Significance
Bacterial pathogens can use a syringe-like structure to inject virulence proteins (effectors) directly into host cells. The YopJ/HopZ superfamily of effectors found in animal and plant pathogens can modify host kinase proteins to suppress host immunity. In the model plant Arabidopsis, HopZ1a is recognized by the resistance protein ZAR1 to induce a robust immune response that blocks pathogen growth. Here, we show that the HopZ1a effector from the plant pathogen Pseudomonas syringae targets the Arabidopsis nonfunctional pseudokinase ZED1 and that ZED1 is required for recognition of HopZ1a by ZAR1. We hypothesize that HopZ1a targets kinases to promote pathogen virulence, and Arabidopsis ZED1 evolved as a decoy to trap HopZ1a in the ZAR1 complex for recognition by the plant immune system.
Keywords: ZED1-related kinase, ZRK, hypersensitive response
Abstract
Plant and animal pathogenic bacteria can suppress host immunity by injecting type III secreted effector (T3SE) proteins into host cells. However, T3SEs can also elicit host immunity if the host has evolved a means to recognize the presence or activity of specific T3SEs. The diverse YopJ/HopZ/AvrRxv T3SE superfamily, which is found in both animal and plant pathogens, provides examples of T3SEs playing this dual role. The T3SE HopZ1a is an acetyltransferase carried by the phytopathogen Pseudomonas syringae that elicits effector-triggered immunity (ETI) when recognized in Arabidopsis thaliana by the nucleotide-binding leucine-rich repeat (NB-LRR) protein ZAR1. However, recognition of HopZ1a does not require any known ETI-related genes. Using a forward genetics approach, we identify a unique ETI-associated gene that is essential for ZAR1-mediated immunity. The hopZ-ETI-deficient1 (zed1) mutant is specifically impaired in the recognition of HopZ1a, but not the recognition of other unrelated T3SEs or in pattern recognition receptor (PRR)-triggered immunity. ZED1 directly interacts with both HopZ1a and ZAR1 and is acetylated on threonines 125 and 177 by HopZ1a. ZED1 is a nonfunctional kinase that forms part of small genomic cluster of kinases in Arabidopsis. We hypothesize that ZED1 acts as a decoy to lure HopZ1a to the ZAR1–resistance complex, resulting in ETI activation.
The plant immune system can be divided into two major branches that share commonalities with animal innate immunity. Pattern recognition receptor (PRR)-triggered immunity (PTI) is activated by the recognition of conserved microbial molecules called microbe-associated molecular patterns (MAMPs) by extracellular PRRs (1), similar to the recognition of MAMPs by animal Toll-like receptors (2). Effector-triggered immunity (ETI) is activated by the recognition of pathogen-derived effector proteins by intracellular NB-LRR proteins that share structural features with animal nucleotide-binding domain, leucine-rich repeat containing (NLR) proteins (3). The ETI response overlaps significantly with PTI but is accelerated, amplified, and often accompanied by the hypersensitive cell death response (HR; refs. 4 and 5). Recognition of effector proteins can occur directly where the effector protein binds directly to the NB-LRR protein, or indirectly, in which case both the effector and NB-LRR proteins bind to a common host protein. In the latter case, ETI is initiated by the NB-LRR protein in response to an effector-induced modification to the host protein (6). In some cases, the host protein monitored by the NB-LRR may represent a true virulence target of the effector protein, whereas in other cases, this protein may be a nonfunctional decoy of the true virulence target maintained by the host for the purposes of pathogen surveillance (7).
The continual arms race between host and pathogen has directed the coevolution of host innate immunity with bacterial virulence strategies. The type III secretion system (T3SS) is a predominant virulence mechanism used by many Gram-negative bacterial pathogens to cause disease in eukaryotic hosts (8). The T3SS delivers type III secreted effector (T3SE) proteins into host cells where they can promote the infection process by suppressing host immunity and/or participating in the pathogen life cycle.
The YopJ/HopZ/AvrRxv T3SE superfamily is evolutionary diverse and found in both animal and plant pathogens (9, 10). Yersinia pestis YopJ, the archetypal member of this superfamily, acetylates serine or threonine residues in the activation loops of members of the mitogen-activated protein kinase kinase (MAPKK) and MAP kinase kinase kinase superfamilies, thereby suppressing innate immunity (11–15). In the plant pathogen Pseudomonas syringae, the HopZ1 subfamily is represented by three closely related alleles, HopZ1a, HopZ1b, and HopZ1c, that diversified under pressure from the host immune system (10). The most phylogenetically basal representative of this subfamily, HopZ1a, is recognized in Arabidopsis by the ZAR1 NB-LRR protein (16, 17) as well as by unidentified proteins in rice, Nicotiana benthamiana, sesame, and soybean (10). Like YopJ, HopZ1a is an acetyltransferase and can promote pathogen growth in Arabidopsis plants lacking the ZAR1 NB-LRR protein. The virulence and immune-eliciting functions of HopZ1a both require the cysteine residue in the acetyltransferase catalytic triad (16, 17). It is likely that ZAR1 evolved to recognize an ancestral virulence activity of HopZ1a; however, the molecular relationship between virulence and immunity-eliciting functions remain to be established (17, 18).
Here, we describe a forward genetic screen designed to characterize the genetic requirements of ZAR1-mediated immunity by identifying Arabidopsis mutant plants that lacked a HopZ1a-induced HR response. We named these mutants hopZ-ETI deficient (zed) and mapped zed1 to At3g57750 by Illumina-based next-generation sequencing. Different zed1 point mutants identified from our mutant screen or a tDNA insertion line in At3g57750 all lack recognition of HopZ1a. PTI and basal defenses are unaffected in zed1; however, zed1 exhibits a loss of HopZ1a-mediated ETI. We show that ZED1 interacts directly with HopZ1a, as well as the N-terminal coiled-coil (CC) domain of ZAR1. ZED1 is an uncharacterized pseudokinase and is acetylated on threonines 125 and 177 by HopZ1a. We hypothesize that ZAR1 is activated in response to ZED1 acetylation by HopZ1a and speculate that ZED1 may be a decoy of the true HopZ1a virulence target because HopZ1a retains its virulence function in zed1 Arabidopsis plants.
Results
ZED1 Is Required for HopZ1a-Triggered Immunity.
