Significance
In the last decade there has been an exponential increase in knowledge about the genetic basis of complex human traits. It is not clear, however, to what extent this knowledge can be used as a starting point for drug identification, one of the central hopes of the human genome project. Here, we report that by using genomic information related to aversive memory—a trait central to posttraumatic stress disorder—we identified several potential drug targets and compounds. In a subsequent pharmacological study with one of the identified compounds, we found a drug-induced reduction of aversive memory. These findings indicate that genomic information can be used as a starting point for the identification of memory-modulating compounds.
Keywords: emotional memory, pharmacology, PTSD
Abstract
In the last decade there has been an exponential increase in knowledge about the genetic basis of complex human traits, including neuropsychiatric disorders. It is not clear, however, to what extent this knowledge can be used as a starting point for drug identification, one of the central hopes of the human genome project. The aim of the present study was to identify memory-modulating compounds through the use of human genetic information. We performed a multinational collaborative study, which included assessment of aversive memory—a trait central to posttraumatic stress disorder—and a gene-set analysis in healthy individuals. We identified 20 potential drug target genes in two genomewide-corrected gene sets: the neuroactive ligand–receptor interaction and the long-term depression gene set. In a subsequent double-blind, placebo-controlled study in healthy volunteers, we aimed at providing a proof of concept for the genome-guided identification of memory modulating compounds. Pharmacological intervention at the neuroactive ligand–receptor interaction gene set led to significant reduction of aversive memory. The findings demonstrate that genome information, along with appropriate data mining methodology, can be used as a starting point for the identification of memory-modulating compounds.
Recent advances in human genetics have led to an unprecedented rate of discovery of genes related to complex human disease, including neuropsychiatric disorders (1–3). The human genome–based gain of knowledge is certainly expected to have a large impact on drug discovery in complex human disease (4–6). It is, however, still not clear to what extent this knowledge can be used as a starting point for the identification of druggable molecular pathways of complex traits (7), including mental disorders (8).
Genomewide association studies (GWASs) using single-marker statistics have been very successful in identifying trait-associated single-gene loci (9). It is, however, widely accepted that single marker–based analyses have limited power to identify the genetic basis of a given trait, as for example, many loci will fail to reach stringent genomewide significance threshold, despite the fact that they may be genuinely associated with the trait. Triggered by statistical approaches for the analysis of gene expression, gene set–based analytical methods have recently become available. These methods aim at identifying biologically meaningful sets of genes associated with a certain trait, rather than focusing on a single GWAS gene locus (10). By taking into account prior biological knowledge, gene set–based approaches examine whether test statistics for a group of related genes have consistent deviation from chance (10). As shown recently in studies on autism (11), bipolar disorder (12, 13), attention deficit hyperactivity disorder (ADHD) (14), and schizophrenia (15), such approaches can convincingly identify convergent molecular pathways relevant to neuropsychiatry. Importantly, the identification of groups of functionally related genes is likely to facilitate drug discovery, because the most significant single loci from a GWAS might not be the best candidates for therapeutic intervention (7, 10).
In the present study, we focused on emotionally aversive memory—a trait central to anxiety disorders such as posttraumatic stress disorder (PTSD) (16–23). Strong memory for emotionally arousing events can be seen as a primarily adaptive phenomenon, which helps us to remember vital information (e.g., dangerous situations). In case of an extremely aversive event, however, this mechanism can also lead to intrusive and persistent traumatic memories, thereby contributing to the development and symptoms of PTSD (18–22). Symptoms related to aversive memory include intrusive daytime recollections, traumatic nightmares, and flashbacks in which components of the event are relived. Aversive memory is a genetically complex trait as shown both in healthy subjects and in traumatized individuals (17, 23). Furthermore, we recently reported evidence suggesting a genetic link between the predisposition to build strong aversive memories and the risk for PTSD (16).
Based on these observations, we developed a process (Fig. 1) aimed at identifying gene sets related to human aversive memory, followed by a pharmacological intervention study as proof-of-concept for the genome-guided identification of memory-modulating drugs.
Fig. 1.
Drug discovery process.
Results
Step 1 (Gene Set Enrichment Analysis and Hypothesis-Testing Sample).
