Significance
Activation of the immune system within the nervous system is widely found in neurodegenerative diseases, including ALS. In mice that develop fatal paralytic disease from ALS-causing superoxide dismutase (SOD1) mutants, motor neurons activate expression of C1q, the initiating component of the classic complement system. As C1q and complement play a central role in developmental synapse elimination, disease-linked activation has suggested that it drives motor neuron denervation. Instead, suppressing C1q induction by gene deletion is shown to enhance loss, not retention, of synapses, whereas elimination of global complement activation by C1q or C3 gene deletions leave onset and progression of paralytic disease unaffected. Thus, C1q induction and complement activation are not significant contributors to SOD1 mutant-mediated ALS disease mechanism in mice.
Keywords: amyotrophic lateral sclerosis, motoneuron, synaptic density, gender differences, neuroinflammation
Abstract
Accumulating evidence from mice expressing ALS-causing mutations in superoxide dismutase (SOD1) has implicated pathological immune responses in motor neuron degeneration. This includes microglial activation, lymphocyte infiltration, and the induction of C1q, the initiating component of the classic complement system that is the protein-based arm of the innate immune response, in motor neurons of multiple ALS mouse models expressing dismutase active or inactive SOD1 mutants. Robust induction early in disease course is now identified for multiple complement components (including C1q, C4, and C3) in spinal cords of SOD1 mutant-expressing mice, consistent with initial intraneuronal C1q induction, followed by global activation of the complement pathway. We now test if this activation is a mechanistic contributor to disease. Deletion of the C1q gene in mice expressing an ALS-causing mutant in SOD1 to eliminate C1q induction, and complement cascade activation that follows from it, is demonstrated to produce changes in microglial morphology accompanied by enhanced loss, not retention, of synaptic densities during disease. C1q-dependent synaptic loss is shown to be especially prominent for cholinergic C-bouton nerve terminal input onto motor neurons in affected C1q-deleted SOD1 mutant mice. Nevertheless, overall onset and progression of disease are unaffected in C1q- and C3-deleted ALS mice, thus establishing that C1q induction and classic or alternative complement pathway activation do not contribute significantly to SOD1 mutant-mediated ALS pathogenesis in mice.
ALS is an adult-onset neurodegenerative disease characterized by degeneration and death of brain and spinal cord motor neurons. No current treatment slows disease progression more than modestly. Almost invariably, ALS leads to death within 1 to 5 y after onset (1). As familial and sporadic ALS cases are clinically nearly indistinguishable, insights from the 10% of cases of familial forms are likely to help understand the more common sporadic forms. The two most prevalent genetic causes are dominant missense mutations in the ubiquitously expressed superoxide dismutase 1 (SOD1) gene and the recently identified intronic hexanucleotide repeat expansions in the C9ORF72 gene (2, 3).
To date, the models most closely resembling human ALS are mice expressing mutant human SOD1 that develop a progressive paralysis characterized by spinal cord motor neuron loss, whereas comparable expression of WT human SOD1 does not generate any neuronal death (1, 4). A plethora of pathological mechanisms have been proposed, including misfolded protein aggregation, glutamate excitotoxicity, mitochondrial damage, and deregulation of RNA metabolism (1, 5). In addition, insights from mutant SOD1 mice have demonstrated that toxicity acts in a non–cell-autonomous manner, involving mutant-dependent damage within motor neurons to drive disease onset (6) and within neighboring astrocytes and microglia to drive more rapid disease progression (6, 7).
Independent of the origin and nature of the actual toxicity, a common feature of ALS is a robust neuroinflammatory response that is present in human ALS and mutant SOD1 mice (1, 8). Accumulating evidence from SOD1 mutant-expressing mice suggests that modulating this neuroinflammatory response can have beneficial or deleterious effects on motor neuron degeneration and thus contributes further to the non–cell-autonomous nature of mutant SOD1-mediated toxicity (1, 8). The neuroinflammatory response is produced by the innate and adaptive immune systems, with activation of resident microglia (9) as well as infiltration of peripheral lymphocytes (10, 11) and (although controversial) monocytes (12, 13). Functional studies in SOD1 mutant mice have shown that suppressing neurotoxic microglial effector systems can reduce motor neuron loss (6, 14), whereas suppressing infiltration of CD4+ T lymphocytes unexpectedly revealed a neuroprotective action (10, 11).
Besides its cellular component, an additional effector of the immune system is the powerful, protein-based complement system (15, 16). This system consists of nine protein components (C1–C9) and additional regulatory proteins that comprise a protease-cascade (C1–C5) that culminates in assembly and activation of the C5b–C9 membrane attack complex, which is used to lyse pathogens and target cells (Fig. S1).
C1q, formed by the three C1q α-, β-, and γ-chains (encoded by the C1qa/b/c genes), represents the initiating component of the classic complement pathway (C1→C4/C2/C3→C5) (17, 18) that is typically triggered by C1q recognizing an antibody-coated target, whereas C3 is central to the classic and the alternative complement pathway (C3→C3/B→C5) that is triggered by spontaneous C3 proteolysis (Fig. S1). Although most complement components are produced by the liver, many different cell types, including CNS microglia, specific neuronal populations, and astrocytes, can produce especially C1q, C3, and C4 complement components (16, 19, 20).
In postmortem ALS spinal cord tissues, a robust complement activation has been detected at RNA and protein levels (21–23), leading to increased amounts of activated complement components in CSF (24) and serum (25) of patients with ALS. In addition, RNA profiling suggested direct induction of C1q complement components in laser-microdissected postmortem ALS motor neurons (26). With respect to ALS mouse models, our own recent laser microdissection-assisted RNA profiling of motor neurons isolated from two different SOD1 mutant ALS mouse lines (SOD1G37R and SOD1G85R) identified the mRNAs encoding all three C1q subunits to be induced early during the disease in both lines (27). Similarly, we (27) and others (28) have detected C1q protein deposition on the motor neuron cell surface throughout symptomatic phases of mutant SOD1-expressing mice, consistent with the ability to produce locally a complement cascade.
Regarding the later steps in complement activation, divergent outcomes have been reported. Deletion of the gene encoding C4 (which is part of the classic complement pathway) did not affect disease course in high-expressing mutant SOD1G93A mice (29). However, the C3-dependent alternative complement pathway activation would still be possible (Fig. S1). Conversely, pharmacological blockage of the C5a receptor in the SOD1G93A mutant rat was reported to provide a beneficial effect on survival (30), although the specificity of drug-induced inhibition of C5a in the rat has not directly been validated. Furthermore, deletion of C4 is a poor test for the role of C1q induction in ALS, as C1q can act independently of the downstream classic complement components via several proposed C1q receptors expressed on diverse cell types, including on microglia (17, 18).
Concerning the functions of C1q on its own and independent of the downstream complement cascade, it has been suggested that C1q can directly recognize apoptotic cells to enhance clearance by phagocytes and modulate their cytokine release (31–33). Indeed, a C1q-mediated general complement activation in the affected spinal cord and a local neuronal induction of C1q acting as a stress signal to modulate microglial responses could impact motor neuron degeneration in ALS. Several in vitro studies have shown that, in the absence of other complement components, C1q can modulate phagocytosis by monocytic cells and enhance clearance of apoptotic neurons by microglia, acting as a direct opsonization agent for phagocytes (31–33). Just this situation has been suggested for an in vivo model of retinitis pigmentosa, in which C1q deletion led to reduced apoptotic cell clearance, independent of C3 (34). C1q can also modulate subsequent cytokine production by activated microglia, monocytes, and macrophages, including reducing specific proinflammatory signals (IL-1β, IL-6, and TNF-α) (31, 32, 35), suggesting a general anti-inflammatory activity of C1q by combining rapid clearance of apoptotic cells, followed by reducing deleterious inflammation. Furthermore, C1q addition to pure cultured rat primary cortical neurons led to an up-regulation of genes with neuroprotective actions (36). These effects of C1q are most likely produced via C1q receptors, although their exact identity is still controversial (17). The sum of this evidence suggests that early C1q induction (independent of consecutive downstream complement activation) could have beneficial effects during neurodegeneration.
