Significance
Development of cytotoxic T lymphocytes (CTLs) from activated CD8+ T cells is a key step of the antiviral immune response and is marked by the up-regulation of lytic molecules (perforin, granzymes). How this process is regulated at the posttranscriptional level is still largely unknown. Here we report that Dicer and microRNAs (miRNAs) restrict the expression of lytic molecules in mouse and human CTLs, and describe a unique signaling network that controls the expression of perforin, eomesodermin, and the IL-2Rα chain (CD25) downstream of IL-2 and inflammatory signals through miR-139 and miR-150 in differentiating CTLs.
Keywords: CD8+ T-cell response, posttranscriptional regulation
Abstract
Acquisition of effector properties is a key step in the generation of cytotoxic T lymphocytes (CTLs). Here we show that inflammatory signals regulate Dicer expression in CTLs, and that deletion or depletion of Dicer in mouse or human activated CD8+ T cells causes up-regulation of perforin, granzymes, and effector cytokines. Genome-wide analysis of microRNA (miR, miRNA) changes induced by exposure of differentiating CTLs to IL-2 and inflammatory signals identifies miR-139 and miR-150 as components of an miRNA network that controls perforin, eomesodermin, and IL-2Rα expression in differentiating CTLs and whose activity is modulated by IL-2, inflammation, and antigenic stimulation. Overall, our data show that strong IL-2R and inflammatory signals act through Dicer and miRNAs to control the cytolytic program and other aspects of effector CTL differentiation.
Differentiation of naïve CD8+ T cells into effector and memory cytotoxic T lymphocytes (CTLs) is driven by antigen (Ag) exposure and inflammation and orchestrated by the induction of specific transcription factors (1). How CTL differentiation is controlled at the posttranscriptional level is still largely unknown. MicroRNAs (miRNA, miR) constitute one of the main mechanisms of posttranscriptional regulation of protein levels; it is estimated that 30–90% of the mouse and human transcriptome is controlled by miRNAs (2). Primary miRNA transcripts (pri-miRNAs) are cleaved in the nucleus by the microprocessor (Drosha–DGR8) complex and further processed in the cytoplasm by the RNaseIII enzyme Dicer into their mature form (2, 3). In the hematopoietic system, several miRNAs are expressed in a stage- and cell-specific manner (4, 5). Deletion of Dicer at the double-negative or double-positive stage of thymocyte development severely impairs the differentiation and survival of αβ+ thymocytes (6) and of peripheral T cells, respectively (7). Global miRNA loss impairs the survival of antigen-specific effector CTLs during viral or bacterial infections (8, 9), and recently the miR-17-92 cluster and miR-155 (10, 11) have been shown to control the differentiation of mouse CTLs during antiviral and antitumor responses.
We previously showed that T-cell receptor (TCR) stimulation of naïve CD8+ T cells followed by expansion in 100 U/mL (high) IL-2 yielded cells that displayed the characteristics of effector CTLs (i.e., high perforin and granzyme B expression), whereas TCR stimulation followed by culture in 10 U/mL (low) IL-2 gave rise to cells with the surface and functional features of memory CTLs (including expression of the IL-7Rα receptor CD127) (12). In naïve T cells, perforin (Prf1) mRNA is clearly detectable, but this is not accompanied by protein expression (12); similarly, resting natural killer (NK) cells express Prf1 mRNA but not perforin protein (13). The posttranscriptional control of perforin and granzyme B expression in human NK cells is regulated by miRNAs that directly target the perforin or granzyme B 3′-UTR (14, 15).
Here we have used acute in vitro models of Dicer deletion or depletion to study the consequences of a global loss of mature miRNAs in CTLs. We find that Dicer-deficient CTLs resemble wild-type effector CTLs obtained by differentiation in the presence of high doses of IL-2 and inflammation in their increased expression of lytic molecules and effector cytokines. We identify five miRNAs that are down-regulated by inflammation in CTLs; of these, miR-139, and to a lesser extent miR-342, regulate perforin expression, whereas miR-150 regulates the expression of the IL-2 receptor α-chain (CD25). We show that strong IL-2 receptor and inflammatory signals down-regulate Dicer expression through a posttranscriptional mechanism, suggesting that CTL differentiation is modulated by changes in the expression level or activity of components of the miRNA machinery. Overall, our results point to the existence of a regulatory pathway downstream of extracellular signals (IL-2, inflammation, and Ag stimulation) that controls CTL differentiation through Dicer and miRNAs.
Results
Dicer Restrains Perforin Expression in Differentiating CTLs.