We recruited a homogenous sample of 1,802 Swiss healthy young adults who performed cognitive tasks quantifying emotional memory performance (see SI Materials and Methods for detailed sample and phenotype description). Aversive memory performance (i.e., enhanced free recall performance for previously shown pictures with negative emotional valence) (17) was the trait of interest and was analyzed as a continuous variable. SNP P values for association with aversive memory were obtained under two genetic models (i.e., additive and dominant, see SI Materials and Methods for detailed description). Only intragenic SNPs were considered. These P values served as input for subsequent identification of gene sets related to aversive memory.
We applied the improved gene set enrichment method (i-GSEA4GWAS) (24), which evaluates whether the distribution of genes sharing a biochemical or cellular function is different from a distribution of a ranked genomewide list of genes (see SI Materials and Methods for detailed description). Four significant [i.e., genomewide false discovery rate (FDR) corrected] gene sets related to aversive memory were detected (Table 1): the VEGF gene set, the IL-1 receptor (IL-1R) gene set, the long-term depression gene set, and the neuroactive ligand receptor interaction gene set. By using the identical settings in this sample, we also applied a second gene set algorithm, GSA-SNP (25), to assess which of these four sets are detected by another gene set method. A recent comparison of both algorithms on identical samples showed that GSA-SNP tends to be more conservative than i-GSEA4GWAS (26). The long-term depression and the neuroactive ligand–receptor interaction gene sets were detected as significant (PFDRcorr = 0.00009 and PFDRcorr = 0.006, respectively), whereas this was not the case for the VEGF and IL-1R gene sets (PFDRcorr = 0.5 and PFDRcorr = 0.08, respectively).
Table 1.
Gene sets related to aversive memory (testing sample)
| Pathway | Pathway URL | Gene set P (nominal) | Gene set P (FDR) | Significant gene set genes (N) | Mapped gene set genes (N) | All gene set genes (N) |
| VEGF pathway | biocarta.com/pathfiles/ h_VEGFPATHWAY.asp | <0.001 | 0.011 | 10 | 21 | 28 |
| IL1R pathway | biocarta.com/pathfiles/ h_IL1RPATHWAY.asp | <0.001 | 0.013 | 13 | 24 | 33 |
| HSA04730 LONG-TERM DEPRESSION | genome.jp/dbget-bin/www_bget? pathway+hsa04730 | <0.001 | 0.019 | 33 | 67 | 76 |
| HSA04080 NEUROACTIVE LIGAND RECEPTOR INTERACTION | genome.jp/dbget-bin/www_bget? pathway+hsa04080 | <0.001 | 0.036 | 69 | 175 | 254 |
All gene set genes, all genes of a given gene set, i.e., mapped and unmapped genes; mapped gene set genes, gene set genes containing at least one intragenic SNP and therefore included in the i-GSEA analysis; significant gene set genes, genes mapped with at least one of the top 5% of all nominally significant SNPs (Materials and Methods).
Step 2 (Gene Set Enrichment Analysis and Replication Sample).
In an additional population of 781 Swiss healthy young adults, who underwent the identical phenotyping and genotyping procedure as the step 1 sample, we also performed gene set enrichment analysis with the identical settings as in step 1. Gene set analysis replicated (at the identical genome-wide FDR < 0.05 level) both gene sets of the first sample, i.e., the neuroactive ligand–receptor interaction set and the long-term depression set (SI Materials and Methods). A total of 20 replicated genes (Table S1) within these gene sets were significantly associated with aversive memory in both the hypothesis-testing (step 1; Table S1) and the replication (step 2; Table S1) samples. These genes served as input for the subsequent step 3.
Step 3 (Drug Selection).