C1q induction and complement activation is also a general aspect of pathogenesis of other major neurodegenerative diseases and has been found in Alzheimer’s and Parkinson diseases (16). Perhaps most provocatively for an involvement in ALS pathogenesis, C1q has also been suggested to play a central role in facilitating developmental synapse elimination (37–39), and hence could play a similar role in the denervation of affected motor neuron units that represents an early phase of disease in SOD1 mutant-mediated pathogenesis in mice (40, 41). Recognizing these multiple potential mechanistic contributions of C1q induction as well as global contributions of general complement pathway activation to neuronal degeneration in ALS, we have now determined how pathogenesis in SOD1-mediated ALS is affected in mice when C1q induction as well as classic C1q- or alternative C3-dependent complement cascades are suppressed by gene deletions.
Results
C1q Induction and Classic Complement Pathway Activation in SOD1 Mutant ALS Mice.
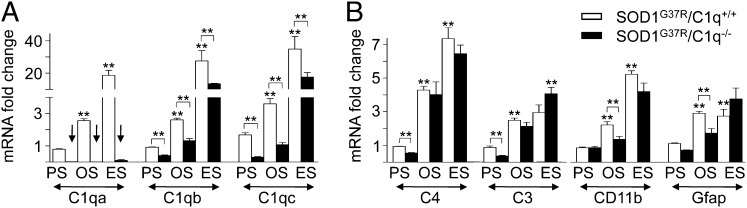
To assess if C1q induction and activation of the classic complement pathway contribute to pathogenesis, we cross-bred C1q-deleted mice (42) with mutant SOD1G37R ALS mice (which have a typical survival of 6 mo). We used C1qa KO mice (C1q−/−), which cannot form a mature C1q complex and therefore suppress activation of the classic complement pathway (42). Real-time RT-PCR of RNA from lumbar spinal cord tissues [at presymptomatic (8 wk of age), onset (18 wk of age, the average weight peak), and end stage (complete hindlimb paralysis, an average at 6 mo of age) time points] revealed that mRNAs encoding all three C1q subunits (C1qa/b/c), as well as two downstream complement components (C4, C3), were strongly induced beginning at onset. Induction correlated temporally with microglial and astrocytic activation markers (CD11b/Itgam and Gfap; Fig. 1 A and B), consistent with previous reports of induction of C1q in spinal cords of other SOD1 mutant ALS mouse lines (43–45). No similar complement induction was found in mice expressing high levels of WT human SOD1 (SOD1WT) and which do not develop disease. As expected, C1qa mRNA induction was absent in SOD1G37R/C1q−/− mice, whereas accumulation of the C1qb/c subunits was strongly reduced (Fig. 1A). Furthermore, C1q deletion led to delayed inductions of C4 and C3, as well as of microglial CD11b/Itgam and astrocytic Gfap mRNAs (Fig. 1B).
Fig. 1.
Early induction of C1q and classic complement components in spinal cords of ALS mice. (A and B) Complement components and glial activation markers were assessed in ALS mice (open bars), and their induction levels compared with ALS mice lacking C1q (the initiating component of the classic complement pathway; filled bars). By using quantitative RT-PCR, lumbar spinal cord mRNA levels of all three C1q genes (C1qa/b/c) (A) and downstream complement components (C4, C3) (B) were assessed in mutant SOD1G37R mice (open bars), presymptomatically (PS; at 8 wk of age), at onset (OS; 18 wk of age, the average weight peak), and end stage (ES; complete hindlimb paralysis, average of 6 mo of age) and correlated to microglial (CD11b/Itgam) and astrocytic (Gfap) activation (B). Compared with control (6-mo-old) SOD1WT samples, early induction of C1q and classic complement components were detected in mutant SOD1G37R mice and paralleled glial activation (A and B). In C1q-deleted ALS mice (filled bars), loss of C1qa was confirmed (arrows, A). Mice deleted for C1qa are unable to form a functional C1q polypeptide, and induction levels of C1qb/c genes were strongly reduced. C1q deletion in ALS mice led to a transient delay in induction of downstream complement components (C4, C3) as well as glial activation markers (microglial CD11b/Itgam and astrocytic Gfap) (B). (**P < 0.01, Student t test; n = 4 mice per genotype and disease stage; error bars represent SEM).
Global Neuroinflammatory Response and Neuronal Loss Are Unchanged in C1q-Deleted SOD1 Mutant ALS Mice.
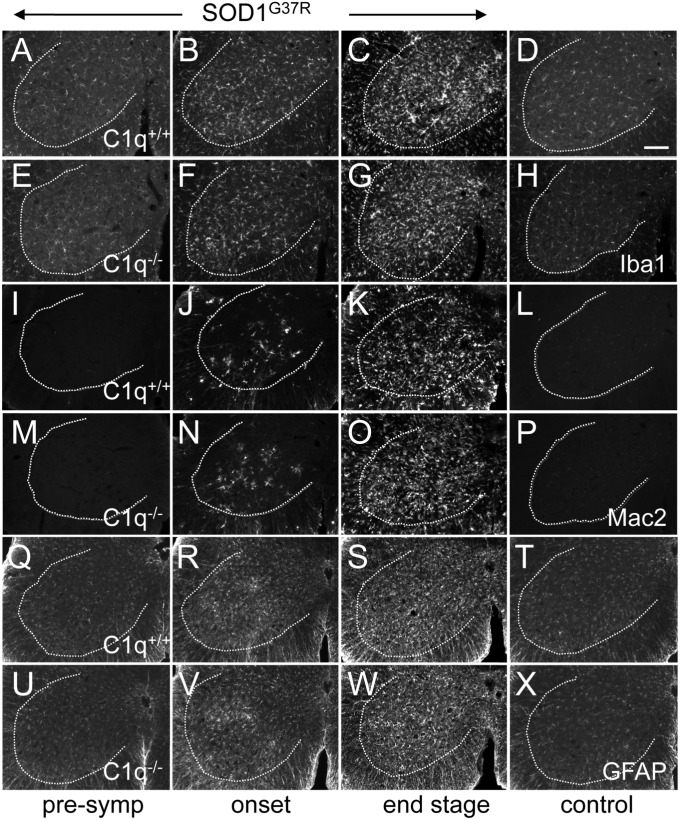
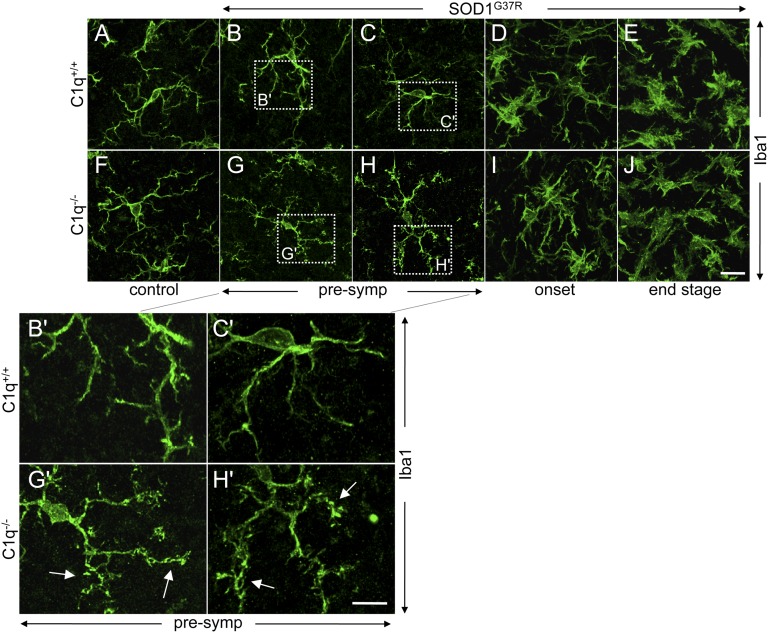
C1q deletion did not influence the global neuroinflammatory response in SOD1G37R mice, as measured by immunohistochemical staining for activated microglia (Iba1/Aif1 and Mac2/Lgals3) and astrocytes (GFAP) in affected lumbar spinal cords (Fig. 2). Similarly, it did not change the time course of overall microglial or astrocytic activation (which initiated before onset) in SOD1G37R ALS mice (Fig. 2). Nevertheless, absence of C1q in ALS mice provoked increased numbers of bulbous termini on microglial processes that was especially prominent at presymptomatic disease stages (Fig. 3), consistent with signs of increased phagocytic activity (46).