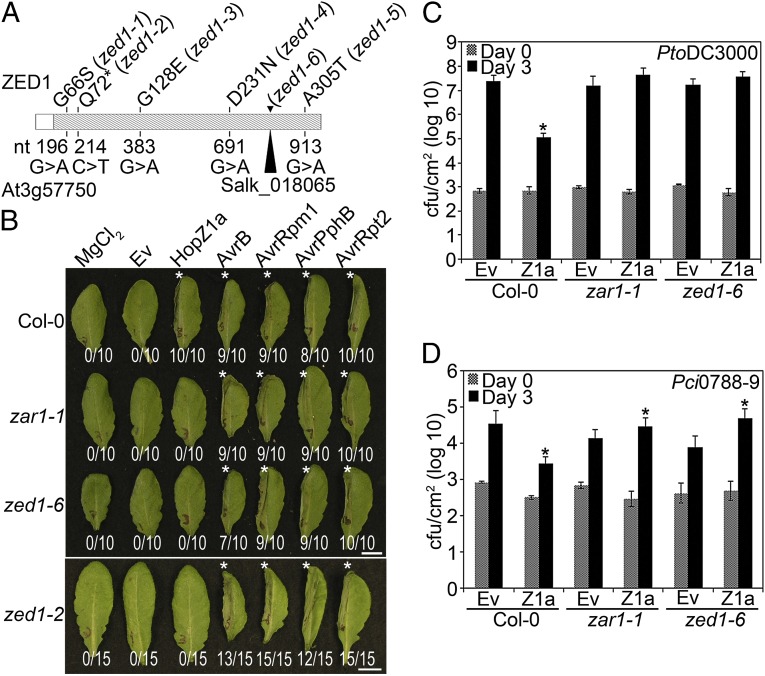
To identify new genes involved in Arabidopsis innate immunity, we designed a forward genetic screen for mutants that no longer exhibited a HopZ1a-induced HR (16) (SI Materials and Methods). One mutant was mapped to At3g57750 using next-generation mapping (NGM) methodology, and a tDNA insertion line in this gene (SALK_018065) lacked a HopZ1a-induced HR (zed1-6; Fig. 1 A and B and Fig. S1). Sanger sequencing of the At3g57750 gene in our point mutant zed1-2 confirmed that this gene was mutated, resulting in a change from a glutamine to an early stop codon at amino acid 72, as predicted from the NGM analysis (Fig. 1A). Sanger sequencing of other zed mutants identified additional residues in At3g57750 that contributed to the recognition of HopZ1a (Fig. 1A).
Fig. 1.
zed1 mutants are affected in the predicted kinase domain, are specifically impaired in the recognition of HopZ1a, and exhibit increased virulence of HopZ1a. (A) ZED1 sequence schematic, with hatched region representing the predicted kinase domain. The nucleotide changes identified in the EMS screen are shown below the schematic, whereas the corresponding amino acid mutations are indicated above. The zed1-6 mutation resulted in a premature stop 71 codons after the start codon. The insertion point of the Salk tDNA insertion line is also shown. (B) The left halves of Col-0, zar1-1, zed1-2, or zed1-6 leaves were infiltrated with 10 mM MgCl2 or PtoDC3000 carrying empty vector (Ev), HopZ1a, AvrB, AvrRpm1, AvrPphB, or AvrRpt2. The bacteria were pressure-infiltrated into the leaves at 5 × 107 cfu/mL Photos were taken at 22 h after infiltration. The number of leaves showing an HR is indicated below the leaf. HRs are marked with an asterisk. (C) Col-0, zar1-1, or zed1-6 plants were infiltrated with PtoDC3000 carrying empty vector (Ev) or HopZ1a, and bacterial counts were determined 1 h after infection (Day 0) and 3 d after infection (Day 3). Two-tailed homoschedastic t tests were performed to test for significant differences (*) between Ev control strain and the strain carrying HopZ1a. Error bars indicate the SD from the mean of 10 samples. Growth assays were performed at least three times. (D) Col-0, zar1-1, or zed1-6 were infiltrated with Pci0788-9 carrying Ev or HopZ1a, as in C.
Role of ZED1 in Plant Immunity.
We carried out infectivity assays to test whether ZED1 contributes to PTI and/or ETI responses. To determine whether ZED1 plays a role in PTI, we infiltrated wild-type Col-0, zar1-1, or zed1-6 plants with the T3SS-deficient mutant PtoDC3000∆hrcC, which cannot suppress PTI. PtoDC3000∆hrcC grew to equivalent densities in the Col-0, zar1-1, and zed1-6 backgrounds, supporting that neither ZAR1 nor ZED1 are involved in mediating PTI (Fig. S2). Furthermore, wild-type PtoDC3000 did not show altered growth on the zed1-6 mutant relative to wild-type Col-0, supporting that the loss of ZED1 does not alter overall basal defense (Fig. 1C; ref. 1).
We investigated whether the role of ZED1 in ETI was specific to HopZ1a recognition by determining whether the recognition of any other T3SEs was impaired in zed1. All of the zed1 ethylmethylsulfonate (EMS) mutants showed a specific loss of the HopZ1a-induced HR, whereas each of AvrRpt2, AvrRpm1, AvrB, and AvrPphB induced an HR in these plants (Fig. 1B and Fig. S3). The zed1-6 tDNA insertion lines also showed a specific loss of the HopZ1a-induced HR like the zar1-1 NB-LRR gene knockout (Fig. 1B; ref. 17), demonstrating the specificity of ZED1 in HopZ1a recognition.
We then confirmed that the loss of HopZ1a HR displayed a commensurate loss of resistance by performing bacterial growth assays with the virulent strain PtoDC3000 carrying empty vector (Ev) or hopZ1a in wild type Col-0, or mutant zar1-1 or zed1-6 plants. As previously observed, HopZ1a triggered a strong defense response in Col-0, leading to ∼2 log reduction in bacterial growth after 3 d (Fig. 1C; ref. 16). This defense response is lost in the zar1-1 line, and the PtoDC3000(hopZ1a) strain is able to grow to similar levels as PtoDC3000(Ev) (Fig. 1C; ref. 17). Similarly, in the zed1-6 knockout, PtoDC3000(hopZ1a) growth is similar to PtoDC3000(Ev) (Fig. 1C). These results specifically link ZED1 to HopZ1a-induced ETI.
HopZ1a Virulence Is Retained in zed1 Plants.