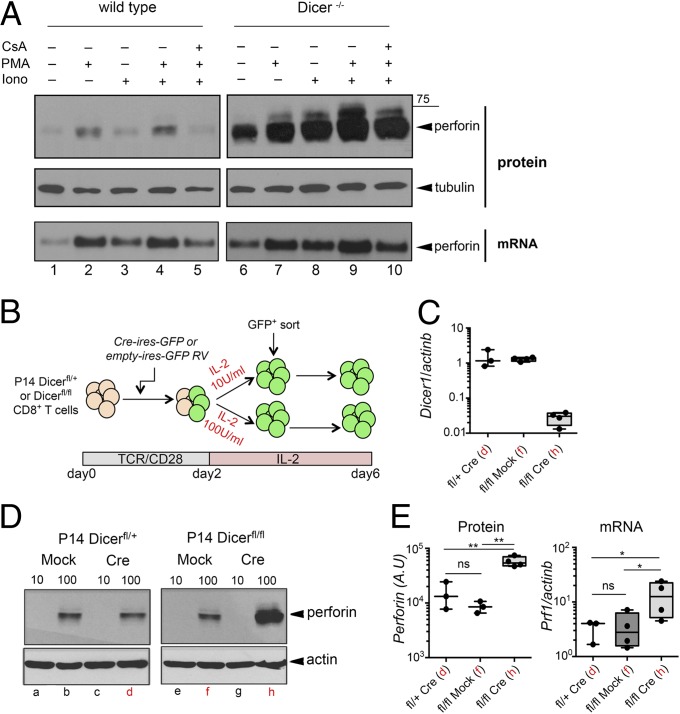
To analyze the phenotype of Dicer−/− CTLs, we activated CD8+ T cells from Dicerfl/fl:CD4-Cre mice and control Dicerfl/fl mice (7) with anti-CD3 and anti-CD28 Abs for 2 days, followed by culture in 100 U/mL IL-2 for 4 additional days, to yield in vitro generated effector CTLs (cells cultured in 10 U/mL IL-2 are termed “memory CTLs”) (12). Resting effector CTLs express perforin, which is further increased upon TCR restimulation. We examined perforin expression in resting effector CTLs from control and Dicer−/− mice, either resting or after 6-h restimulation with phorbol 12-myristate 13-acetate (PMA), ionomycin, or both (used to bypass any defects of proximal TCR signaling due to miRNA loss). The basal expression of perforin by wild-type CTLs was increased upon stimulation with PMA or PMA/ionomycin and was blocked by pretreatment with the calcineurin inhibitor cyclosporin A (CsA) (Fig. 1A). Dicer−/− CTLs expressed dramatically increased amounts of perforin protein relative to control CTLs, both in the resting state and upon restimulation (Fig. 1A). This up-regulation of perforin was accompanied by only a modest increase in Prf1 mRNA (Fig. 1A). Similar results were obtained when naïve CD8+ T cells from P14 TCR transgenic [whose TCR is specific for the lymphocytic choriomeningitis virus (LCMV) gp33 peptide (16)] Dicerfl/fl:CD4-Cre and control P14 Dicerfl/+:CD4Cre mice (17) were differentiated into effector and memory CTLs (Fig. S1A). Therefore, increased perforin expression was not the consequence of activation of peripheral T cells in Dicer−/− mice.
Fig. 1.
Regulation of perforin expression in Dicer−/− CTLs. (A) Perforin protein (Western blot) and mRNA (Northern blot) levels analyzed in effector CTLs differentiated in vitro from Dicerfl/fl (WT) and Dicerfl/fl:CD4-Cre (Dicer−/−) mice. On day 6, CTLs were restimulated with PMA, ionomycin, or both, in the presence or absence of CsA. Results are representative of three independent experiments. (B) Scheme of Cre-mediated Dicer deletion in activated CD8+ T cells from P14+:Dicerfl/fl mice. (C) Real-time PCR analysis of Dicer mRNA in control (P14+:Dicerfl/+-Cre or P14+:Dicerfl/fl-mock) and Dicer−/− effector CTLs analyzed on day 6. Values are normalized to the P14+:Dicerfl/+-Cre sample. Red letters on the x axis refer to Western blot lanes in D. Mean ± SE for three or four different experiments is shown. (D) Western blot analysis of perforin expression in resting control (P14+:Dicerfl/+-Cre, P14+:Dicerfl/+-mock–transduced, and P14+:Dicerfl/fl-mock–transduced) and Dicer−/− (P14+:Dicerfl/fl-Cre) effector and memory CTLs. Lanes marked with a red letter represent samples subsequently used for RNA sequencing. Results are representative of two (IL-2 10 U) or four (IL-2 100 U) experiments. (E) Quantification of perforin protein and mRNA levels in samples corresponding to lanes marked by red letters in D. Results from three or four experiments are averaged. *P < 0.05; **P < 0.01; ns, not significant.
Acute Deletion of Dicer in Activated CD8+ T Cells Promotes Effector CTL Differentiation.
To control for cells that have escaped Cre-mediated Dicer deletion in vivo as well as for possible compensatory effects (7), Dicer deletion was induced in activated CD8+ T cells isolated from P14+:Dicerfl/fl and P14+:Dicerfl/+ mice by retroviral introduction of Cre recombinase (Fig. 1 B and C). Transduced P14+:Dicerfl/fl CTLs that had undergone Cre-mediated Dicer deletion displayed strong up-regulation of perforin protein (Fig. 1D, compare lane h with lanes b, d, and f) and CD69 (Fig. S1B), consistent with previous reports of increased CD69 expression in Dicer−/− CTLs (8). The increase in perforin protein was paralleled by a smaller increase in Prf1 mRNA levels (Fig. 1E). We also profiled several activation and differentiation markers on control and Dicer−/− CTLs (Fig. S1B): Expression of CD44 and CD127 was unchanged, CD62L expression was slightly increased, LAG3 was strongly down-modulated, and expression of Eomes (eomesodermin), a T-box transcription factor that regulates perforin expression in CTLs (18) and is required for memory CD8+ T-cell survival (19), was modestly reduced in Dicer−/− CTLs. Dicer deletion in activated CD8+ T cells did not increase granzyme B expression (Fig. S1B). Upon restimulation, a substantial increase in IL-10 levels was also observed in Dicer−/− CTLs (Fig. S1C).
Together, these results indicate that Dicer deletion in activated CD8+ T cells recapitulates the effects of constitutive Dicer deletion in terms of perforin and CD69 up-regulation, and that the overall effect of Dicer and miRNAs is to restrict activation and the acquisition of effector functions by CTLs.
Effects of Dicer Knockdown on Human CD8+ T-Cell Differentiation.