Fig. 2.
Time course of global glial activation is unchanged in spinal cords of C1q-deleted ALS mice. (A–X) Immunofluorescence stainings for two microglial [Iba1/Aif (A–H), Mac2/Lgals3 (I–P)] and one astrocytic (GFAP) (Q–X) activation markers. No signs of glial activation were present in 6-mo-old control SOD1WT mice with normal C1q content (D, L, and T) or in 6-mo-old control mice deleted in C1q (H, P, and X). In mutant SOD1G37R ALS mice, overall glial activation was detectable at onset (B, J, and R) (at 18 wk of age, the average weight peak) and increased further at end stage (C, K, and S) (complete hindlimb paralysis,average of 6 mo of age); however, this was independent of whether C1q was present (A–C, I–K, and Q–S) or absent (E–G, M–O, and U–W) (n = 4 mice per genotype and disease stage). (Scale bar: D, 100 μm.)
Fig. 3.
C1q-dependent morphological changes of microglia during disease course of ALS mice. (A–J) Confocal images of immunofluorescence stainings for the microglial marker Iba1. (A and F) Control 6-mo-old SOD1WT (A) or C1q-deleted mice (F) showed microglia with a resting phenotype and long branched processes. (B and C, G and H) In presymptomatic (at 8 wk of age) mutant SOD1G37R ALS mice, C1q-deletion led to an increased number of bulbous termini (arrows, G’ and H’, Insets) on microglial processes (compare B′ and C′ for ALS mice with normal C1q-content vs. G′ and H′ for C1q-deleted ALS mice). (D, E, I, and J) In mutant SOD1G37R ALS mice at onset (at 18 wk of age, the average weight peak) and end stage (complete hindlimb paralysis, average of 6 mo of age) time points, microglia had acquired a classic activated morphology with retracted processes, but overall morphology was independent of C1q deletion (n = 4 mice per genotype and disease stage). (Scale bars: J, 20 µm; H′, 10 µm.)
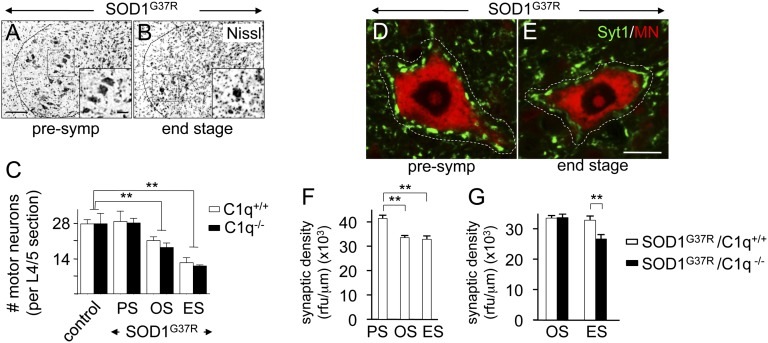
Next, the influence of C1q deletion on SOD1 mutant-induced motor neuron loss was determined by counting Nissl-positive motor neurons (diameter >20 μm) in lumbar spinal cord ventral horns (Fig. 4 A and B). This quantification revealed that control SOD1WT and C1q−/− mice showed equal numbers of motor neurons, demonstrating that C1q deletion did not affect motor neuron development (Fig. 4C). However, absence of C1q and downstream complement activation in SOD1 mutant mice did not significantly change the initial number or speed of overall loss of motor neurons throughout disease, which, in SOD1G37R/C1q+/+ mice, reached 24% (±5.5, SEM) and 56% (±7.2, SEM) at onset and end stage, respectively (P < 0.01, Student t test; Fig. 4C).
Fig. 4.
Motor neuron loss is unchanged but loss of presynaptic density onto motor neurons is more prounounced in C1q-deleted ALS mice. (A–C) Motor neuron loss. Shown are Nissl stainings with enlargements (Insets) of lumbar spinal cord ventral horns to identify motor neurons in mutant SOD1G37R ALS mice presymptomatically (A) and at end stage (B). (C) Quantification indicated equal numbers of large lumbar (L4/5) motor neurons in (6-mo-old) control SOD1WT and C1q-deleted mice whereas significant motor neuron loss was measured at onset and end stage in SOD1G37R ALS mice, but independent of the prescence of C1q. (Scale bar: A, 100 µm.) (D–G) Loss of general presynaptic density onto large lumbar (L4/5) ventral horn motor neurons during disease course. Confocal images of double-immunofluorescence stainings for the general presynaptic marker Syt1 (green) and for a fluorescent Nissl stain (MN; red) indicated significant disease-linked reduction of presynaptic density (measured between the perikaryon and the dotted white line) on affected motor neurons, when comparing presymptomatic (D) vs. end-stage (E) mutant SOD1G37R ALS mice. (F) Quantification of the loss in presynaptic density [measured as relative fluorescence units (rfu) of Syt1 per micrometer of motor neuron contour length] observed in D and E. (G) Quantification showing increased loss after onset in presynaptic density in the absence of C1q (**P < 0.01, Student t test; n = 4 mice per genotype and disease stage; error bars indicate SEM). ES, end stage (complete hindlimb paralysis, average of 6 mo of age); OS, onset (18 wk of age, average weight peak); PS, presymptomatic (8 wk of age). (Scale bar: E, 15 μm.)
Increased Loss of Presynaptic Density on Motor Neurons in C1q-Deleted ALS Mice.
Provocative recent evidence from Stevens and Barres has indicated that C1q and classic complement pathway activation are necessary for developmental synapse elimination in the mouse visual system and that this developmental program is reactivated during adult neurodegeneration in mouse glaucoma models (37, 47). Thus, we determined if C1q deletion influenced the rate of synaptic degeneration around affected spinal cord motor neurons in SOD1G37R mutant ALS mice (Fig. 4 D–G).