We previously demonstrated that HopZ1a confers a growth advantage in the absence of ZAR1 when it is delivered by P. syringae pathovar cilantro (Pci0788–9), a strain closely related to a virulent strain of P. syringae pathovar maculicola (PmaES4326) (17). To determine whether HopZ1a would also confer a growth advantage in zed1, we infiltrated wild-type Col-0 or the zar1-1 or zed1-6 mutants with Pci0788–9 carrying hopZ1a or empty vector. Pci0788–9 (hopZ1a) grew to significantly higher densities compared with Pci0788–9(Ev) in both zed1-6 and zar1-1 (17) (Fig. 1D). This result demonstrates that HopZ1a retains its virulence function in plants lacking ZED1 and, importantly, indicates that HopZ1a must have additional targets beyond ZED1. This result is also consistent with previous work showing that HopZ1a can destabilize microtubule networks and suppress cell wall-mediated defenses (18).
ZED1 Interacts with HopZ1a and ZAR1.
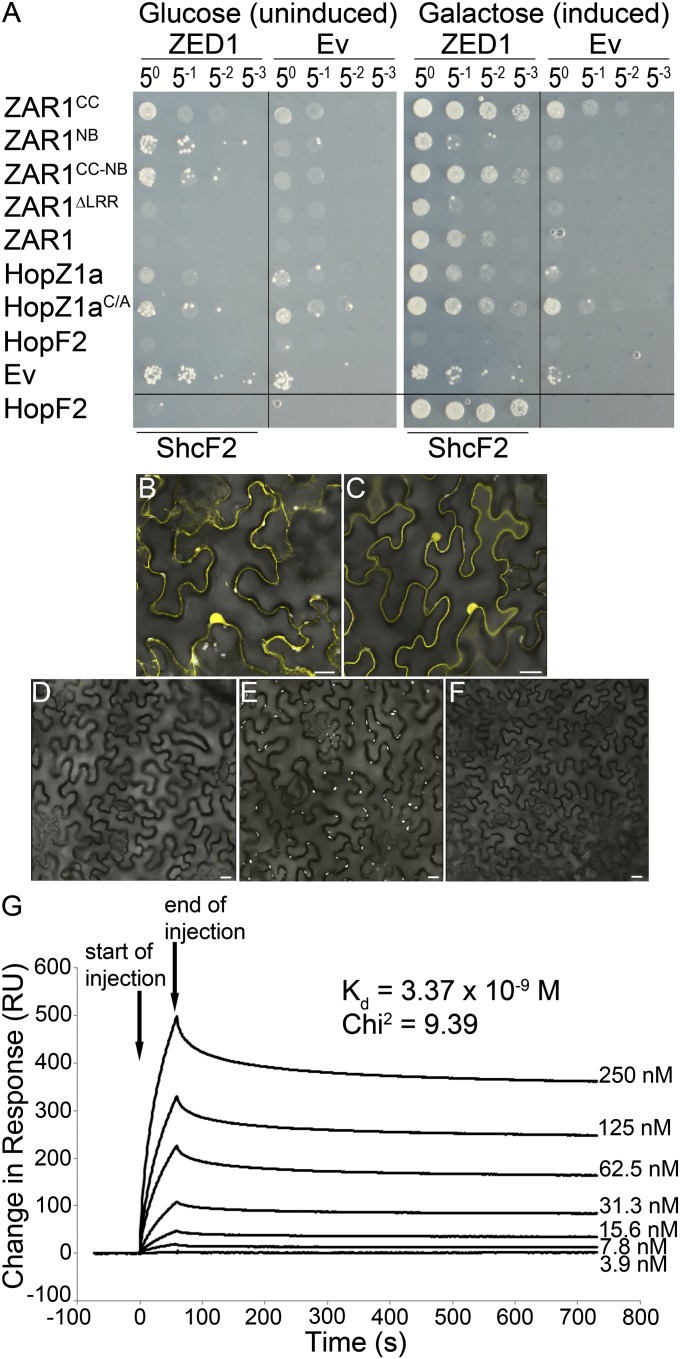
We first tested for direct interactions among ZED1, HopZ1a, and ZAR1 by using the LexA yeast two-hybrid system. We constructed five ZAR1 clones, one full-length (ZAR11-853) and four truncations containing combinations of the coiled-coil domain (CC), nucleotide-binding site (NB), and leucine-rich repeats (LRR) of ZAR1 fused to the DNA binding domain (see SI Materials and Methods for details). Wild-type HopZ1a or the catalytic mutant HopZ1aC216A (hereafter HopZ1aC/A) was also cloned as a fusion to the LexA DNA-binding domain. Catalytically inactive enzymes have been found to exhibit stabilized interactions with substrates (19, 20). ZED1 was cloned as a fusion with the B42 activation domain. ZED1 interacted directly with HopZ1a and slightly more strongly with HopZ1aC/A, indicating the interaction is independent of the catalytic cysteine residue of HopZ1a (Fig. 2A). ZED1 also interacted with ZAR1CC, ZAR1CC-NB, and the full-length ZAR1, but not ZAR1NB or ZAR1∆LRR (Fig. 2A). This is similar to other NB-LRR proteins whose N-terminal domains have been shown to interact with T3SE targets and downstream signaling components (21, 22). The ZED1–ZAR1 interaction was strongest using the ZAR1CC domain alone, compared with ZAR1CC-NB or full-length ZAR1; therefore, the CC domain is sufficient for ZAR1–ZED1 interactions. The lack of interaction with some of the ZAR1 truncations was not due to a lack of protein expression (Fig. 2A and Fig. S4A). We used the PtoDC3000 T3SE HopF2PtoDC3000 (hereafter HopF2) as a specificity control and found that it did not interact directly with ZED1, indicating that the ZED1 interaction is specific for HopZ1a. However, HopF2 did interact strongly with its chaperone ShcF2PtoDC3000, as previously observed (23).
Fig. 2.