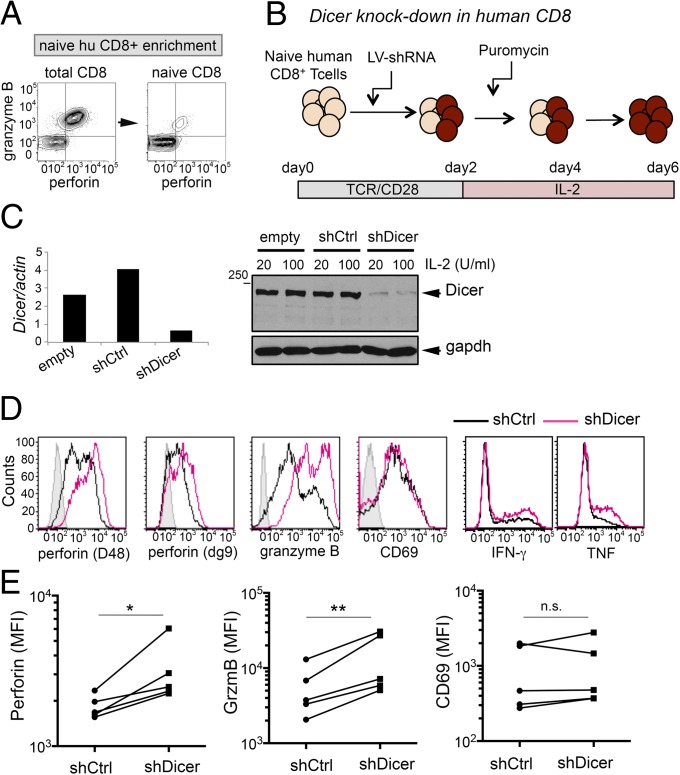
To investigate the role of Dicer in human CD8+ T cells, we enriched naïve CD8+ T cells from peripheral blood so that the starting population expressed very low levels of perforin and granzyme B (Fig. 2A), activated them with anti-CD3 plus anti-CD28 Ab, transduced them with lentiviral vectors containing shRNA against Dicer (shDicer) or scramble (shCtrl), and cultured them under puromycin selection for an additional 3 days in low or high IL-2 (Fig. 2B). Dicer was depleted at both mRNA and protein levels (Fig. 2C). In cells cultured with high IL-2, Dicer depletion resulted in strong up-regulation of granzyme B and increased expression of perforin, assessed with two different monoclonal antibodies to human perforin, D48 (20) and δg9 (Fig. 2 D and E). Restimulation with PMA and anti-CD3 resulted in a mild increase in IFN-γ and TNF expression (Fig. 2D). CD69 was not significantly up-regulated by Dicer knockdown in human CTLs (Fig. 2 D and E).
Fig. 2.
Effect of Dicer knockdown on human CTL differentiation. (A) Representative expression of perforin and granzyme B before and after naïve CD8+ T-cell enrichment from human peripheral blood mononuclear cells (PBMCs). (B) Experimental scheme of Dicer knockdown mediated by shRNA-encoding lentiviral (LV) vectors in human CTLs. (C) Real-time PCR (Left) and Western blot (Right) analysis of DICER mRNA and protein expression in human CTLs transduced with empty LV vector or LV encoding a control shRNA (shCtrl) or shRNA targeting Dicer (shDicer) and cultured with the indicated doses of IL-2 for 6 days. (D) Flow cytometric analysis of perforin (detected with the indicated antibody clones), granzyme B, and CD69 in resting human CTLs, transduced with shCtrl or shDicer. IFN-γ and TNF expression was analyzed after restimulation with anti-CD3 plus PMA. (E) Summary of the effect of Dicer depletion on perforin, granzyme B, and CD69 expression in five independent donors. *P < 0.05; **P < 0.01; MFI, mean fluorescence intensity; n.s., not significant.
In summary, depletion of Dicer in human CD8+ T cells recapitulates many of the effects observed in mouse cells in which the Dicer gene has been deleted either in thymocytes or activated peripheral CD8+ T cells. Up-regulation of perforin, CD69, and IL-10 is more pronounced in mouse CD8+ T cells, whereas up-regulation of granzyme B is evident mainly in human CD8+ T cells.
IL-2 and Inflammatory Signals Modulate Perforin and Dicer Expression.
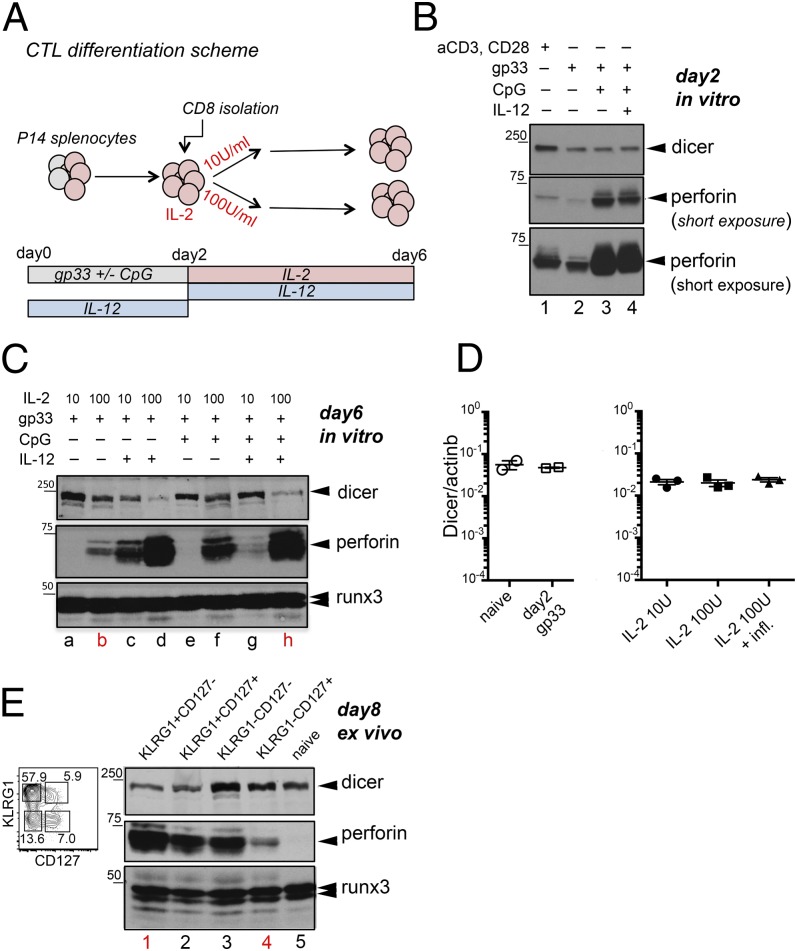
We previously showed that strong IL-2Rα signaling sustains perforin expression in CTLs, whereas inflammatory signals (CpG, IL-12) counteract the expression of perforin mRNA (12). To examine the influence of inflammation on perforin protein expression, we stimulated splenocytes from P14+:TCRα−/− mice with gp33 peptide with or without unmethylated CpG oligonucleotides, which activate B cells through TLR9; on day 2, CD8+ T cells were isolated and expanded for an additional 4 days with high or low IL-2 in the presence or absence of IL-12 (Fig. 3A). Perforin protein was absent in naïve CD8+ T cells (12) (Fig. 3E, lane 5) and slightly induced by stimulation of total splenocytes with gp33 or of purified CD8+ T cells with anti-CD3 plus anti-CD28 for 2 days (Fig. 3B, lanes 1 and 2). CpG strongly up-regulated perforin expression in CD8+ T cells on day 2 poststimulation (Fig. 3B, compare lanes 2 and 3). When added together with CpG, IL-12 did not further increase perforin expression after 2 days of stimulation (Fig. 3B, compare lanes 3 and 4) but, at a later time point (day 6), IL-12 markedly increased perforin protein levels (Fig. 3C, compare lanes a and b with lanes c and d). In high–IL-2 cultures, addition of IL-12, CpG, or both strikingly increased perforin expression (Fig. 3C, compare lanes b, d, f, and h); in contrast, in low–IL-2 cultures, the presence of CpG had no effect by itself, but suppressed the increase in perforin expression induced by IL-12 (Fig. 3C, compare lanes a, c, and g).