Here we aimed at providing a proof-of-concept for human genetics-guided identification of memory-modulating drugs. Therefore, we selected from the 20 candidate targets only gene products with already existing therapeutic compounds. Ingenuity Pathway Analysis (IPA; Ingenuity Systems) indicated the existence of therapeutic compounds for 15 of the candidate targets (Table S2). To facilitate the subsequent step 4 (pharmacological intervention study), we applied following priority criteria to these compounds with respect to their suitability in the context of a pharmacological intervention study in Switzerland: (i) existing registration as therapeutic compound by the Swiss Agency for Therapeutic Products (Swissmedic) and availability on the Swiss market; (ii) acceptable side effects profile [i.e., no major side effects such as nephrotoxicity, neurotoxicity or myelotoxicity (e.g., anticancer drugs were not considered)]; and (iii) favorable application mode (e.g., oral administration was prioritized over i.v. or s.c. medication). Application of these priority criteria to the list of existing compounds (Table S2) resulted in nine compounds/compound groups (corresponding to nine genes; Table 2).
Table 2.
Compounds/compound groups fulfilling the feasibility criteria
| Drug/drug group | Genes |
| Antidepressants (e.g., mirtazapine) | HTR2A |
| Antihistamines (e.g., diphenhydramine) | CHRM2, CHRM3, CHRM5, HRH1 |
| Antimuscarinics (e.g., fesoterodine) | CHRM2, CHRM3, CHRM5 |
| Aprepitant | TACR1 |
| Ergot alkaloids (e.g., ergotamine) | HTR2A |
| Fasoracetam | GRM1, GRM5 |
| Memantine | GRIN2A |
| Antipsychotics | CHRM2, CHRM3, CHRM5, GRIN2A, HRH1, HTR2A |
| Opioids (e.g., morphine) | GRIN2A |
Note on the feasibility criteria: IPA (Ingenuity Systems indicated the existence of therapeutic compounds for 15 of the candidate targets (see Table S1 for all candidate targets). The following priority criteria were applied with respect to suitability in the context of a pharmacological intervention study in Switzerland: 1, existing registration as therapeutic compound by the Swiss Agency for Therapeutic Products (Swissmedic) and availability on the Swiss market; 2, acceptable side effects profile [i.e., no major side effects such as nephrotoxicity, neurotoxicity or myelotoxicity (e.g., anticancer drugs were not considered)]; and 3, favorable application mode (e.g., oral administration was prioritized over i.v. or s.c. medication). Application of these priority criteria to the list of existing compounds (see Table S2 for all existing compounds) resulted in nine compounds/compound groups (corresponding to nine genes, shown in this table).
Step 4 (Pharmacological Intervention Study).
To facilitate the selection of one drug for a proof-of-concept study, we studied which of the nine high priority genes are also linked to highly aversive (traumatic) memory in a population of 349 severely traumatized genocide survivors (16) (SI Materials and Methods). After correction for multiple comparisons, one gene (HRH1) was found to be associated with traumatic memory (Table S3). Therefore, compounds targeting the histamine 1 receptor (encoded by HRH1) were given highest priority for this proof-of-concept intervention study. Two drug groups are known to interact with the histamine 1 receptor: antipsychotics (e.g., olanzapine) and antihistamines (e.g., diphenhydramine). Antipsychotics were considered to have an unfavorable side effect profile relatively to antihistamines. Therefore, an antihistamine known to cross the blood–brain barrier (diphenhydramine) was selected for the subsequent intervention study. To our knowledge, the effect of diphenhydramine on emotional memory has not been studied to date.
The study with the HRH1 antagonist diphenhydramine was carried out using a double-blind, placebo-controlled, cross-over design in 40 healthy, young (23.0 ± 0.8 y; 21 males) volunteers of European ancestry. Subjects received a single oral administration of diphenhydramine 50 mg or placebo in a cross-over design with 7 ± 3 d between visits (for flow diagram, see Fig. S1). Three hours after administration of the study medication, subjects underwent comprehensive emotional and cognitive testing (SI Materials and Methods).
As expected, diphenhydramine significantly increased sleepiness (as measured with a visual analog scale during the test phase, P < 0.001; Table S4). Therefore, drug-induced sleepiness, which reflects the activity of centrally acting antihistamines (27), was used as a between-subjects factor in the statistical analysis of drug effects on memory (SI Materials and Methods).