In a first approach, we assessed global synaptic density by using a rather general presynaptic marker, synaptotagmin-1 (Syt1), likely present in the terminals of all three major inputs (glutamatergic, GABAergic, and cholinergic) onto motor neurons, and measured total Syt1 fluorescence intensity juxtaposed to the perikarya of large ventral horn motor neurons (identified by fluorescent Nissl stain). By using this method, a small but significant loss of presynaptic densities (19.1 ± 2.0%, SEM; P = 0.004, Student t test) was identified on surviving motor neurons when comparing end-stage to presymptomatic mutant SOD1G37R mice (Fig. 4 D–F), consistent with similar loss previously reported for the aggressive SOD1G93A mutant ALS line (40). Absence of C1q and complement activation produced no difference at disease onset in Syt1 presynaptic density in compairing SOD1G37R mutant ALS mice with and without C1q (Fig. 4G). However, by end stage, absence of C1q led to an increased loss of Syt1 presynaptic density juxtaposed around motor neurons, resulting in a significant difference between SOD1G37R/C1q+/+ and SOD1G37R/C1q−/− mice (18.9 ± 2.2%, SEM; P = 0.005, Student t test; Fig. 4G). This led to a further decrease of an additional 20% in presynaptic density by end stage in C1q-deleted SOD1 mutant mice compared with the SOD1 mutant mice with normal C1q content (Fig. 4G). Thus, C1q activity potentiates maintenance, not loss, of synaptic connections during ALS-like disease progression.
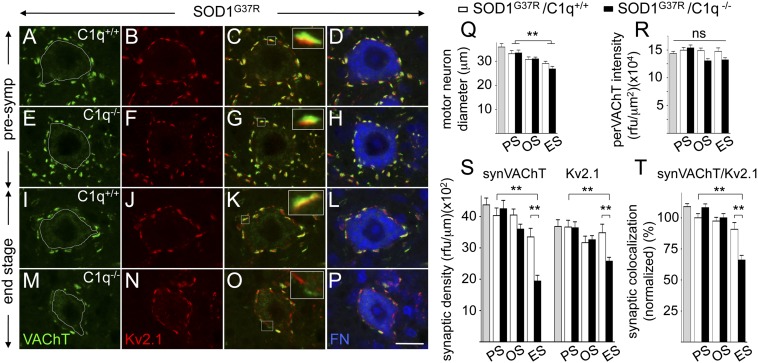
In a second approach, we asked which subtype of synaptic input could be affected by C1q deletion. To achieve this, antibodies were identifed that recognized pre- or postsynaptic components under conditions compatible with the tissue fixation conditions used with our mouse cohorts. The most prominent cholinergic input onto ventral horn motor neurons, which takes the form of the large and easily identifiable C-boutons (48), are exceptionally well suited to assess detailed pre/postsynaptic marker analysis. C-bouton nerve terminals mostly originate from a small population of V0c interneurons localized around the central canal (48). We used well-documented antibodies against presynaptic vesicular acetylcholine transferase (VAChT), which is localized to the C-bouton nerve terminals of the cholinergic V0c interneurons and postsynaptic Kv2.1 (a potassium channel; closely associated to m2-type muscarinic receptors), which is localized in motor neurons to the postsynaptic density of C-boutons (48). Total fluorescent intensities of presynaptic VAChT and closely apposed postsynaptic Kv2.1 densities were measured on large ventral horn motor neuron perikarya (identified by fluorescent Nissl stain; Fig. 5). First, we detected a decrease (16 ± 5.1%, SEM; P = 0.017, Student t test) in overall motor neuron diameter at end stage compared with presymptomatic SOD1G37R mutant ALS mice, but independent of the prescence of C1q (Fig. 5Q). Next, and in line with recent data from SOD1G93A mutant ALS mice (49, 50), C-bouton nerve terminal input (apposed VAChT/Kv2.1 synaptic densities) on surviving large ventral horn motor neurons remained relatively stable during disease in SOD1G37R mice expressing C1q (Fig. 5 S and T). [At end stage, a trend (that did not reach statistical significance) was measured for loss of presynaptic VAChT, possibly part of the general presynaptic loss (measured using Syt1 in Fig. 4F).] At presymptomatic and onset disease stages, C1q deletion did not lead to significant changes in synaptic VAChT/Kv2.1 densities (Fig. 5 S and T). Surprisingly, and consistent with what we already found for the global presynaptic analysis (Fig. 4 D–G), at end-stage C1q deletion led to a significant loss of VAChT/Kv2.1 synaptic densities, resulting in a significant difference of 40% for VAChT and 25% for Kv2.1 between SOD1G37R/C1q+/+ and SOD1G37R/C1q−/− mice (42 ± 9.9%, SEM; P = 0.0001, Student t test; and 26 ± 8.7%, SEM; P = 0.0045, Student t test, for VAChT and Kv2.1, respectively; Fig. 5S).
Fig. 5.
C1q deletion leads to a preferential loss of cholinergic C-bouton nerve terminals onto motor neurons in affected SOD1 mutant-expressing ALS mice. (A–P) Confocal images of triple-immunofluorescence stainings to assess cholinergic C-bouton nerve terminals on large lumbar (L4/5) ventral horn motor neurons during disease course in SOD1G37R ALS mice with (A–D and I–L) or without (E–H and M–P) C1q at presymptomatic (A–H) and end-stage (I–P) time points. C-boutons were identified by presynaptic VAChT (A, E, I, and M; green) and apposed postsynaptic Kv2.1 (B, F, J, and N; red) densities, whereas motor neurons were marked by fluorescent Nissl (FN, blue; D, H, L, and P). Apposition/colocalization of VAChT/Kv2.1 are shown as yellow overlap (C, G, K, O) with enlargements (Insets) showing an individual C-bouton. Although ALS mice with normal C1q content did not show major loss of C-boutons on motor neuron during disease (compare A–D vs. I–L), C1q-deleted ALS mice developed a loss of C-boutons at disease end stage (compare I–L vs. M–P). (Q and R) Analysis of large surviving motor neurons at three disease stages in SOD1G37R ALS mice revealed a slight shrinkage of diameter (Q) (independent of C1q), whereas perikaryal VAChT fluorescence intensity (designated perVAChT within the dotted white line; A, E, I, and M) stayed relatively constant (R). (S and T) Quantification of C-bouton synaptic density analysis on motor neurons indicated that C1q deletion led to a strong loss of presynaptic VAChT-positive input (designated synVAChT) and corresponding Kv2.1-positive postsynaptic densities on motor neurons [measured as relative fluorescence units (rfu) of synVAChT and Kv2.1 per micrometer of contour length] at end stage in SOD1G37R ALS mice (S). Detailed analysis indicated that C-boutons were not simply lost, but [as a result of greater synVAChT than Kv2.1 loss (S)] developed reduced VAChT/Kv2.1 apposition/colocalization (measured as the ratio of fully apposed to total VAChT/Kv2.1 synaptic densities) (T), consistent with reduced synaptic integrity. (**P < 0.01, Student t test; ns, nonsignificant; n = 4 mice per genotype and disease stage; error bars represent SEM). Gray bars in Q–T: 6-mo-old control, nontransgenic mice. ES, end stage (complete hindlimb paralysis, average of 6 mo of age); OS, onset (18 wk of age, average weight peak); PS, presymptomatic (at 8 wk of age). (Scale bar: P, 25 μm.)