Fig. 3.
Inflammation regulates perforin and Dicer protein expression in differentiating CTLs. (A) Scheme of the in vitro differentiation system used to generate memory-like or effector CTLs from mouse P14 transgenic CD8+ T cells. (B) Perforin and Dicer expression measured by Western blot in activated CD8+ T cells 2 days after stimulation. Lane 1, lysates from CD8+ T cells stimulated for 2 days with anti-CD3 plus anti-CD28; lanes 2–4, lysates of CD8+ T cells isolated from splenocyte cultures 2 days after stimulation with gp33 ± CpG and IL-12 as indicated. (C) Perforin and Dicer expression measured by Western blot in CD8+ T cells activated with gp33 or gp33 plus CpG and differentiated for 6 days in the presence of high or low IL-2, with or without IL-12. Runx3 is used as a loading control. Results are representative of three independent experiments. (D) Real-time PCR analysis of Dicer mRNA levels in (Left) naïve and day 2 activated CD8+ T cells or (Right) day 6 CTLs derived from gp33-stimulated splenocytes and cultured with low or high IL-2, or IL-2 plus CpG and IL-12. (Mean +/− standard error.) (E) (Left) Sorting strategy of effector and memory CTL precursors based on the expression of CD127 and KLRG1, after gating on CD44high CD8+ T cells, in LCMV-infected B6 mice. (Right) Western blot analysis of perforin and Dicer expression in naïve CD8+ T cells and in the four populations sorted according to KLRG1 and CD127 expression. Results are representative of two experiments with 10–14 mice each. Lanes marked with a red letter or number represent samples subsequently used for RNA sequencing.
We also observed that Dicer itself was slightly down-regulated in CD8+ T cells purified from whole-splenocyte cultures after stimulation for 2 days with gp33 or gp33 plus CpG, relative to cells stimulated through TCR/CD28 but in the absence of antigen-presenting cells (Fig. 3B, compare lane 1 with lanes 2–4). On day 6, a clear down-regulation of Dicer protein, but not mRNA, was evident in CTLs differentiated with high IL-2 in the presence of CpG and IL-12 (Fig. 3 C and D). Additionally, inflammation coupled to strong IL-2 signals induced up-regulation of effector cytokines (IFN-γ, TNF, and IL-10) and CD25 and down-regulation of Eomes (Fig. S2).
To investigate whether inflammation modulates perforin and Dicer expression in vivo, we isolated four populations of CTLs based on KLRG1 and CD127 expression from mice infected with LCMV-Armstrong on day 8 postinfection (Fig. 3E, Left). KLRG1 and CD127 (IL-7Rα chain) are used to distinguish CD8+ T cells that have differentiated in vivo into fully armed effector CTLs from those that will preferentially generate long-lived memory cells (21). The two cell types, KLRG1+CD127– effector cells and KLRG1–CD127+ memory precursors, are thought to embark on these different differentiation routes because of exposure to higher versus lower levels of inflammatory stimuli (22). Notably, KLRG1+CD127– effector CD8+ T cells expressed higher amounts of perforin and lower amounts of Dicer protein compared with KLRG1–CD127+ memory precursor cells (Fig. 3E, Right, compare lanes 1 and 4). This inverse ratio was not observed in all populations; in fact, KLRG1–CD127– cells, which are known to contain precursors of both effector and memory CD8+ T cells (22), expressed high levels of both Dicer and perforin (Fig. 3E), suggesting that this population may contain cells that are Dicerhighperforinlow and vice versa. Overall, these data show that Dicer deletion recapitulates some of the effects exerted by strong IL-2 signaling and inflammatory stimuli on CTL differentiation, and suggest that IL-2 and inflammation may regulate the expression of miRNAs that restrain effector protein expression.