ZED1 interacts with HopZ1a and ZAR1. (A) ZED1 was constructed as an activation-domain (AD) fusion protein and was tested for its interaction with DNA-binding domain (BD)-fused ZAR1, HopZ1a, the catalytic mutant HopZ1aC/A, and HopF2PtoDC3000 (hereafter HopF2) in the LexA yeast two-hybrid system. HopF2 and ShcF2PtoDC3000 were used as positive controls because they are known to strongly interact (23). (B–F) BiFC of HopZ1aC/A, ZED1, and ZAR1. Agrobacterium carrying HopZ1aC/A::nYFP, ZAR1::nYFP, ZED1::cYFP, and MLO2∆1–280::nYFP or cYFP were mixed in equivalent optical densities and pressure-infiltrated into the leaves of N. benthamiana. HopZ1aC/A was used because HopZ1a causes an HR in N. benthamiana. Protein expression was induced by using 30 µM dexamethasone. Leaf sections were imaged by using a Leica SP3 confocal scanning microscope 24–48 h after induction. (Scale bar: 20 µm.) Coexpressed pairs are as follows. (B) ZAR1::nYFP and ZED1::cYFP. (C) HopZ1aC/A::nYFP and ZED1-cYFP. (D) MLO2∆1–280::nYFP and ZED1::cYFP. (E) ZAR1::nYFP and MLO2∆1–280::cYFP. (F) HopZ1aC/A::nYFP and MLO2∆1–280::cYFP. (G) Surface plasmon resonance analyses of the HopZ1a/ZED1 interaction. His-HopZ1a was immobilized to the surface of a Biacore CM5 sensor chip, and His-ZED1 was flowed over the bound surface at increasing protein concentration to calculate a dissociation constant (Kd). ZED1 displays very high-binding affinity (Kd = 3–5 nM) to His-HopZ1a (χ2 of 9.39). Arrows indicate the start and end of His-ZED1 injection.
To test for in planta interactions, we used Agrobacterium-mediated transient expression and bimolecular fluorescence microscopy (BiFC). ZED1, ZAR1, HopZ1aC/A, and MLO2Δ1–280 were cloned as in-frame fusions to the N terminus of YFP (nYFP) or the C terminus of YFP (cYFP), and expressed under a dexamethasone-inducible promoter. HopZ1aC/A was used in these experiments because HopZ1a elicits an HR when expressed alone in N. benthamiana (10, 16). MLO2Δ1–280 was used as a negative control, because we have previously shown that it interacts directly with HopZ2 but not with other members of the HopZ family (24). We observed bright fluorescence in leaf sections when ZAR1::nYFP was coexpressed with ZED1::cYFP (Fig. 2B), as well as the reciprocal combination (Fig. S4B). Fluorescence was observed in structures reminiscent of the nucleus, endoplasmic reticulum, and plasma membrane. No fluorescence was observed between MLO2Δ1–280::nYFP and ZED1::cYFP (Fig. 2D), ZAR1::nYFP and MLO2Δ1–280::cYFP (Fig. 2E), or the reciprocal combination (Fig. S4 D and E) at these time points. We also observed bright fluorescence in leaf sections, primarily in structures reminiscent of the nucleus and plasma membrane, when HopZ1aC/A::nYFP was coexpressed with ZED1::cYFP (Fig. 2E), as well as the reciprocal combination (Fig. S4C). No fluorescence was observed between HopZ1aC/A::nYFP and MLO2Δ1–280::cYFP (Fig. 2F), or the reciprocal combination (Fig. S4F), which is consistent with our previous yeast two-hybrid studies (24).
We used surface plasmon resonance technology to test for a direct interaction in vitro between recombinant purified His-tagged ZED1 and immobilized HopZ1a. His-ZED1 bound with very high affinity (Kd = 3–5 nM) to His-HopZ1a in a dosage-dependent manner (Fig. 2G). The nature of the purification tag was not responsible for the interaction because ZED1 also bound with high affinity (Kd = 3–5 nM) to GST-tagged HopZ1a (Fig. S4G). The ZED1–HopZ1a interaction was specific because GST alone did not bind His-HopZ1a or GST-HopZ1a (Fig. S4H).
ZED1 Is Acetylated by HopZ1a.
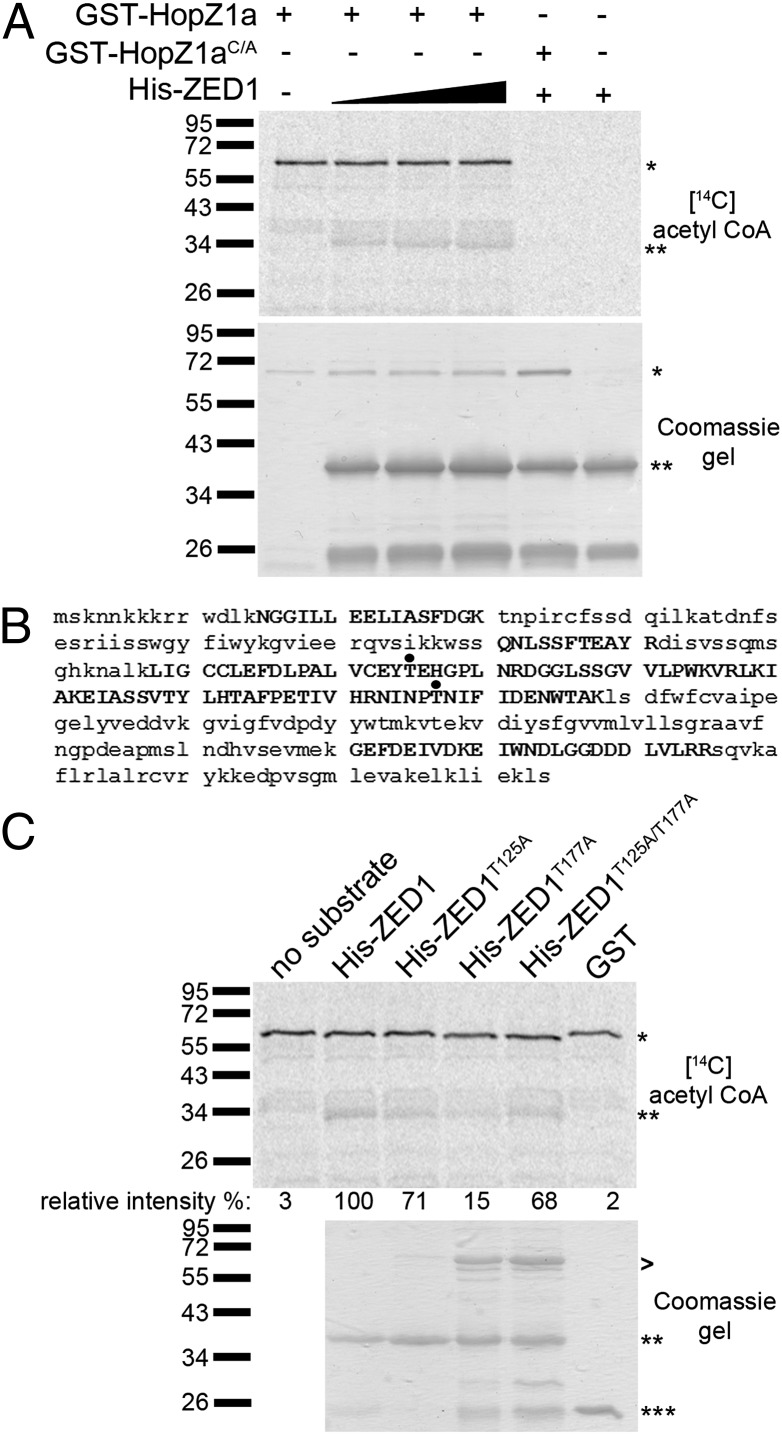
Previously, we showed that HopZ1a is an acetyltransferase that is activated by phytic acid (18). To test whether HopZ1a directly acetylates ZED1, we carried out in vitro acetylation assays with recombinant GST-HopZ1a and His-ZED1, which was expressed and purified from Escherichia coli. As observed (18), GST-HopZ1a autoacetylated in the presence of phytic acid, whereas the catalytic mutant GST-HopZ1aC/A did not. In addition, HopZ1a transacetylated His-ZED1 (Fig. 3A). His-ZED1 acetylation was not observed with GST-HopZ1aC/A, indicating that the catalytic cysteine residue in HopZ1a is essential for ZED1 acetylation.