Whereas SOD1G37R mice with normal C1q retained almost all of their Kv2.1 (95 ± 7.4%, SEM; P = 0.5943, Student t test) and VAChT (83 ± 6.7%, SEM; P = 0.0694, Student t test) synaptic densities, C1q deletion resulted in 70% of Kv2.1 (69 ± 3.5%, SEM; P = 0.0001, Student t test) and only 50% of VAChT (48 ± 4.6%, SEM; P = 0.0001, Student t test) synaptic densities remaining on motor neurons of end-stage SOD1G37R/C1q−/− mice, indicating a preferential C1q-dependent loss of presynaptic VAChT nerve terminal input (Fig. 5S). Indeed, when analyzing the ratio of fully apposed to total VAChT/Kv2.1 synaptic densities (as a rough measure of pre-/postsynaptic integrity), no change was detected during disease in SOD1G37R mice with normal C1q content, whereas a 35% reduction (34 ± 5.5%, SEM; P = 0.0001, Student t test) was identified in endstage C1q-deleted SOD1G37R mice (compared with end-stage SOD1G37R/C1q+/+ mice; Fig. 5T). Of note, the measured C1q-dependent loss of VAChT presynaptic density (designated “synVAChT”), was not linked to an overall decrease in perikaryal VAChT fluorescence intensity (designated “perVAChT”) of surviving motor neurons (Fig. 5R), further supporting C1q-dependent loss of C-bouton nerve terminal inputs within end-stage ALS mice.
C1q Induction and Global Complement Pathway Activation Do Not Contribute to Overall Neurotoxicity in SOD1 Mutant ALS Mice.
We next tested if the C1q-dependent retention of synaptic connections between upper and lower motor neurons affected disease onset, progression, or survival of SOD1G37R mutant ALS mice. The well-established paradigm of weight loss from denervation-induced muscle atrophy was used to provide an unbiased measure of disease onset (27, 51) (defined by peak weight achieved before denervation-dependent muscle atrophy), progression to an early symptomatic disease phase correlating with muscle wasting and hindlimb weakness (defined by 10% weight loss; reached at ∼24 wk of age in SOD1G37R mice), and end stage defined by complete hindlimb paralysis, culminating in paralysis so severe that the animal was unable to right itself when placed on its side.
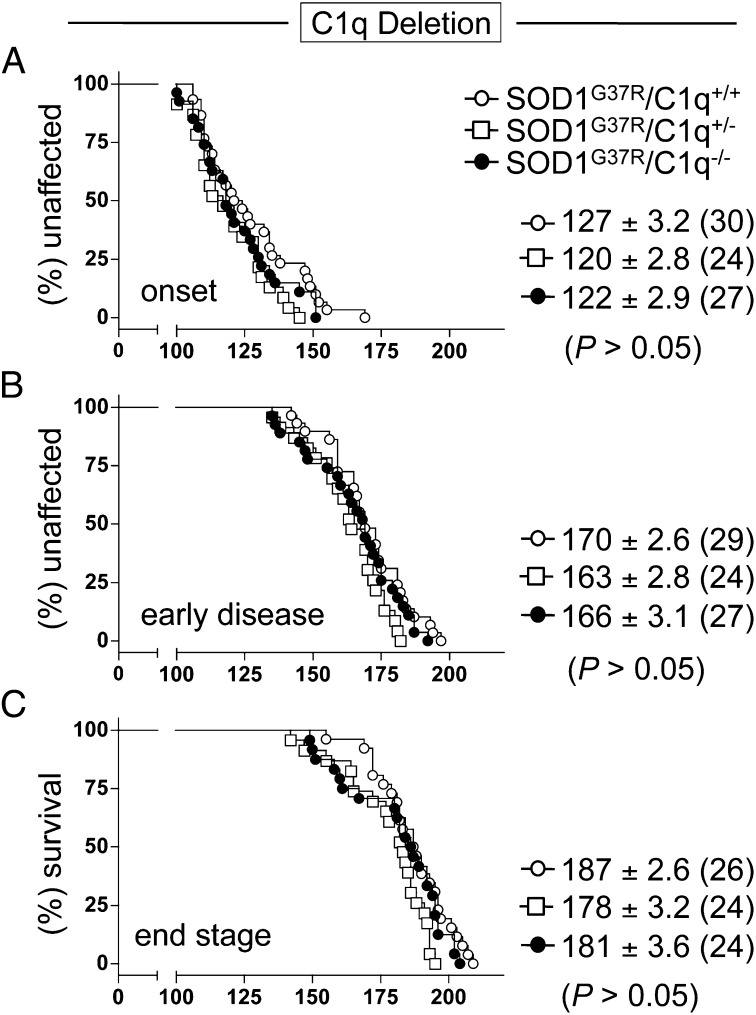
Despite strong and early C1q induction in affected motor neurons (27, 52–54) and total spinal cords (Fig. 1A), as well as induction of other components of the classic complement pathway (Fig. 1B) (27, 28, 43–45), elimination of classic complement activation by C1q deletion did not significantly change disease onset, progression, or survival (Fig. 6 and Table S1). SOD1G37R/C1q+/+ and SOD1G37R/C1q−/− mice reached onset at 127 d (±3.2; n = 30) and 122 d (±2.9; n = 27), respectively (Fig. 6A), whereas early disease was reached at 170 d (±2.6; n = 29) and 166 d (±3.1; n = 27), respectively (Fig. 6B). Similarly, SOD1 mutant mice with C1q (SOD1G37R/C1q+/+) survived 187 d (±2.6; SEM ; n = 26), whereas C1q-deleted ALS mice (SOD1G37R/C1q−/−) survived 181 d (±3.6; n = 24; Fig. 6C).
Fig. 6.
C1q induction and its activation of the classic complement pathway do not contribute to overall disease mechanism in SOD1G37R mutant ALS mice. (A–C) Kaplan–Meier plots of ages (in days) at which onset (weight peak) (A), early disease (10% weight loss) (B), or end stage (complete hindlimb paralysis) (C) were reached in mutant SOD1G37R mice with normal (C1q+/+, ○), partially (C1q+/−, □) or fully deleted (C1q−/−, ●) C1q content. Average (±SEM) ages (in days) of the different disease stages are shown, and animal numbers are indicated in brackets (equally sex-mixed). No statistical significant difference could be detected among the three genotypes [P = 0.19/P = 0.26 (A), P = 0.12/P = 0.34 (B), and P = 0.06/P = 0.27 (C), log-rank and Student t test; Table S1].
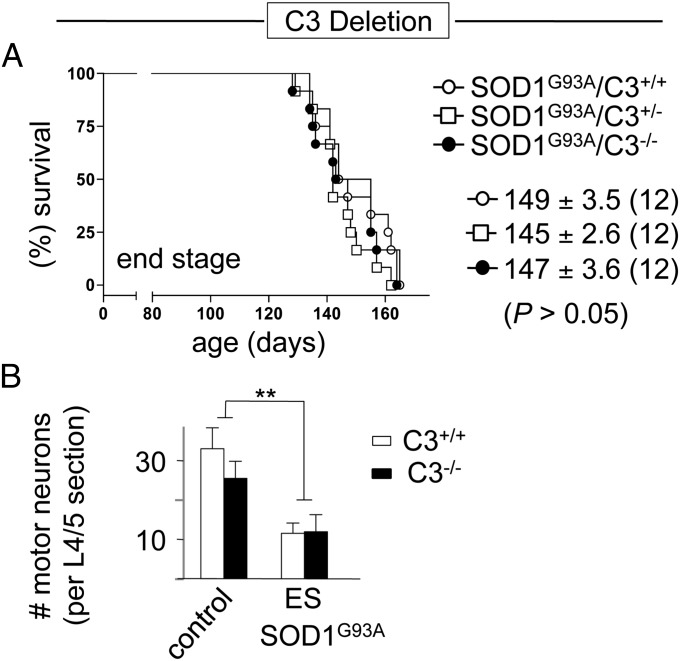
Although genetic deletion of C1q unambigously assessed the role of C1q-dependent classic complement pathway activation, the strong C3 induction we measured in affected ALS mouse spinal cords (Fig. 1B) could be evidence of an alternative pathway activation (Fig. S1). Therefore, we also genetically deleted C3 in SOD1 mutant ALS mice to block classic (C1q/C3-driven) and alternative (exclusively C3-driven) pathways. Overall survival was unchanged in SOD1G93A mutant ALS mice with C3 (149 d ± 3.6, SEM; n = 12) or without C3 (147 d ± 3.6, SEM ; n = 12; P = 0.50, log-rank and Student t test; Fig. 7A). Similarly, there was no C3 dependency on SOD1 mutant-induced motor neuron number loss (Fig. 7B), indicating that global complement pathway activation in SOD1 mutant mice does not contribute to overall ALS toxicity.