Inflammation Regulates the Expression of miRNAs Involved in CTL Differentiation and Function.
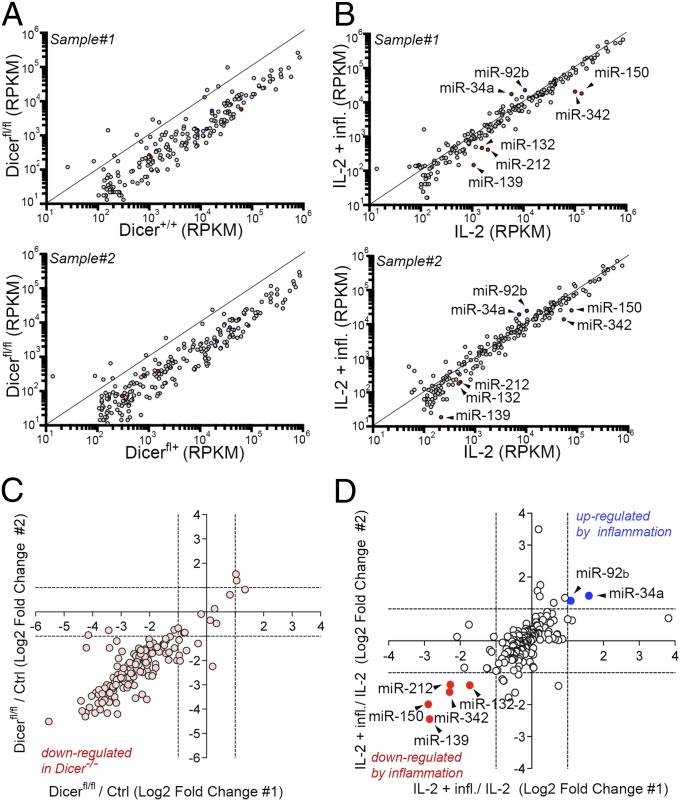
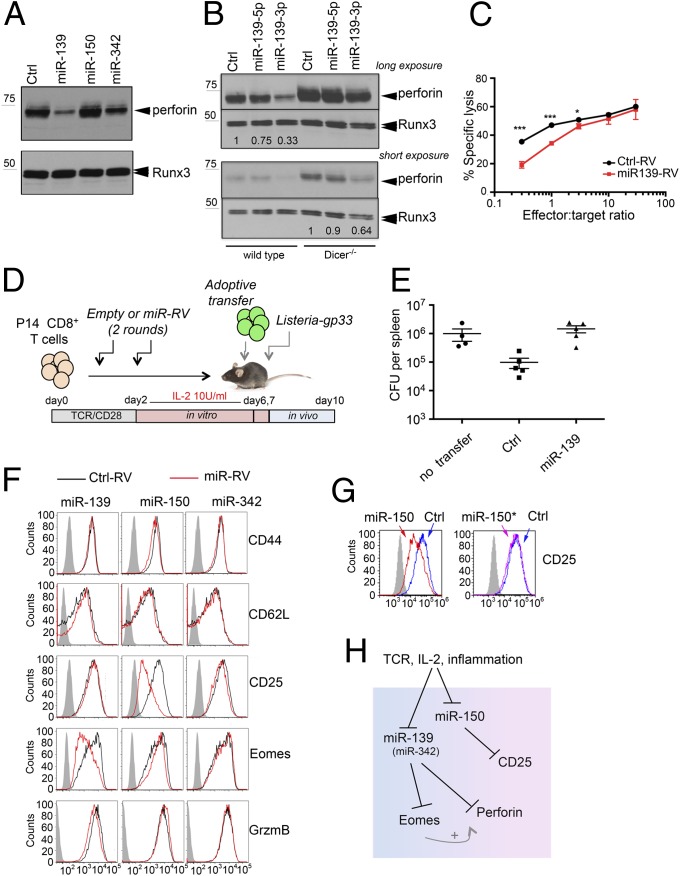
To identify miRNAs that may regulate the expression of perforin and other proteins in CTLs, we used next-generation sequencing to profile small RNAs from control and Dicer−/− CTLs (Fig. 1D, lanes d, f, and h), wild-type CTLs generated in vitro with or without inflammation (Fig. 3C, lanes b and h), and effector and memory CTL precursors differentiated in vivo and sorted on day 8 post-LCMV infection (Fig. 3E, lanes 1 and 4). Acute Dicer deletion caused a global reduction of mature miRNAs over a 5-day period (Fig. 4 A and C). Only seven miRNAs showed a greater than twofold difference between CTLs differentiated in vitro in the presence of inflammation and CTLs differentiated without inflammation; of these, two were up-regulated and five were down-regulated by inflammation (Fig. 4 B and D and Dataset S1). We focused on the three miRNAs that were most strongly down-regulated by inflammation. Of these, only miR-139 and miR-342 were also down-regulated in KLRG1+CD127– effector cells compared with CD127+KLRG1– memory precursor cells isolated from day 8 LCMV-infected mice (Dataset S1). To investigate the functions of these miRNAs, we used retroviral vectors encoding pri-miRNA sequences to transduce activated P14 CD8+ T cells. pri-miRNAs have to be processed by the cellular miRNA machinery, which avoids the huge overexpression usually observed upon transfection of mature miR mimics. We found that miR-139 strongly inhibited perforin expression, both in resting effector CTLs (Fig. 5A) and in memory CTLs restimulated with PMA and anti-CD3 (Fig. S3A). miR-342 had a similar, but much less pronounced, effect on perforin expression in effector CTLs, whereas miR-150 had no effect (Fig. 5A). Real-time analysis showed that transduction with pri-miR-139 retroviral vector induced a substantial increase in miR-139-3p expression (Fig. S3B, Left), whose levels became comparable to those of the miR-139-5p “guide” strand detected in naïve and memory CD8+ T cells (Fig. S4). To identify which miRNA strand (the guide or “star” strand) was responsible for reduced perforin expression, we transfected miRNA mimics corresponding to the miR-139-5p or -3p strand in differentiating CTLs; miR-139-3p down-regulated perforin expression in wild-type CTLs (Fig. 5B, Left) and partially reversed the increase in perforin expression observed in Dicer−/− CTLs (Fig. 5B, Right). The down-regulation of perforin induced by miR-139 correlated with a slight but significant reduction in the ability of miR-139–transduced P14 CTLs to kill gp33-loaded target EL4 cells (Fig. 5C). Because CTLs can kill via multiple mechanisms, including the engagement of death receptors on target cells, we verified by perforin depletion that granule-mediated lysis was required to kill target cells in our system (Fig. S3C).