Fig. 3.
HopZ1a acetylates ZED1. (A Upper) Purified GST-HopZ1a or GST-HopZ1aC/A (∼70 kDa) and 6xHis-ZED1 (∼41 kDa) were incubated with phytic acid in the presence of [14C] acetyl-CoA. Samples were separated on a 9% polyacrylamide gel, and 14C incorporation was visualized by Phosphoimager. *, GST-HopZ1a or GST-HopZ1aC/A; **, His-ZED1. (A Lower) Coomassie gel of proteins. (B) LC-MS/MS analysis of immunoprecipitated FLAG-tagged ZED1 proteins in yeast coexpressing ZED1 and HopZ1a. Peptides recovered in LC-MS/MS analysis are shown in capital letters. Acetylated threonines are shown with a black dot. (C Upper) Purified GST-HopZ1a and 6xHis-ZED1, 6xHis-ZED1T125A, 6xHis-ZED1T177A, or 6xHis-ZED1T125A/T177A were incubated with phytic acid in the presence of [14C] acetyl-CoA. Samples were separated on a 9% polyacrylamide gel, and 14C incorporation was visualized by Phosphoimager. *, GST-HopZ1a; **, His-ZED1 or mutants; ***, GST. The signal intensity for each 14C-labeled band was quantified by using ImageJ and is shown below the Coomassie gel. The maximum signal intensity was arbitrarily set at 100% for wild-type His-ZED1, and other signal intensities are relative to this value. (C Lower) Coomassie gel of His-ZED1, His-ZED1 mutant, and GST proteins as a loading control. GST-HopZ1a is loaded as shown in A. >, contaminating proteins in the protein preparation.
We identified HopZ1a-mediated acetylation sites on ZED1 via an in vivo acetylation assay by coexpressing HopZ1a and ZED1 in yeast followed by immunoprecipitation (IP) and liquid chromatography tandem mass spectrometry (LC-MS/MS). When ZED1 was coexpressed with HopZ1a, we observed two peptides (peptide A: ZED1 108–145 aa, and peptide B: ZED1 173–188 aa) with a 42 Da mass increase corresponding to the size of an acetyl group (Fig. 3B and Fig. S5 A and B). Further analysis revealed that threonine 125 (T125) in peptide A and threonine 177 (T177) in peptide B of ZED1 were modified by acetylation when coexpressed with HopZ1a, whereas T125 and T177 were not acetylated in the presence of catalytically inactive HopZ1aC/A in yeast (Fig. S5 C–F and I). We cannot exclude the possibility that additional threonines may be acetylated, because we obtained 42% coverage of the ZED1 protein from LC-MS/MS (Fig. 3B and Fig. S5I).
To confirm that T125 and T177 on ZED1 are acetylated, we constructed and purified recombinant protein for the single mutants (ZED1T125A or ZED1T177A) and the double mutant (ZED1T125A/T177A) and tested the proteins in our in vitro acetylation assay. The ZED1T125A substrate had a slight reduction of ZED1 acetylation (29% less signal intensity than wild-type ZED1), whereas the ZED1T177A substrate showed a strong reduction of ZED1 acetylation (85% less signal intensity than wild-type ZED1) (Fig. 3C). The ZED1T125A/T177A substrate resulted in a slight reduction (32%) in ZED1 acetylation, comparable to the ZED1T125A substrate (Fig. 3C). The ZED1T125A/T177A mutant protein had nonadditive effects on acetylation, perhaps by affecting the conformation of the protein and enhancing alternative acetylation sites. Similar observations have been made in kinase receptors that do not show an additive reduction in phosphorylation in double mutants versus single mutants of phosphorylated residues (25). Acetylation was specific to ZED1 and HopZ1a as GST alone was not acetylated (Fig. 3C).
ZED1 Is a Pseudokinase.