Fig. 7.
Unaltered survival and motor neuron loss in mutant SOD1-expressing ALS mice deleted in C3 demonstrating no contribution of an alternative complement activation pathway to toxicity in this ALS model. (A) Kaplan–Meier plot of ages (in days) at which end stage (complete hindlimb paralysis) was reached in mutant SOD1G93A mice with normal (C3+/+, ○) or partially (C3+/−, □) or fully deleted (C3−/−, ●) C3 content. Average (±SEM) ages (in days) of the disease end stages are shown, and animal numbers are indicated in brackets (equally sex-mixed). No statistically significant difference was detected among the three genotypes [P = 0.50/P = 0.23 (A), log-rank and Student t test]. (B) Quantification of motor neuron loss indicated equal numbers of large lumbar (L4/5) motor neurons in (5 mo old) nontransgenic control and C3-deleted mice. Significant motor neuron loss measured at end stage was independent of the prescence of C3 (**P < 0.01, Student t test; n = 4 mice per genotype and disease stage; error bars indicate SEM). ES, end stage (complete hindlimb paralysis).
Modest Protective Effect of C1q Induction on Disease Course in Male SOD1 Mutant ALS Mice.
In a recent study assessing the role of the neuroinflammation-related chemokine receptor CX3CR1 (the fraktalkine receptor), gene deletion (of Cx3cr1) was reported to very modestly change disease course, but only in male mutant SOD1 mice (55). Thus, we tested for similarly subtle C1q-dependent changes by reanalysis of the dataset in a sex-specific manner. No sex differences in the baseline mutant SOD1G37R mouse line with normal C1q content were identified, producing almost identical survival and disease progression ages between male and female mice (Fig. S2 and Table S1). Additionally, reduction or deletion in C1q did not influence disease progression or survival in female SOD1G37R mice (with an end stage of 184 d for all three genotypes, SOD1G37R/C1q+/+, SOD1G37R/C1q+/−, and SOD1G37R/C1q−/− ; n = 12–14 each; Fig. S3 A–C and Table S1). However, reduction in C1q in male SOD1G37R/C1q+/− mice modestly accelerated disease onset by 2 wk (15 d; P = 0.03, log-rank and Student t test), with a corresponding reduction of survival by 2 wk (13 d; P = 0.04, log-rank and Student t test; Fig. S3 D–F and Table S1). Complete deletion of C1q (SOD1G37R/C1q−/−) did not further accelerate onset (with an 11 d earlier onset and end stage; P = 0.13 and P = 0.18, respectively, log-rank and Student t test; Fig. S3 D–F and Table S1). The C1q and mutant SOD1 mice were on identical C57BL/6J genetic backgrounds, thereby eliminating confounding genetic influences beyond the mutant SOD1 gene and deletion of one or both alleles of C1q. These results suggest that C1q induction can have a modest protective effect on delaying disease onset in the context of sex-specific influences.
Discussion
We have previously identified strong induction of C1q, the initiating component of the classic complement pathway, in laser-microdissected motor neurons of SOD1 mutant ALS mice (27), consistent with an actual stress signal that could modulate local responses, including inflammatory ones. Other groups have also identified C1q induction in motor neurons of other ALS-linked SOD1 mutant-expressing mice (52, 53), including a study suggesting that C1q induction is more prominent in vulnerable relative to resistant motor neuron pools (54). Similarly, we and others have detected direct C1q protein deposition on affected motor neurons in ALS mice (27, 28), consistent with activation of the classic complement pathway, and C1q protein depositions were reported at the neuromuscular junctions in SOD1 mutant mice (44). Finally, in addition to C1q specifically induced within motor neurons, our present data (Fig. 1), along with other studies (27, 28, 30, 43–45), have detected global complement pathway activation in spinal cords (C1q/C4/C3/C5) and along peripheral nerves (C3) (29) of different SOD1 mutant ALS mouse and rat lines.
With respect to human ALS pathology, there is strong evidence for C1q induction and global complement pathway activation, revealed by the detection of increased amounts or activated forms of diverse complement components (C1q/C2/C4/C3/membrane attack complex) in affected postmortem spinal cord tissue as well as serum (C3c) and CSF (C4d/C3c) (18–22). Consistent with our findings (27) and those of others (28, 52–54) from isolated motor neurons of SOD1 mutant ALS mice, detailed analyses of a recent laser microdissection-based RNA profiling approach of human ALS postmortem tissue revealed evidence for induction of C1q components (C1QB/C) directly within affected motor neurons (table S2 in ref. 26). Interestingly, as those motor neurons were isolated from lumbar regions in relatively early stages of degeneration, this suggests motor neuronal C1q induction to be an early event in the neurodegenerative cascade in human disease (26).
These findings have implicated pathological C1q induction as a contributor to neurodegeneration in ALS. Further support came also from the recent discovery that C1q induction in retinal ganglion neurons is implicated in developmental synaptic pruning of the mouse visual system, by acting as a tag to mediate microglial-driven synaptic elimination (37, 39). Such synaptic localized C1q has been reported in mouse models of glaucoma, and C1q deletion provided protection from glaucoma (38, 47). Likewise, strong C1q induction has been reported in brains of Alzheimer’s disease mice, with C1q deletion leading to reduced neuropathology, consistent with C1q induction driving neurodegenerative conditions in general (56). However, our finding that suppression of C1q induction in ALS mice did not protect against motor neuron degeneration, nor against synaptic loss, rather suggests that any disease contribution of C1q induction is dependent on the precise neurodegnerative condition or neuronal systems implicated, and a proposed deleterious role, with respect to neuroinflammation and synaptic degeneration, cannot be easily generalized. Consistent with this idea, recent findings from Parkinson disease mouse models did not find protection against dopaminergic neurodegeneration after suppression of C1q induction (57).
Our evidence from mice that develop fatal ALS-like paralysis from expression of mutant SOD1 does identify a transient benefit from C1q expression in delaying changes in microglial morphology that probably reflect an increased activation state (Fig. 3). Similarly, during disease, C1q deletion led to an increased loss, not retention, in mutant SOD1-driven reduction in presynaptic density on lower motor neurons (Fig. 4G). The role of C1q in brain function is clearly complex, with established functions (together with C3) in synaptic elimination during development (37) but also unique roles (with C1q on its own, independent of C3) in normal brain aging, as suggested by a very recent study from Stephan et al. that found a general increase of parenchymal C1q protein levels in aged mouse brains (20). This increase was not, however, associated with increased synaptic elimination, but a role in circuit plasticity (and if anything, rather associated with enhanced synapse numbers) (20). Interestingly, our analysis of double pre-/postsynaptic stainings suggested that C1q deletion especially affected VOc interneuron-derived modulatory cholinergic inputs, detected by a reduction of (VAChT-positive) C-bouton nerve terminals on surviving ALS motor neurons (Fig. 5 S and T). As loss of the corresponding postsynaptic (Kv2.1-positive) density was less pronounced, it is possible that C1q could play a protective role against mutant SOD1 mutant-mediated toxicity in the corresponding cholinergic VOc interneurons, especially as no major loss of C-boutons on motor neurons was reported in ALS mice with normal C1q content (49, 50).