The primary outcome variable (i.e., aversive memory) was normally distributed (Shapiro–Wilk test: P > 0.5). We observed a significant effect of diphenhydramine on free recall performance for aversive pictures as early as 5 min after picture presentation (P < 0.05; Table 3) and also after a longer delay (90 min after picture presentation, P = 0.008; Table 3). The effect size of diphenhydramine on free recall of aversive pictures (90-min delay) was medium (Cohen’s d = 0.42). The effect of diphenhydramine on 90-min delayed free recall of aversive pictures was also significant after correcting for sex (P = 0.006), age (P = 0.009), treatment order (P = 0.012), and body weight (P = 0.009). There was no main effect of treatment order (P = 0.647) on recall performance and no significant order × treatment interaction (P = 0.266).
Table 3.
Drug effects on free recall of pictures
| Memory type | Diphenhydramine mean (SE) | Placebo mean (SE) | P |
| Short-delay (5 min) free recall of pictures | |||
| Positive pictures | 13.53 (0.49) | 14.32 (0.58) | 0.110 |
| Neutral pictures | 9.03 (0.55) | 10.36 (0.53) | 0.032 |
| Aversive pictures | 13.15 (0.55) | 14.41 (0.44) | 0.014* |
| Long-delay (90 min) free recall of pictures | |||
| Positive pictures | 13.92 (0.55) | 14.56 (0.54) | 0.197 |
| Neutral pictures | 9.40 (0.62) | 10.36 (0.63) | 0.114 |
| Aversive pictures | 13.68 (0.60) | 14.92 (0.47) | 0.008* |
Cross-over study with 40 healthy subjects. P indicates nominal P values.
Significance after Bonferroni correction for the three valence groups.
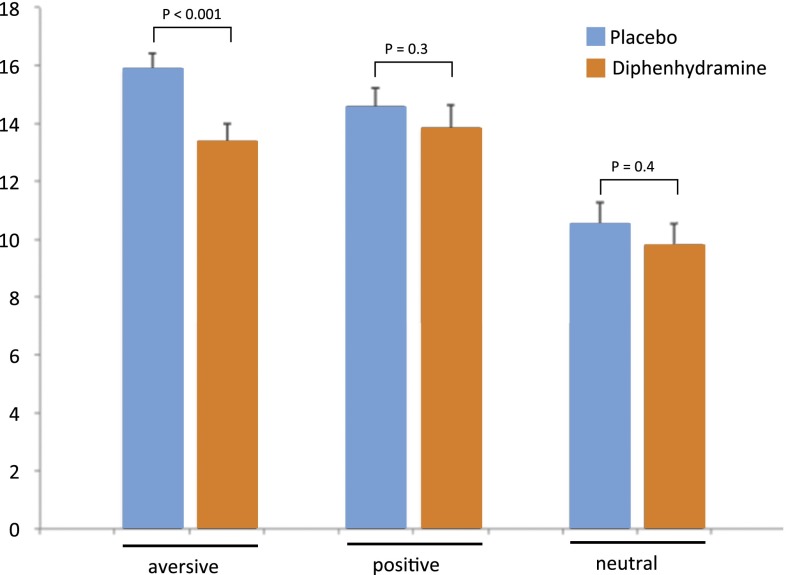
The statistical model including treatment (placebo, diphenhydramine) and all three valence groups (positive, neutral, aversive) as independent variables in this repeated-measures ANOVA and sleepiness as between-subject factor revealed a significant main effect of valence (P < 0.001), a significant main effect of treatment (P = 0.023), and a significant treatment × valence × sleepiness interaction (P = 0.02). Post hoc analyses revealed that diphenhydramine reduced recall of aversive information significantly (P < 0.001; Fig. 2 and Table S5) in subjects who reported diphenhydramine-induced sleepiness, but not significantly (P > 0.9) in subjects who did not report diphenhydramine-induced sleepiness (SI Materials and Methods). There were no significant drug effects on the recall of positive or neutral pictures in the entire sample (Table 3) and also no significant drug effects on the recall of positive or neutral pictures after stratification for sleepiness (high sleepiness/positive pictures: P = 0.328; no sleepiness/positive pictures: P = 0.411; high sleepiness/neutral pictures: P = 0.384; no sleepiness/neutral pictures: P = 0.181). These findings show that diphenhydramine did not affect memory for neutral or positive pictures despite its sedative effect. Thus, diphenhydramine-induced sleepiness did not interfere with general mnemonic processes. This finding is in line with previous studies, which consistently report that diphenhydramine does not affect nonemotional, episodic memory (28–31).