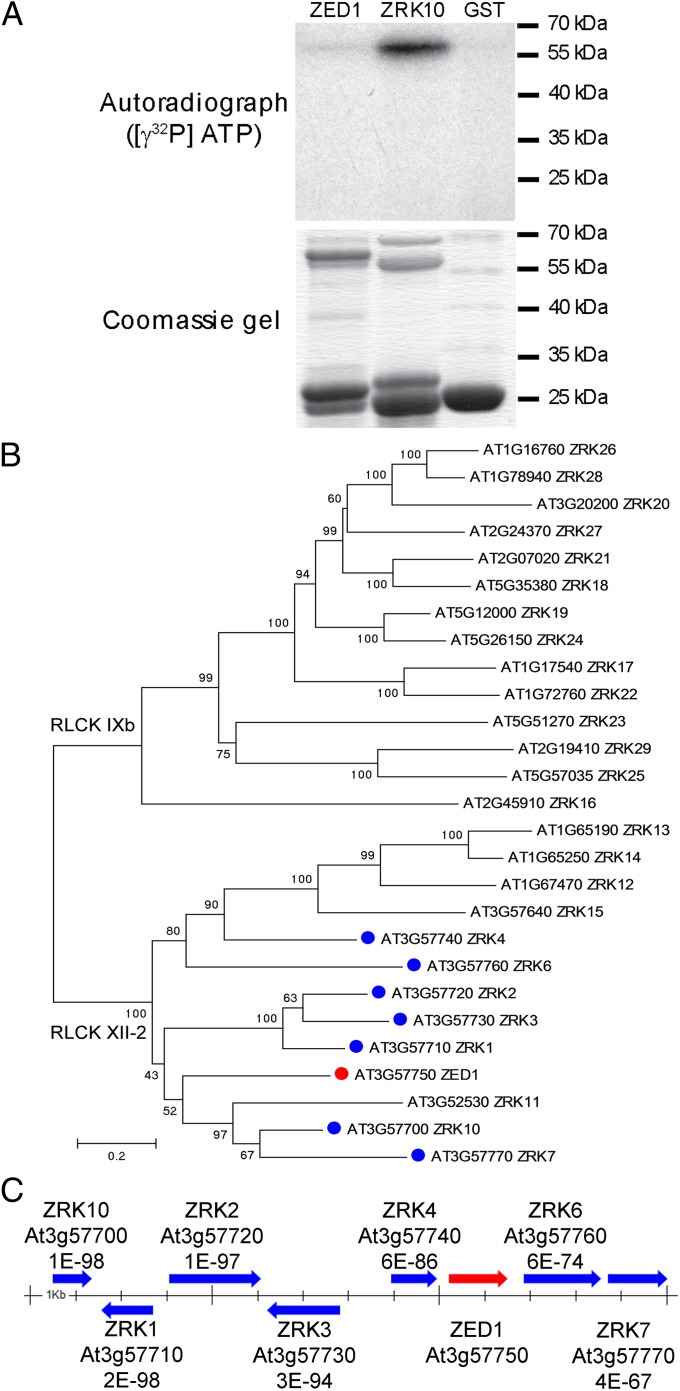
A conserved domain analysis of the predicted ZED1 protein indicated that it contained a protein kinase domain, consisting of ATP binding, and proton acceptor motifs (Fig. S6A). We assayed for ZED1 kinase activity via in vitro phosphorylation assays with recombinant His-tagged ZED1 protein expressed and purified from E. coli. His-ZED1 did not demonstrate autophosphorylation activity by using magnesium and/or manganese as cofactors (Fig. S7A). Because the substrate of ZED1 is not known, we also tested the generic kinase substrates histone H1 or myelin basic protein (MBP) in our assay but could not detect any kinase activity (Fig. S7B). We also did not detect any phosphorylation of GST-HopZ1a by His-ZED1 (Fig. S7A). To investigate whether the nature of the purification tag was responsible for the lack of activity, we further tested a recombinant GST-tagged ZED1 and showed that the GST-ZED1 protein was also not functional in all assays (Fig. 4A).
Fig. 4.
ZED1 is a pseudokinase that is part of a genomic cluster of related kinases. (A) ZRK10 (At3g57700) is a functional kinase, whereas ZED1 is not. (Upper) Purified GST-ZED1, GST-ZRK10, or GST was incubated in the presence of [γ-32P]ATP. Samples were separated on a 12% polyacrylamide gel and visualized by autoradiography. (Lower) Coomassie gel of proteins. (B) Phylogenetic analysis of the ZED1 (At3g57750) amino acid sequence and close homologs found in A. thaliana ecotype Col-0. These proteins correspond to kinase subfamilies RLCK XII-2 and RLCK IXb. ZED1 is indicated with a red dot. The colocalized, tandemly-arrayed ZRKs are indicated with a blue dot. The tree was constructed by neighbor joining and is congruent in general structure to trees constructed by maximum likelihood and Bayesian approaches. Numbers above the nodes are bootstrap scores indicating phylogenetic confidence. Only those scores >50 are shown. The scale bar indicated evolutionary distance based on a JTT substitution model. The tree is midpoint rooted, but the position corresponds to the root when more divergent kinase proteins are included in the analysis (Fig. S9). (C) ZED1 (At3g57750) is part of a cluster of eight closely-related ZRKs on A. thaliana chromosome 3. The arrows indicate the relative size and orientation of the genes in the cluster. Each gene is labeled with its locus tags and the blastp expect values relative to ZED1. The scale bar in the center is demarcated in 1-kb increments.
To confirm that our reaction conditions can detect kinase activity, we also tested At3g57700, which is one of seven closely related putative kinases that form a genomic cluster with ZED1 (Fig. 4). At3g57700 shares 48.7% amino acid identity and 63.8% amino acid similarity with ZED1. Importantly, we were able to detect kinase activity with At3g57700 (Fig. 4A). A SnRK kinase (At2g30360) was also active in our assay, further demonstrating that our kinase assay conditions are robust (Fig. S7 A and B).
Prosite analysis of the ZED1 protein sequence indicated that ZED1 lacks the critical aspartate residue in the “HRD” motif, which serves as a base acceptor to achieve proton transfer, whereas At3g57700 has an aspartate at that site (Fig. S6B). These data support that ZED1 is not a functional kinase.
Discussion
ZED1 is an ETI-associated protein required for ZAR1-mediated immunity induced by the P. syringae T3SE HopZ1a. ZED1 directly interacts with HopZ1a in yeast and in vitro (Fig. 2 A and H and Fig. S4 F and G), with the ZAR1CC domain in yeast (Fig. 2A), and with HopZ1a and ZAR1 in planta (Fig. 2 B and C and Fig. S4 B and C). ZED1 is acetylated by HopZ1a at threonines 125 and 177 (Fig. 3 and Fig. S5). Intriguingly, ZED1 appears to be a pseudokinase that is physically located in a genomic cluster of highly similar receptor-like cytoplasmic kinases in the Arabidopsis genome (Fig. 4 and Fig. S6). Because no biochemical function has been ascribed to ZED1, and the loss of the protein affects HopZ1a avirulence, but not virulence (Fig. 1), we hypothesize that ZED1 acts as a decoy to trigger ZAR1-dependent defense responses in response to HopZ1a acetylation. The alternative guard model implies that the NB-LRR guardee (i.e., ZED1) is a virulence target of the effector protein in the absence of recognition (6, 7). This model is not supported by our findings because ZED1 does not play a role in PTI or basal defense (Fig. 1C and Fig. S2), and HopZ1a retains its virulence function in the absence of ZED1 (Fig. 1D).