With respect to general complement pathway activation in SOD1 mutant mice, strong C3 induction in ALS mice suggested a possible C1q-independent pathway for complement activation. However, deletion of C3 in ALS model mice did not affect disease course (Fig. 7). Thus, despite the many implications of C1q activation as a component of ALS pathogenesis, our present study demonstrates that neither C1q induction nor activation of the global complement cascade (initiated by the classic C1q driver or an alternative one acting through C3) contributes significantly to ALS-like mutant SOD1-mediated toxicity in mice.
Materials and Methods
Animals.
Previously in the laboratory-generated transgenic mutant SOD1G37R (line 42) ALS mice (27) and C1qa KO mice, initially generated by Marina Botto (Imperial College London, London, United Kingdom) (42, 58), as well as SOD1G93A (Gur/J; Jackson Laboratory) and C3 KO mice (Jackson Laboratory) were used, and all were on a pure C57BL/6J genetic background. Additional details are provided in SI Materials and Methods.
Survival Analysis.
Mice were weighed weekly as an objective and unbiased measure of disease course, as described previously (27, 51). Additional details are provided in SI Materials and Methods.
Synaptic Density Analysis.
On six serially collected L4/5 lumbar spinal cord sections, total fluorescence intensities of the presynaptic markers Syt1 (Synaptic Systems) and VAChT (Millipore/Merck Germany) and the postsynaptic marker Kv2.1 (Millipore/Merck) around 25 to 50 randomly chosen large (>25 μm in diameter) NeuroTrace (Invitrogen) positive motor neurons (per animal) were measured by using confocal microscopy (with 60× objectives; n = 4 mice per genotype and disease stage, equally sex-mixed). Starting from the plane with a clear NeuroTrace-positive nuclear stain, individual synaptic fluorescence intensities (closely juxtaposed to the neuronal perikarya) were summed within a total of 11 confocal sections 0.30 μm thick (±5 sections above and below the middle plane) by using ImageJ/Fiji (National Institutes of Health). Synaptic densities were calculated as the total accumulated fluorescence intensity of synaptic stain per micrometer of (Nissl-positive) motor neuron contour length. Additional details are provided in SI Materials and Methods.
Disease stage-specific tissue collection, real-time RT-PCR analysis of spinal cord inflammation, immunohistochemical analysis of spinal cord inflammation, and motor neuron counts in spinal cords are described in SI Materials and Methods. All PCR primer sequences used are listed in Table S2.
Supplementary Material
Acknowledgments
We thank Drs. B. Stevens, B. Barres, and M. Botto for essential discussions about the complement system; Dr. M. Mallat for help with microglial morphology analysis; Dr. A. Dauphin for his ImageJ scripts used for synaptic density analysis; and Dr. M. Botto (Imperial College London) for giving us permission to use her C1q-deleted mice that were provided by B. Barres (Stanford Univeristy). This work was supported by National Institutes of Health Grant NS 27036 (to D.W.C.), a grant from the Packard Center for ALS Research (to D.W.C.), a Swiss National Science Foundation Fellowship (to C.S.L.), a Neuropole de Recherche Francilien Fellowship (to C.S.L.), a Marie-Curie International Reintegration Grant (to S.B.), and a Muscular Dystrophy Association Grant (to S.B.). Salary support for D.W.C. was provided by the Ludwig Institute for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318309110/-/DCSupplemental.
References
- 1.Boillée S, Vande Velde C, Cleveland DW. ALS: A disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52(1):39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Renton AE, et al. ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21(6):904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19(R1):R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boillée S, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312(5778):1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 7.Yamanaka K, et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11(3):251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbeito AG, Mesci P, Boillée S. Motor neuron-immune interactions: The vicious circle of ALS. J Neural Transm. 2010;117(8):981–1000. doi: 10.1007/s00702-010-0429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall ED, Oostveen JA, Gurney ME. Relationship of microglial and astrocytic activation to disease onset and progression in a transgenic model of familial ALS. Glia. 1998;23(3):249–256. doi: 10.1002/(sici)1098-1136(199807)23:3<249::aid-glia7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 10.Chiu IM, et al. T lymphocytes potentiate endogenous neuroprotective inflammation in a mouse model of ALS. Proc Natl Acad Sci USA. 2008;105(46):17913–17918. doi: 10.1073/pnas.0804610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beers DR, Henkel JS, Zhao W, Wang J, Appel SH. CD4+ T cells support glial neuroprotection, slow disease progression, and modify glial morphology in an animal model of inherited ALS. Proc Natl Acad Sci USA. 2008;105(40):15558–15563. doi: 10.1073/pnas.0807419105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butovsky O, et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J Clin Invest. 2012;122(9):3063–3087. doi: 10.1172/JCI62636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu IM, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013;4(2):385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu DC, Ré DB, Nagai M, Ischiropoulos H, Przedborski S. The inflammatory NADPH oxidase enzyme modulates motor neuron degeneration in amyotrophic lateral sclerosis mice. Proc Natl Acad Sci USA. 2006;103(32):12132–12137. doi: 10.1073/pnas.0603670103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veerhuis R, Nielsen HM, Tenner AJ. Complement in the brain. Mol Immunol. 2011;48(14):1592–1603. doi: 10.1016/j.molimm.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander JJ, Anderson AJ, Barnum SR, Stevens B, Tenner AJ. The complement cascade: Yin-Yang in neuroinflammation—neuro-protection and -degeneration. J Neurochem. 2008;107(5):1169–1187. doi: 10.1111/j.1471-4159.2008.05668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohlson SS, Fraser DA, Tenner AJ. Complement proteins C1q and MBL are pattern recognition molecules that signal immediate and long-term protective immune functions. Mol Immunol. 2007;44(1-3):33–43. doi: 10.1016/j.molimm.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 18.Nayak A, Ferluga J, Tsolaki AG, Kishore U. The non-classical functions of the classical complement pathway recognition subcomponent C1q. Immunol Lett. 2010;131(2):139–150. doi: 10.1016/j.imlet.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Gasque P, Dean YD, McGreal EP, VanBeek J, Morgan BP. Complement components of the innate immune system in health and disease in the CNS. Immunopharmacology. 2000;49(1-2):171–186. doi: 10.1016/s0162-3109(00)80302-1. [DOI] [PubMed] [Google Scholar]
- 20.Stephan AH, et al. A dramatic increase of C1q Protein in the CNS during normal aging. J Neurosci. 2013;33(33):13460–13474. doi: 10.1523/JNEUROSCI.1333-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sta M, et al. Innate and adaptive immunity in amyotrophic lateral sclerosis: Evidence of complement activation. Neurobiol Dis. 2011;42(3):211–220. doi: 10.1016/j.nbd.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Grewal RP, Morgan TE, Finch CE. C1qB and clusterin mRNA increase in association with neurodegeneration in sporadic amyotrophic lateral sclerosis. Neurosci Lett. 1999;271(1):65–67. doi: 10.1016/s0304-3940(99)00496-6. [DOI] [PubMed] [Google Scholar]