Fig. 4.
Effects of Dicer deletion and inflammation on the miRNA profile of mouse CTLs. (A) Expression (reads per kb per million; RPKM) of individual miRNAs in control (Dicer+/+, Upper; Dicer+/fl, Lower) and Dicer−/− CTLs, differentiated with 100 U/mL IL-2 for 6 days. Two biological replicates are shown. (B) Expression (RPKM) of individual miRNAs in wild-type P14 CTLs differentiated for 6 days in 100 U/mL IL-2 or 100 U/mL IL-2 plus IL-12 and CpG. miRNAs down-regulated (red) or up-regulated (blue) by inflammation (greater than twofold change in both biological replicates) are shown. (C) Log-twofold change of miRNAs between Dicer−/− and control CTLs. Each dot represents a single miRNA; dots falling in the lower left quadrant are down-regulated in Dicer−/− compared with controls. (D) As in C, except that miRNAs in wild-type effector CTLs from P14+TCRα−/− mice obtained by culture in 100 U/mL IL-2 with and without inflammation are compared. Each dot represents a single miRNA; dots in the lower left quadrant (red) and in the upper right quadrant (blue) are down-regulated and up-regulated more than twofold by inflammation, respectively.
Fig. 5.
Control of effector CTL differentiation by miR-139, -150, and -342. (A) Perforin expression analyzed by Western blot in CTLs transduced with empty retrovirus (RV) (Ctrl) or RV containing the indicated pri-miR and cultured for 6 days in 100 U/mL IL-2. Runx3 is used as a loading control. Results are representative of three experiments. (B) Perforin expression in wild-type and Dicer−/− CTLs transfected with miRNA mimics, either nontargeting (Ctrl) or corresponding to the miR-139-5p or -3p strand. (C) Cytotoxicity assay performed using day 6 effector CTLs, either mock-transduced (Ctrl-RV) or transduced with miR-139-RV. Each point represents the average ± SD of three samples. Results are representative of two experiments, one with three mice per condition and the other with two mice. *P < 0.05; ***P < 0.001. (D) Schematic representation of the Listeria protection assay, with transfer of in vitro generated memory-like CTLs transduced with empty (Ctrl) or miR-139–expressing RV. (E) Quantification of Listeria CFUs per spleen 3 days after infection. “No transfer,” mice that did not receive any cell transfer. Each dot represents a single mouse. Results are representative of three independent experiments, each with four to six mice per group. (Mean +/− standard error.) (F) Activated CD8+ T cells were transduced as in A, and expression of the indicated markers was analyzed on day 6. The results are representative of at least three experiments. (G) CD25 expression in CTLs from P14 transgenic mice transfected with miR-150 or miR-150* miRNA mimics. (H) Schematic model of how TCR, IL-2, and inflammatory signals modulate the expression of perforin, Eomes, and CD25 through miR-139, -150, and -342 in CTLs.
To further investigate the consequences of miR-139 overexpression, we used a Listeria protection assay that is partially dependent on perforin expression by CTLs (23, 24). P14 transgenic CD8+ T cells were transduced with control or miR-139–expressing retrovirus, differentiated in vitro into memory CTLs, and transferred into recipient B6 mice, which were then infected with a nonlethal dose of Listeria-gp33 (Fig. 5D). Although the recipient mice are able to mount an endogenous response against Listeria, at early time points the transferred antigen-specific CD8+ T cells confer protection by limiting the spread of bacteria into the spleen and liver. Analysis of the bacterial load in the spleen on day 3 postinfection showed that whereas mock-transduced P14 CD8+ T cells were able to reduce Listeria colony-forming units (CFUs) per spleen by about 10-fold compared with unprotected mice (no cell transfer), miR-139–expressing P14 CD8+ T cells showed reduced ability to protect (Fig. 5E).
Microinspector (http://bioinfo1.uni-plovdiv.bg/cgi-bin/microinspector) and TargetScan (www.targetscan.org) predict the existence of a binding site for miR-139-3p in the perforin 3′-UTR as well as for miR-139-5p in the Eomes 3′-UTR. Indeed, we found that Eomes was down-regulated by miR-139 (Fig. 5F and Fig. S3D), suggesting that miR-139 controls effector CTL differentiation at multiple levels. Moreover, miR-150, but not the other two miRNAs tested, caused a strong down-regulation of CD25 expression in CTLs differentiated with high IL-2 (Fig. 5F); by transfecting miR mimics corresponding to the guide miR-150 or the star miR-150 strand (miR-150*), we traced this effect to the guide miR-150 strand (Fig. 5G). miR-139-5p, miR-139-3p, and miR-150 were all high in naïve and memory CD8+ T cells, which do not express perforin, relative to their levels in day 2 gp33-stimulated CD8+ T cells and day 6 effector CTLs, which express high levels of both perforin and CD25 (Fig. S4 A and B).