Fig. 2.
Diphenhydramine-induced reduction of aversive memory in the high sleepiness group (n = 20). A single administration of diphenhydramine (50 mg) compared with placebo significantly reduced delayed recall of aversive, but not of positive or neutral, pictures (Table S5). Diphenhydramine did not affect valence or arousal ratings (P > 0.2) of the presented pictures. Error bars represent SEM.
Furthermore, we did not observe a significant (P = 0.26) correlation between sleepiness and aversive memory in the placebo condition, although large interindividual variability in sleepiness was observed in that group [3.06 ± 2.12 (mean ± SD); range, 0–8, on a visual analog scale from 0 to 10]. This finding is in line with a recent study investigating the acute effects of the sedative benzodiazepine lorazepam and the stimulating drug methylphenidate on aversive memory. Despite producing opposite effects on sleepiness, both lorazepam and methylphenidate reduce the impact of emotionally arousing aversive material on memory, indicating a dissociation between drug effects on aversive memory and drug effects on sleepiness (32). Furthermore, a study with the sedative lorazepam found drug-induced memory impairments independent of emotional valence (33). Finally, a study with melatonin, a drug that also induces sleepiness, did not find drug-induced effects on memory performance in healthy humans (34). Together, these findings indicate that the effects of diphenhydramine on aversive memory were not mediated by sleepiness.
There were no significant drug effects on attention or fluid intelligence as measured in the present study (P ≥ 0.3; Table S4). Diphenhydramine did also not affect valence or arousal ratings in the picture task (P ≥ 0.195; Table S6) or state anxiety levels (P = 0.38; Table S4). Diphenhydramine did not affect significantly emotionally neutral memory tasks such as verbal recall, figural recall, and n-back working memory (P > 0.05). Furthermore, diphenhydramine did not affect significantly depression scores or number of reported side effects (P > 0.05), indicating that in the present study the single dose of the drug did not affect general cognitive functions, mood, or well-being.
Discussion
The present study demonstrates the potential of the use of genomic information for the identification of drugs influencing emotionally aversive memory. Specifically, genomewide gene set analyses pointed to HRH1 (encoding histamine receptor H1) as one of many possible drug targets. A subsequent double-blind, placebo-controlled intervention study in healthy volunteers revealed that single-dose administration of the antihistamine diphenhydramine led to significant reduction of aversive memory recall but not of general mnemonic functions. Although SNP-based association studies represent a valuable tool for the identification of druggable molecules, it is important to note that genetic signals arising from marker SNPs generally do not allow inferences about functional consequences at the mRNA or protein expression level, nor do they predict the direction of drug effects. Consequently, subsequent molecular and pharmacological studies in animals or humans are needed to corroborate the genetic association results and to define the direction of effect of a drug identified through SNP-based association.
It is important to stress that in the present study the priority criteria for the compound tested in the intervention study were weighted toward feasibility. For example, only compounds registered in Switzerland and available in the Swiss market were considered in this study. Consequently, a number of interesting compounds (Table S2) are also potentially efficacious and may be considered as candidates for following clinical trials. For example, the drug discovery approach described herein also identified morphine as a drug with potential aversive memory-modulating properties (Table S2). Interestingly, the use of morphine after combat injury was recently associated with reduced risk for the development of PTSD (35).
The present study also demonstrates the feasibility and importance of gene set–based analytical methods, which identify biologically meaningful groups of genes associated with a certain trait, rather than focusing on single GWAS gene loci (10). Two replicated gene sets related to aversive memory were identified in the present study: the long-term depression gene set and the neuroactive ligand–receptor interaction gene set. Both sets include genes intimately involved in learning and memory processes with well-established roles in crucial neuronal traits such as plasticity and synaptic function.