- 23.Kawamata T, Akiyama H, Yamada T, McGeer PL. Immunologic reactions in amyotrophic lateral sclerosis brain and spinal cord tissue. Am J Pathol. 1992;140(3):691–707. [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuboi Y, Yamada T. Increased concentration of C4d complement protein in CSF in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 1994;57(7):859–861. doi: 10.1136/jnnp.57.7.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldknopf IL, et al. Complement C3c and related protein biomarkers in amyotrophic lateral sclerosis and Parkinson’s disease. Biochem Biophys Res Commun. 2006;342(4):1034–1039. doi: 10.1016/j.bbrc.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 26.Rabin SJ, et al. Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum Mol Genet. 2010;19(2):313–328. doi: 10.1093/hmg/ddp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobsiger CS, Boillée S, Cleveland DW. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc Natl Acad Sci USA. 2007;104(18):7319–7326. doi: 10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeuchi S, et al. Induction of protective immunity by vaccination with wild-type apo superoxide dismutase 1 in mutant SOD1 transgenic mice. J Neuropathol Exp Neurol. 2010;69(10):1044–1056. doi: 10.1097/NEN.0b013e3181f4a90a. [DOI] [PubMed] [Google Scholar]
- 29.Chiu IM, et al. Activation of innate and humoral immunity in the peripheral nervous system of ALS transgenic mice. Proc Natl Acad Sci USA. 2009;106(49):20960–20965. doi: 10.1073/pnas.0911405106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodruff TM, et al. The complement factor C5a contributes to pathology in a rat model of amyotrophic lateral sclerosis. J Immunol. 2008;181(12):8727–8734. doi: 10.4049/jimmunol.181.12.8727. [DOI] [PubMed] [Google Scholar]
- 31.Fraser DA, Laust AK, Nelson EL, Tenner AJ. C1q differentially modulates phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells. J Immunol. 2009;183(10):6175–6185. doi: 10.4049/jimmunol.0902232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraser DA, Pisalyaput K, Tenner AJ. C1q enhances microglial clearance of apoptotic neurons and neuronal blebs, and modulates subsequent inflammatory cytokine production. J Neurochem. 2010;112(3):733–743. doi: 10.1111/j.1471-4159.2009.06494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Färber K, et al. C1q, the recognition subcomponent of the classical pathway of complement, drives microglial activation. J Neurosci Res. 2009;87(3):644–652. doi: 10.1002/jnr.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphries MM, et al. C1q enhances cone photoreceptor survival in a mouse model of autosomal recessive retinitis pigmentosa. Eur J Hum Genet. 2012;20(1):64–68. doi: 10.1038/ejhg.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benoit ME, Clarke EV, Morgado P, Fraser DA, Tenner AJ. Complement protein C1q directs macrophage polarization and limits inflammasome activity during the uptake of apoptotic cells. J Immunol. 2012;188(11):5682–5693. doi: 10.4049/jimmunol.1103760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benoit ME, Tenner AJ. Complement protein C1q-mediated neuroprotection is correlated with regulation of neuronal gene and microRNA expression. J Neurosci. 2011;31(9):3459–3469. doi: 10.1523/JNEUROSCI.3932-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131(6):1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 38.Stephan AH, Barres BA, Stevens B. The complement system: An unexpected role in synaptic pruning during development and disease. Annu Rev Neurosci. 2012;35:369–389. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- 39.Chung WS, Barres BA. The role of glial cells in synapse elimination. Curr Opin Neurobiol. 2012;22(3):438–445. doi: 10.1016/j.conb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zang DW, Lopes EC, Cheema SS. Loss of synaptophysin-positive boutons on lumbar motor neurons innervating the medial gastrocnemius muscle of the SOD1G93A G1H transgenic mouse model of ALS. J Neurosci Res. 2005;79(5):694–699. doi: 10.1002/jnr.20379. [DOI] [PubMed] [Google Scholar]
- 41.Lobsiger CS, Garcia ML, Ward CM, Cleveland DW. Altered axonal architecture by removal of the heavily phosphorylated neurofilament tail domains strongly slows superoxide dismutase 1 mutant-mediated ALS. Proc Natl Acad Sci USA. 2005;102(29):10351–10356. doi: 10.1073/pnas.0503862102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Botto M, et al. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat Genet. 1998;19(1):56–59. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- 43.Olsen MK, et al. Disease mechanisms revealed by transcription profiling in SOD1-G93A transgenic mouse spinal cord. Ann Neurol. 2001;50(6):730–740. doi: 10.1002/ana.1252. [DOI] [PubMed] [Google Scholar]
- 44.Heurich B, et al. Complement upregulation and activation on motor neurons and neuromuscular junction in the SOD1 G93A mouse model of familial amyotrophic lateral sclerosis. J Neuroimmunol. 2011;235(1-2):104–109. doi: 10.1016/j.jneuroim.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 45.Fukada Y, et al. Gene expression analysis of the murine model of amyotrophic lateral sclerosis: Studies of the Leu126delTT mutation in SOD1. Brain Res. 2007;1160:1–10. doi: 10.1016/j.brainres.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 46.Davalos D, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8(6):752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 47.Howell GR, et al. Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J Clin Invest. 2011;121(4):1429–1444. doi: 10.1172/JCI44646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zagoraiou L, et al. A cluster of cholinergic premotor interneurons modulates mouse locomotor activity. Neuron. 2009;64(5):645–662. doi: 10.1016/j.neuron.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pullen AH, Athanasiou D. Increase in presynaptic territory of C-terminals on lumbar motoneurons of G93A SOD1 mice during disease progression. Eur J Neurosci. 2009;29(3):551–561. doi: 10.1111/j.1460-9568.2008.06602.x. [DOI] [PubMed] [Google Scholar]
- 50.Herron LR, Miles GB. Gender-specific perturbations in modulatory inputs to motoneurons in a mouse model of amyotrophic lateral sclerosis. Neuroscience. 2012;226:313–323. doi: 10.1016/j.neuroscience.2012.09.031. [DOI] [PubMed] [Google Scholar]
- 51.Lobsiger CS, et al. Schwann cells expressing dismutase active mutant SOD1 unexpectedly slow disease progression in ALS mice. Proc Natl Acad Sci USA. 2009;106(11):4465–4470. doi: 10.1073/pnas.0813339106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perrin FE, et al. No widespread induction of cell death genes occurs in pure motoneurons in an amyotrophic lateral sclerosis mouse model. Hum Mol Genet. 2005;14(21):3309–3320. doi: 10.1093/hmg/ddi357. [DOI] [PubMed] [Google Scholar]
- 53.Ferraiuolo L, et al. Microarray analysis of the cellular pathways involved in the adaptation to and progression of motor neuron injury in the SOD1 G93A mouse model of familial ALS. J Neurosci. 2007;27(34):9201–9219. doi: 10.1523/JNEUROSCI.1470-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12(5):627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 55.Cardona AE, et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9(7):917–924. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- 56.Fonseca MI, Zhou J, Botto M, Tenner AJ. Absence of C1q leads to less neuropathology in transgenic mouse models of Alzheimer’s disease. J Neurosci. 2004;24(29):6457–6465. doi: 10.1523/JNEUROSCI.0901-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Depboylu C, et al. Upregulation of microglial C1q expression has no effects on nigrostriatal dopaminergic injury in the MPTP mouse model of Parkinson disease. J Neuroimmunol. 2011;236(1-2):39–46. doi: 10.1016/j.jneuroim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 58.Mitchell DA, et al. C1q deficiency and autoimmunity: The effects of genetic background on disease expression. J Immunol. 2002;168(5):2538–2543. doi: 10.4049/jimmunol.168.5.2538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.