists (to K.O.-N.) and by research grants from JST, CREST (to H. Karasuyama), JST, PRESTO (to S.N.), and the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to H. Karasuyama).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311668110/-/DCSupplemental.
References
- 1.Orkin SH, Zon LI. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto M, et al. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 1990;4(10):1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- 3.Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285(41):31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80(3):575–581. [PubMed] [Google Scholar]
- 5.Romeo PH, et al. Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature. 1990;344(6265):447–449. doi: 10.1038/344447a0. [DOI] [PubMed] [Google Scholar]
- 6.Martin DI, Zon LI, Mutter G, Orkin SH. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990;344(6265):444–447. doi: 10.1038/344444a0. [DOI] [PubMed] [Google Scholar]
- 7.Zon LI, et al. Expression of mRNA for the GATA-binding proteins in human eosinophils and basophils: Potential role in gene transcription. Blood. 1993;81(12):3234–3241. [PubMed] [Google Scholar]
- 8.Migliaccio AR, Rana RA, Vannucchi AM, Manzoli FA. Role of GATA-1 in normal and neoplastic hemopoiesis. Ann N Y Acad Sci. 2005;1044:142–158. doi: 10.1196/annals.1349.019. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25(4):1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito E, et al. Erythroid transcription factor GATA-1 is abundantly transcribed in mouse testis. Nature. 1993;362(6419):466–468. doi: 10.1038/362466a0. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93(22):12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16(13):3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi S, et al. Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J Biol Chem. 1997;272(19):12611–12615. doi: 10.1074/jbc.272.19.12611. [DOI] [PubMed] [Google Scholar]
- 14.Yu C, et al. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195(11):1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majewski IJ, et al. A mutation in the translation initiation codon of Gata-1 disrupts megakaryocyte maturation and causes thrombocytopenia. Proc Natl Acad Sci USA. 2006;103(38):14146–14151. doi: 10.1073/pnas.0606439103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDevitt MA, Shivdasani RA, Fujiwara Y, Yang H, Orkin SH. A “knockdown” mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proc Natl Acad Sci USA. 1997;94(13):6781–6785. doi: 10.1073/pnas.94.13.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harigae H, et al. Differential roles of GATA-1 and GATA-2 in growth and differentiation of mast cells. Genes Cells. 1998;3(1):39–50. doi: 10.1046/j.1365-2443.1998.00166.x. [DOI] [PubMed] [Google Scholar]
- 18.Migliaccio AR, et al. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. J Exp Med. 2003;197(3):281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–174. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- 20.Galli SJ. Mast cells and basophils. Curr Opin Hematol. 2000;7(1):32–39. doi: 10.1097/00062752-200001000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Falcone FH, Haas H, Gibbs BF. The human basophil: A new appreciation of its role in immune responses. Blood. 2000;96(13):4028–4038. [PubMed] [Google Scholar]
- 22.Min B. Basophils: What they ‘can do’ versus what they ‘actually do’. Nat Immunol. 2008;9(12):1333–1339. doi: 10.1038/ni.f.217. [DOI] [PubMed] [Google Scholar]
- 23.Sullivan BM, Locksley RM. Basophils: A nonredundant contributor to host immunity. Immunity. 2009;30(1):12–20. doi: 10.1016/j.immuni.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Schroeder JT. Basophils beyond effector cells of allergic inflammation. Adv Immunol. 2009;101:123–161. doi: 10.1016/S0065-2776(08)01004-3. [DOI] [PubMed] [Google Scholar]
- 25.Sokol CL, Medzhitov R. Emerging functions of basophils in protective and allergic immune responses. Mucosal Immunol. 2010;3(2):129–137. doi: 10.1038/mi.2009.137. [DOI] [PubMed] [Google Scholar]
- 26.Karasuyama H, Mukai K, Obata K, Tsujimura Y, Wada T. Nonredundant roles of basophils in immunity. Annu Rev Immunol. 2011;29:45–69. doi: 10.1146/annurev-immunol-031210-101257. [DOI] [PubMed] [Google Scholar]