To test whether the identification of these brain-related gene sets is a function of the core phenotype under study, i.e., aversive memory, we conducted an additional study on a brain-unrelated trait: rheumatoid arthritis (RA: Tables S7, S8, and S9). Specifically, we applied the identical search strategy (i.e., gene set enrichment analysis and Ingenuity-based compound search) to a large dataset (n = 25,500) of patients with RA and controls (36). SNP-to-gene assignment and correction for multiple comparisons were done as reported for the aversive memory study. Gene set enrichment analysis retrieved six genomewide-corrected gene sets related to RA, including sets relevant to inflammation, antigen presentation, and cell adhesion (Tables S7 and S8). Interestingly, most of the existing compounds identified are characterized by immunomodulatory properties (Table S9), and for several of the retrieved drugs, the primary indication is RA (e.g., Abatacept, Belatacept, Anakinra, Tocilizumab, Adalimumab, Ifliximab, and Etanercept; Table S9). This result demonstrates the face validity of the gene set approach and suggests that the identification of brain-related groups of genes was a function of the core phenotype under study.
Although the present results underscore the importance and potential of genetic studies of human complex traits for the identification of therapeutic compounds, we would like to point out that the approach described herein is one of many possible approximations. Despite current lack of evidence, we hope that future developments related to both the understanding of the human genome and to the progress in biostatistical methodology will improve this concept. First, genetics-guided drug discovery might be improved by the rapid development of affordable whole genome sequencing technologies. Indeed, the vast majority of genetic association studies to date are based on the concept of proxy SNPs. As a consequence, many genes remain uncovered by common SNPs and are thereby a priori excluded from further statistical analysis (Table S1, unmapped genes). Capitalizing on the possibility of whole genome sequencing in large samples might increase the number of genes that are considered within a certain biological pathway. Second, genetic complexity is not only related to the polygenic nature of the traits under study but it also reflects the complexity of the human genome per se, which exerts its influence on the traits not only through simple linear gene effects but also through gene–gene interactions, gene–environment interactions, and epigenetic mechanisms. A better understanding of the fundamental complexity through which genetic variation exerts its effects on complex traits is expected to refine the information needed for genetics-guided drug discovery. Finally, the relation between genetic and phenotypic variability is not expected to follow simple and general rules applicable to every phenotype. Indeed, it is impossible to predict which statistical and bioinformatics approaches will ideally explain the relationship between genetic and phenotypic variability of complex traits. It will be most probably a combination of several approaches specific to distinct phenotypes. Consequently, genetics-guided drug discovery will require a statistical framework that accounts for the complexity of functional molecular networks specific to the phenotype under study. The present approach (i.e., gene set enrichment analysis) seems to be well suited for the target phenotype of this study. However, as more and more methods will become available (10), study designs based on continuously curated gene set databases, multidisciplinary research frameworks, and focusing on the identification of complex genetic structures will prove invaluable for robust and reliable discovery of druggable targets.
Gene set approaches represent important tools for the development of drugs directed against memory-related disorders. Additional studies are warranted to identify an optimal dose–response relation and optimal timing of administration, to investigate the effects of diphenhydramine on the distinct memory phases of memory formation and retrieval, and to study whether the administration timed to traumatic reactivation is able to interfere with reconsolidation in patients suffering from PTSD (37, 38). Our study also provides additional, potentially interesting compounds, which can be tested in following clinical trials.
Materials and Methods
Step 1 involved a homogenous sample of 1,802 (1,211 females and 591 males) Swiss healthy young (22.4 ± 0.1 y; mean ± SE) adults who performed a memory task consisting of freely recalling previously presented aversive, positive, and neutral pictures. In this sample, we applied the improved gene set enrichment method (i-GSEA4GWAS) (24) and a second gene set algorithm GSA-SNP (25). In step 2, we performed a replication study in 781 (484 females and 297 males) Swiss healthy young (22.4 ± 0.1 y) adults. In step 3, we used the Ingenuity Pathway Analysis (IPA; Ingenuity Systems) to identify therapeutic compounds for the identified candidate genes. In step 4, to facilitate the selection of one drug for a proof-of-concept pharmacological study, we studied which of the nine high priority genes are also linked to highly aversive (traumatic) memory in a population of 349 severely traumatized genocide survivors. Finally, we performed a double-blind, placebo-controlled, cross-over pharmacological study in 40 healthy, young (23.0 ± 0.8 y; 21 males) volunteers. Subjects received a single oral administration of diphenhydramine 50 mg or placebo in a cross-over design with 7 ± 3 d between visits. For a detailed description of the study populations, designs, and procedures of steps 1–4, see SI Material and Methods.