If Eomes and CD25 are miRNA targets, as our results suggest, they should be up-regulated at some point during CTL differentiation in Dicer−/− cells. However, CD25 expression was unchanged and Eomes expression was slightly reduced in effector Dicer−/− CTLs, when Dicer was deleted after T-cell priming (Fig. S1B). To examine miRNA-dependent effects that take place in the first 48 h after stimulation, we sorted naïve CD62L+CD44lowYFP+CD8+ T cells from Dicer+/+-Cd4-Cre:R26R and Dicerfl/fl-Cd4-Cre:R26R mice (in which YFP marks all cells that underwent Cre-mediated deletion in vivo), differentiated them into effector CTLs, and analyzed CD25 and CD69 expression. CD69 was up-regulated in Dicer−/− CTLs at all time points tested, whereas CD25 and Eomes were transiently but significantly up-regulated in Dicer−/− CTLs compared with wild-type CTLs on day 2 and days 4 and 5 poststimulation, respectively (Fig. S5). Together, these results show that TCR, IL-2, and inflammatory signals control the expression of molecules crucially involved in CD8+ T-cell function and proliferation—perforin, Eomes, and CD25—not only transcriptionally but also through miRNAs (Fig. 5H), and that the regulatory function of these miRNAs is dynamically regulated during CTL differentiation.
Discussion
In this study, we investigated the contribution of posttranscriptional mechanisms mediated by miRNAs to CTL differentiation. By using a controlled in vitro system, we uncovered a previously unappreciated miRNA-regulated signaling network that controls CTL activation and function downstream of IL-2 and inflammatory signals.
In both human and mouse CD8+ T cells, Dicer controls the expression of miRNAs that directly or indirectly suppressed perforin protein expression under all conditions tested. We show that miR-139 represses the expression of both perforin and Eomes [a direct transcriptional activator of the Prf1 gene (12, 18)] and that miR-342 may cooperate to repress perforin expression. These data highlight two concepts: that a single miRNA can act at multiple levels in the same pathway and that multiple miRNAs, which by themselves may have moderate effects, can converge on a single target. Notably, we observed Eomes up-regulation only when Dicer was deleted in naïve CD8+ T cells but not when it was deleted in activated CD8+ T cells, suggesting that the regulatory pathway linking miR-139 to Eomes may act during early phases of CTL differentiation. At later time points (days 6 and 7), Eomes was down-regulated in Dicer−/− CTLs despite the strong up-regulation of perforin under the same conditions, a scenario reminiscent of the effect of inflammation on wild-type CTLs. Thus, Eomes cannot be the only contributor to perforin up-regulation in Dicer−/− CTLs, and the phenotype of Dicer−/− CTLs most likely results from disturbance of both positive and negative signals. Our results suggest that control of effector CTL differentiation by miRNAs is partially conserved between humans and mice, even though the precise miRNAs and proteins through which this control is exerted can vary depending on the species.
Our finding of increased IL-10 production in Dicer−/− CTLs is consistent with the observation that a significant proportion of IL-10 regulation occurs posttranscriptionally (25). IL-10 produced by influenza-specific CTLs may be part of a self-regulatory mechanism aimed at restraining excessive tissue inflammation, as well as providing, together with IL-21, instructive signals for memory CTL generation (26). In contrast, the effects of Dicer deficiency on IFN-γ production are modest and seem to vary depending on the system and the cells used (27).
We and others have previously shown that the strength of IL-2 signaling is a key determinant of CTL differentiation. Strong IL-2 signals promote effector CTL differentiation and limit their survival in vivo and, conversely, low IL-2 signals are permissive for memory CD8+ T-cell generation (12, 28). The results presented here suggest that inflammation up-regulates CD25 expression in part by repressing miR-150. Our data are consistent with two previous studies that identify miR-150 as being down-regulated in effector compared with memory or naïve CD8+ T cells (29, 30), and clarify that TCR, IL-2 receptor, and inflammatory signals contribute to down-regulate miR-150 during CTL differentiation. Interestingly, miR-150 levels do not differ between KLRG1+CD127– effectors and KLRG1–CD127+ memory precursors on day 8 after LCMV infection; however, CD25 is not expressed by either of these cell populations. Analysis of effector CTLs at earlier time points after LCMV infection shows that miR-150 is lower in day 5 effector CTLs that express CD25 than in day 8 effector CTLs that have down-regulated CD25 (9).
miR-139 follows a similar trend to that of miR-150, except that expression of the 3p strand is extremely low in CTLs derived in vitro with anti-CD3 plus anti-CD28 stimulation and cultured in high IL-2, and virtually undetectable when CTLs are generated by gp33 stimulation followed by culture in high IL-2 or IL-2 plus inflammation. However, miR-139-3p is expressed in naïve as well as in in vitro or in vivo generated memory CTLs. We suggest that miR-139-3p is mainly involved in repressing perforin expression in naïve CD8+ T cells and memory CTLs.
Our findings that Dicer protein expression is suppressed by strong IL-2 receptor signals and inflammation in CTLs, without a corresponding mRNA change, are indicative of posttranscriptional regulation. At least two miRNAs, let-7 (31) and miR-103/107 (32), can target Dicer. Although let-7 is not regulated by inflammation in our system, let-7a-1 is one of the very few miRNAs that are paradoxically up-regulated in Dicer−/− CTLs (from 4- to 11-fold over controls in two biological replicates), raising the possibility that let-7a-1 and Dicer influence each other’s expression. The notion that Dicer levels can be regulated by extracellular stimuli has precedents in other systems. In neurons, the growth factor BDNF modulates Dicer levels, regulating the global miRNA output (33). Likewise, inflammatory or “stress” signals (reactive oxygen species, double-strand RNA, and type I IFNs) have been shown to down-regulate Dicer in cell lines and in mouse spleen (34). Recently, in activated CD4+ T cells, degradation of Argonaute2 was observed, and this correlates with a general decrease in mature miRNA levels (35). Interestingly, in our system, despite a substantial loss of Dicer protein induced by the combination of high-dose IL-2 plus inflammatory stimuli, there are not major consequences for the mature miRNA output; therefore, Dicer levels do not appear to become overtly limiting. However, we did observe a moderate down-regulation of low-expressed miRNAs in CTLs generated with inflammation, suggesting that competition to access the miRNA machinery may constitute an additional level of fine-tuning of mature miRNA levels during CTL differentiation downstream of transcriptional regulation.