Pseudokinases have important roles in regulating active kinases and can act as scaffolding proteins in animals (26, 27). In Arabidopsis, the pseudokinase CORYNE has been shown to mediate stem cell production (28). Pseudokinases defective in phosphotransfer can still bind ATP (29), leading to the suggestion that ATP binding may act as an allosteric switch between active and inactive conformations, without requiring phosphotransfer to a substrate (26). Despite the lack of kinase activity, ZED1 clearly plays a critical role in HopZ1a recognition. We modeled the 3D structure of ZED1 by using the Phyre protein fold recognition server (Fig. S8). Phyre identified Protein Data Bank ID code 2FO0, Homo sapiens tyrosine kinase ABL1, as the best structural match to ZED1. In the 3D model, the putative ATP-binding domains are clustered together, and three of the zed1 point mutations map to this region as well as the acetylated threonines, indicating that ATP binding could be important for ZED1 function (Fig. S8). In addition, the two identified HopZ1a acetylation sites are also located in predicted ATP binding domains, suggesting that acetylation may interfere with ATP binding, similar to the acetylation of MAPKKs by the YopJ family member VopA (Fig. S8; ref. 30).
ZED1 is part of the broad receptor-like kinase (RLK)/Pelle family of protein kinases, which has dramatically expanded in the land plant lineages (31). The RLK/Pelle family is composed of Ser/Thr kinases, and most are similar to transmembrane receptors with an extracellular domain, a transmembrane domain, and an intracellular kinase domain. Based on the phylogeny of the kinase domains, the RLK/Pelle family has been divided into 64 subfamilies, with various extracellular domains. ZED1, however, is not a receptor kinase because it contains only the kinase domain and is thus classified as a receptor-like cytoplasmic kinase (RLCK) belonging to the RLCK XII-2 subfamily (31). Currently, the RLCK XII-2 subfamily has 13 members in A. thaliana (Fig. 4B and Fig. S9), including seven highly similar protein kinases genomically clustered with ZED1 (Fig. 4C) (31). The RLCK IXb subfamily is closely related to RLCK XII-2 subfamily, and we refer to members of both of these subfamilies as ZED1-related kinases (ZRKs; Fig. 4B). Homologs of the RLCK IXb subfamily are also found in other plant species, including the closely related A. lyrata as well as poplar (Populus trichocarpa) and grape (Vitis vinifera). Interestingly, this subfamily has only been identified in dicots and is absent from other vascular land plants unlike most other subfamilies of the RLK/Pelle family (31). The tandemly arrayed ZRKs are numbered according to the second-to-last digit of their ATG number (e.g., At3g57770 = ZRK7) with the exception of At3g57700, which we have numbered ZRK10. The rest of the ZRKs are numbered based on their similarity to ZED1 starting at ZRK11. With this numbering, there is no ZRK5 (ZED1), ZRK8, or ZRK9. The RLCK XII-2 subfamily includes ZRK1–ZRK15, whereas the RLCK IXb includes ZRK16–ZRK29 (Fig. 4B). Intriguingly, the RLCK XII-2 subfamily is only found among plants in the genus Arabidopsis, whereas RLCK IXb clade is broadly distributed among the flowering plants.
Kinases play key roles in T3SE recognition via their interactions with NB-LRR proteins. The PBS1 and Pto RLCK family VII kinases interact with the RPS5 and Prf NB-LRR proteins, respectively (32–34). Similar to the ZED1/ZAR1 interaction, both PBS1 and Pto interact with the N-terminal domain of their corresponding NB-LRR protein (22). However, unlike ZED1, both Pto and PBS1 are functional kinases that display activity in vitro (35, 36). The cleavage of PBS1 kinase by the cysteine protease T3SE AvrPphB (HopAR1) leads to recognition by the RPS5 NB-LRR protein (37), whereas Pto kinase interaction with two unrelated T3SEs, AvrPtoB (HopAB2) and AvrPto, is required for Prf recognition (38). Prf can also associate with at least five additional members of the Pto kinase cluster in planta, suggesting that Prf may be responsible for the recognition of multiple T3SEs (34). In a similar vein, the ZED1 pseudokinase is also part of a small genomic cluster of ZED1-related kinases (ZRKs) (Fig. 4C). If the above pattern holds, the ZAR1 NB-LRR may associate with multiple ZRKs and potentially be responsible for the recognition of multiple T3SEs. Although ZED1 shows no kinase activity, some of the ZRKs are predicted to be functional, which is supported by our finding that ZRK10 (At3g57700) has kinase activity in vitro (Fig. 4).
HopZ1a may have originally evolved to acetylate an immune-related kinase to suppress plant immunity similar to the function of other members of the YopJ superfamily (11, 12, 30, 39, 40). We speculate that HopZ1a-induced virulence involves manipulation of a ZED1-related kinase, in addition to other known virulence targets like microtubules (18). Plants may have responded to this attack on a ZED1-related kinase by duplicating and diversifying the ZRK family and either nonfunctionalizing or evolving ZED1 as a “kinase” trap that lures HopZ1a to the ZAR1 resistance complex to induce ETI. The high-affinity interaction between HopZ1a and ZED1 (Kd = 3–5 nM) would make ZED1 an efficient probe to detect HopZ1a. It remains to be determined whether ZED1 acetylation is the trigger for ZAR1-mediated immunity, whether other members of the ZRK family play a role in plant immunity and/or P. syringae virulence, and whether HopZ1a can influence their activity.
Materials and Methods
Details are described in SI Materials and Methods. We describe our screen for zed mutants and characterization of zed1. The SI includes information on plant material and bacterial strains, cloning, Illumina sequencing, P. syringae infections, protein purification, yeast interaction assays, bimolecular fluorescence microscopy, surface plasmon resonance analysis, immunoblot analysis, mass spectrometry, acetylation assays, kinase assays, phylogenetic analysis, and protein analysis.
Supplementary Material
Acknowledgments
We thank Wasan Abada, Ronald Wu, Jing Huang, and Tiffany Tong for help with mutant screening; Gregory Walker for help with N. benthamiana transient assays; Dr. Ryan Austin for help with NGM; Pauline Fung and Dr. Yunchen Gong for help with the Illumina GAII; Drs. Peter McCourt and Shelley Lumba for the SnRK3.22 clone; Dr. Jianfeng Zhang for cloning At3g57700; Dr. Brenda Andrews for yeast expression vectors; Dr. Dinesh Christendat for the modified pET28a vector; Corinna Felsensteiner for constructing the split-YFP vectors; and the Arabidopsis Biological Research Center for providing seed stocks. This work was supported by Natural Sciences and Engineering Research Council of Canada awards (to D.S.G. and D.D.), a Canada Research Chair in Plant-Microbe Systems Biology (to D.D.) or Comparative Genomics (to D.S.G.); the Centre for the Analysis of Genome Evolution and Function (D.D. and D.S.G.); and US Department of Agriculture Agricultural Research Service Grant 5335-21000-040-00D (to J.D.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315520110/-/DCSupplemental.