- 27.Siracusa MC, Comeau MR, Artis D. New insights into basophil biology: Initiators, regulators, and effectors of type 2 inflammation. Ann N Y Acad Sci. 2011;1217:166–177. doi: 10.1111/j.1749-6632.2010.05918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voehringer D. Protective and pathological roles of mast cells and basophils. Nat Rev Immunol. 2013;13(5):362–375. doi: 10.1038/nri3427. [DOI] [PubMed] [Google Scholar]
- 29.Arock M, Schneider E, Boissan M, Tricottet V, Dy M. Differentiation of human basophils: An overview of recent advances and pending questions. J Leukoc Biol. 2002;71(4):557–564. [PubMed] [Google Scholar]
- 30.Arinobu Y, et al. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci USA. 2005;102(50):18105–18110. doi: 10.1073/pnas.0509148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metcalf D, Ng AP, Baldwin TM, Di Rago L, Mifsud S. Concordant mast cell and basophil production by individual hematopoietic blast colony-forming cells. Proc Natl Acad Sci USA. 2013;110(22):9031–9035. doi: 10.1073/pnas.1307711110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwasaki H, et al. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20(21):3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukai K, et al. Critical role of P1-Runx1 in mouse basophil development. Blood. 2012;120(1):76–85. doi: 10.1182/blood-2011-12-399113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi X, Nishida J, Chaves L, Ohmori K, Huang H. CCAAT/enhancer-binding protein alpha (C/EBPalpha) is critical for interleukin-4 expression in response to FcepsilonRI receptor cross-linking. J Biol Chem. 2011;286(18):16063–16073. doi: 10.1074/jbc.M110.213389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siracusa MC, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 2011;477(7363):229–233. doi: 10.1038/nature10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obata-Ninomiya K, et al. The skin is an important bulwark of acquired immunity against intestinal helminths. J Exp Med. 2013 doi: 10.1084/jem.20130761. 10.1084/jem.20130761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baba Y, et al. GATA2 is a critical transactivator for the human IL1RL1/ST2 promoter in mast cells/basophils: Opposing roles for GATA2 and GATA1 in human IL1RL1/ST2 gene expression. J Biol Chem. 2012;287(39):32689–32696. doi: 10.1074/jbc.M112.374876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yagi R, Tanaka S, Motomura Y, Kubo M. Regulation of the Il4 gene is independently controlled by proximal and distal 3′ enhancers in mast cells and basophils. Mol Cell Biol. 2007;27(23):8087–8097. doi: 10.1128/MCB.00631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henkel G, Brown MA. PU.1 and GATA: Components of a mast cell-specific interleukin 4 intronic enhancer. Proc Natl Acad Sci USA. 1994;91(16):7737–7741. doi: 10.1073/pnas.91.16.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiyama C, et al. GATA-1 is required for expression of FcepsilonRI on mast cells: Analysis of mast cells derived from GATA-1 knockdown mouse bone marrow. Int Immunol. 2005;17(7):847–856. doi: 10.1093/intimm/dxh278. [DOI] [PubMed] [Google Scholar]
- 41.Doyle AD, et al. Expression of the secondary granule proteins major basic protein 1 (MBP-1) and eosinophil peroxidase (EPX) is required for eosinophilopoiesis in mice. Blood. 2013;122(5):781–790. doi: 10.1182/blood-2013-01-473405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JJ, et al. Defining a link with asthma in mice congenitally deficient in eosinophils. Science. 2004;305(5691):1773–1776. doi: 10.1126/science.1099472. [DOI] [PubMed] [Google Scholar]
- 43.Humbles AA, et al. A critical role for eosinophils in allergic airways remodeling. Science. 2004;305(5691):1776–1779. doi: 10.1126/science.1100283. [DOI] [PubMed] [Google Scholar]
- 44.Walsh ER, et al. Strain-specific requirement for eosinophils in the recruitment of T cells to the lung during the development of allergic asthma. J Exp Med. 2008;205(6):1285–1292. doi: 10.1084/jem.20071836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.