Overall, our study shows that IL-2 and inflammatory signals regulate the expression of miRNAs that control CTL differentiation, and specifically the expression of Eomes, perforin, and CD25. We identify miR-139, -150, and to a lesser extent miR-342 as components of this posttranscriptional regulatory mechanism. Although further proteomic analysis will be required to define globally all of the potential targets of these miRNAs in CTLs, our data support the existence of an miRNA-based posttranscriptional program that needs to be shut down or dampened to relieve the silencing of effector molecules and ensure proper effector CTL differentiation.
Materials and Methods
Standard procedures for T-cell isolation and culture, retroviral transduction and transfection, RNA isolation, and real-time PCR are described in SI Materials and Methods. A detailed description of experimental samples used in RNA sequencing experiments is also available in SI Materials and Methods. The method used for sequencing data analysis is outlined in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Riitta Lahesmaa and Harri Lähdesmäki (Turku Centre for Biotechnology) for reviewing the manuscript and for scientific discussion. We thank Ryan Hastie for excellent assistance; Shane Crotty (La Jolla Institute for Allergy and Immunology) for scientific advice; and Wes Gifford and Todd Macfarlan (Salk Institute) for help with library preparation. S.T. and G.J.M. are supported by postdoctoral fellowships from the Cancer Research Institute and the Jane Coffin Childs Memorial Fund, respectively. T.Ä. is supported by a graduate student fellowship from the Finnish Doctoral Programme in Computational Sciences. This work was funded by National Institutes of Health Grants AI70788, CA42471, AI40127, RC4 AI092763 (to A.R.), and AI095634 (to M.E.P.); and by the Academy of Finland Centre of Excellence in Molecular Systems Immunology and Physiology Research (Grant 250114; to A.R.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317191110/-/DCSupplemental.
References
- 1.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12(11):749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monticelli S, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6(8):R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuchen S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32(6):828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong MM, et al. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24(17):1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202(2):261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci USA. 2010;107(50):21629–21634. doi: 10.1073/pnas.1016299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu T, et al. Temporal expression of microRNA cluster miR-17-92 regulates effector and memory CD8+ T-cell differentiation. Proc Natl Acad Sci USA. 2012;109(25):9965–9970. doi: 10.1073/pnas.1207327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudda JC, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38(4):742–753. doi: 10.1016/j.immuni.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gracias DT, et al. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013;14(6):593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pipkin ME, et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32(1):79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fehniger TA, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26(6):798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Kim TD, et al. Human microRNA-27a* targets Prf1 and GzmB expression to regulate NK-cell cytotoxicity. Blood. 2011;118(20):5476–5486. doi: 10.1182/blood-2011-04-347526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P, et al. Identification of resting and type I IFN-activated human NK cell miRNomes reveals microRNA-378 and microRNA-30e as negative regulators of NK cell cytotoxicity. J Immunol. 2012;189(1):211–221. doi: 10.4049/jimmunol.1200609. [DOI] [PubMed] [Google Scholar]
- 16.Pircher H, Bürki K, Lang R, Hengartner H, Zinkernagel RM. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 1989;342(6249):559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 17.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102(31):10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz-Guilloty F, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206(1):51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6(12):1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 20.Hersperger AR, et al. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog. 2010;6(5):e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaech SM, et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4(12):1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 22.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27(2):281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kägi D, Ledermann B, Bürki K, Hengartner H, Zinkernagel RM. CD8+ T cell-mediated protection against an intracellular bacterium by perforin-dependent cytotoxicity. Eur J Immunol. 1994;24(12):3068–3072. doi: 10.1002/eji.1830241223. [DOI] [PubMed] [Google Scholar]
- 24.Messingham KA, Badovinac VP, Harty JT. Deficient anti-listerial immunity in the absence of perforin can be restored by increasing memory CD8+ T cell numbers. J Immunol. 2003;171(8):4254–4262. doi: 10.4049/jimmunol.171.8.4254. [DOI] [PubMed] [Google Scholar]
- 25.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9(4):353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 26.Sun J, Dodd H, Moser EK, Sharma R, Braciale TJ. CD4+ T cell help and innate-derived IL-27 induce Blimp-1-dependent IL-10 production by antiviral CTLs. Nat Immunol. 2011;12(4):327–334. doi: 10.1038/ni.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bezman NA, et al. Distinct requirements of microRNAs in NK cell activation, survival, and function. J Immunol. 2010;185(7):3835–3846. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalia V, et al. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32(1):91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Almanza G, et al. Selected microRNAs define cell fate determination of murine central memory CD8 T cells. PLoS One. 2010;5(6):e11243. doi: 10.1371/journal.pone.0011243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H, et al. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One. 2007;2(10):e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA. 2008;105(39):14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martello G, et al. A microRNA targeting Dicer for metastasis control. Cell. 2010;141(7):1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 33.Huang YW, Ruiz CR, Eyler EC, Lin K, Meffert MK. Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell. 2012;148(5):933–946. doi: 10.1016/j.cell.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46(6):1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bronevetsky Y, et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. 2013;210(2):417–432. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.