References
- 1.Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428(6984):764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6(10):973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 3.Ting JPY, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8(5):372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- 4.Heath MC. Hypersensitive response-related death. Plant Mol Biol. 2000;44(3):321–334. doi: 10.1023/a:1026592509060. [DOI] [PubMed] [Google Scholar]
- 5.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 6.Van der Biezen EA, Jones JDG. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23(12):454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 7.van der Hoorn RAL, Kamoun S. From Guard to Decoy: A new model for perception of plant pathogen effectors. Plant Cell. 2008;20(8):2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galán JE, Collmer A. Type III secretion machines: Bacterial devices for protein delivery into host cells. Science. 1999;284(5418):1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 9.Lewis JD, et al. The YopJ superfamily in plant-associated bacteria. Mol Plant Pathol. 2011;12(9):928–937. doi: 10.1111/j.1364-3703.2011.00719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma WB, Dong FFT, Stavrinides J, Guttman DS. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2006;2(12):e209. doi: 10.1371/journal.pgen.0020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittal R, Peak-Chew SY, McMahon HT. Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci USA. 2006;103(49):18574–18579. doi: 10.1073/pnas.0608995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukherjee S, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312(5777):1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 13.Orth K, et al. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 1999;285(5435):1920–1923. doi: 10.1126/science.285.5435.1920. [DOI] [PubMed] [Google Scholar]
- 14.Paquette N, et al. Serine/threonine acetylation of TGFβ-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc Natl Acad Sci USA. 2012;109(31):12710–12715. doi: 10.1073/pnas.1008203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meinzer U, et al. Yersinia pseudotuberculosis effector YopJ subverts the Nod2/RICK/TAK1 pathway and activates caspase-1 to induce intestinal barrier dysfunction. Cell Host Microbe. 2012;11(4):337–351. doi: 10.1016/j.chom.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Lewis JD, Abada W, Ma WB, Guttman DS, Desveaux D. The HopZ family of Pseudomonas syringae type III effectors require myristoylation for virulence and avirulence functions in Arabidopsis thaliana. J Bacteriol. 2008;190(8):2880–2891. doi: 10.1128/JB.01702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis JD, Wu R, Guttman DS, Desveaux D. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 2010;6(4):e1000894. doi: 10.1371/journal.pgen.1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee AH-Y, et al. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog. 2012;8(2):e1002523. doi: 10.1371/journal.ppat.1002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchetot C, Chagnon M, Dubé N, Hallé M, Tremblay ML. Substrate-trapping techniques in the identification of cellular PTP targets. Methods. 2005;35(1):44–53. doi: 10.1016/j.ymeth.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Overall CM, et al. Protease degradomics: Mass spectrometry discovery of protease substrates and the CLIP-CHIP, a dedicated DNA microarray of all human proteases and inhibitors. Biol Chem. 2004;385(6):493–504. doi: 10.1515/BC.2004.058. [DOI] [PubMed] [Google Scholar]
- 21.Eitas TK, Dangl JL. NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 2010;13(4):472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukasik E, Takken FLW. STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol. 2009;12(4):427–436. doi: 10.1016/j.pbi.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Shan LB, et al. The HopPtoF locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol Plant Microbe Interact. 2004;17(5):447–455. doi: 10.1094/MPMI.2004.17.5.447. [DOI] [PubMed] [Google Scholar]
- 24.Lewis JD, et al. Quantitative Interactor Screening with next-generation Sequencing (QIS-Seq) identifies Arabidopsis thaliana MLO2 as a target of the Pseudomonas syringae type III effector HopZ2. BMC Genomics. 2012;13:8. doi: 10.1186/1471-2164-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langlet C, et al. Contribution of the carboxyl terminus of the VPAC1 receptor to agonist-induced receptor phosphorylation, internalization, and recycling. J Biol Chem. 2005;280(30):28034–28043. doi: 10.1074/jbc.M500449200. [DOI] [PubMed] [Google Scholar]
- 26.Taylor SS, Shaw A, Hu J, Meharena HS, Kornev A. Pseudokinases from a structural perspective. Biochem Soc Trans. 2013;41(4):981–986. doi: 10.1042/BST20130120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16(9):443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Nimchuk ZL, Tarr PT, Meyerowitz EM. An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell. 2011;23(3):851–854. doi: 10.1105/tpc.110.075622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer GH, Garrod S, Woods VL, Jr, Taylor SS. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: Study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol. 2005;351(5):1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Trosky JE, et al. VopA inhibits ATP binding by acetylating the catalytic loop of MAPK kinases. J Biol Chem. 2007;282(47):34299–34305. doi: 10.1074/jbc.M706970200. [DOI] [PubMed] [Google Scholar]
- 31.Lehti-Shiu MD, Shiu SH. Diversity, classification and function of the plant protein kinase superfamily. Philos Trans R Soc B-Biol Sci. 2012;367(1602):2619–2639. doi: 10.1098/rstb.2012.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mucyn TS, et al. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18(10):2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA. 2007;104(7):2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutierrez JR, et al. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61(3):507–518. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- 35.Swiderski MR, Innes RW. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 2001;26(1):101–112. doi: 10.1046/j.1365-313x.2001.01014.x. [DOI] [PubMed] [Google Scholar]
- 36.Chang JH, et al. Functional analyses of the Pto resistance gene family in tomato and the identification of a minor resistance determinant in a susceptible haplotype. Mol Plant Microbe Interact. 2002;15(3):281–291. doi: 10.1094/MPMI.2002.15.3.281. [DOI] [PubMed] [Google Scholar]
- 37.Shao F, et al. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301(5637):1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 38.Lewis JD, Guttman DS, Desveaux D. The targeting of plant cellular systems by injected type III effector proteins. Semin Cell Dev Biol. 2009;20(9):1055–1063. doi: 10.1016/j.semcdb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 39. Fehr D, et al. (2006) AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-kappa B signalling pathway. Microbiology-(UK) 152(9):2809–2818. [DOI] [PubMed]
- 40.Jones RM, et al. Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe. 2008;3(4):233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.