Supplementary Material
Acknowledgments
This work was funded by Swiss National Science Foundation Sinergia Grants CRSIK0_122691 and CRSI33_130080 (to D.J.-F.d.Q. and A. Papassotiropoulos) and by grants from the German Research Foundation (Deutsche Forschungsgemeinschaft; to I.-T.K. and T.E.).
Footnotes
Conflict of interest statement: The study was done in collaboration with and was partially funded by ACImmune SA, Lausanne, Switzerland. P.P., A. Muhs, and A. Pfeifer are employees of ACImmune. A. Papassotiropoulos is scientific consultant of ACImmune. ACImmune SA and University of Basel are coapplicants. A. Papassotiropoulos and D.J.-F.d.Q. are inventors of a patent related to the use of diphenhydramine as memory modulator.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314478110/-/DCSupplemental.
References
- 1.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 2.Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187(2):367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cichon S, et al. Psychiatric GWAS Consortium Coordinating Committee Genomewide association studies: History, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166(5):540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 5.Hyman SE. A glimmer of light for neuropsychiatric disorders. Nature. 2008;455(7215):890–893. doi: 10.1038/nature07454. [DOI] [PubMed] [Google Scholar]
- 6.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol. 2012;30(4):317–320. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 7.Muglia P. From genes to therapeutic targets for psychiatric disorders - what to expect? Curr Opin Pharmacol. 2011;11(5):563–571. doi: 10.1016/j.coph.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 8. Hyman SE (2012) Revolution stalled. Sci Transl Med 4(155): 1–5. [DOI] [PubMed]
- 9.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118(5):1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11(12):843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 11.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmans P, et al. Wellcome Trust Case-Control Consortium Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009;85(1):13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sklar P, et al. Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43(10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stergiakouli E, et al. deCODE Genetics Psychiatric GWAS Consortium Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am J Psychiatry. 2012;169(2):186–194. doi: 10.1176/appi.ajp.2011.11040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Dushlaine C, et al. International Schizophrenia Consortium Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2011;16(3):286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- 16.de Quervain DJ, et al. PKCα is genetically linked to memory capacity in healthy subjects and to risk for posttraumatic stress disorder in genocide survivors. Proc Natl Acad Sci USA. 2012;109(22):8746–8751. doi: 10.1073/pnas.1200857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Quervain DJ, et al. A deletion variant of the alpha2b-adrenoceptor is related to emotional memory in Europeans and Africans. Nat Neurosci. 2007;10(9):1137–1139. doi: 10.1038/nn1945. [DOI] [PubMed] [Google Scholar]
- 18.de Quervain DJ, Margraf J. Glucocorticoids for the treatment of post-traumatic stress disorder and phobias: A novel therapeutic approach. Eur J Pharmacol. 2008;583(2-3):365–371. doi: 10.1016/j.ejphar.2007.11.068. [DOI] [PubMed] [Google Scholar]
- 19.McGaugh JL. The Making of Lasting Memory. Memory and Emotion. London: Weidenfeld and Nicolson; 2003. [Google Scholar]
- 20.Mineka S, Oehlberg K. The relevance of recent developments in classical conditioning to understanding the etiology and maintenance of anxiety disorders. Acta Psychol (Amst) 2008;127(3):567–580. doi: 10.1016/j.actpsy.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Phelps EA, LeDoux JE. Contributions of the amygdala to emotion processing: from animal models to human behavior. Neuron. 2005;48(2):175–187. doi: 10.1016/j.neuron.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 22.Pitman RK. Post-traumatic stress disorder, hormones, and memory. Biol Psychiatry. 1989;26(3):221–223. doi: 10.1016/0006-3223(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 23.Rasch B, et al. A genetic variation of the noradrenergic system is related to differential amygdala activation during encoding of emotional memories. Proc Natl Acad Sci USA. 2009;106(45):19191–19196. doi: 10.1073/pnas.0907425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang K, Cui S, Chang S, Zhang L, Wang J (2010) i-GSEA4GWAS: A web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res 38(Web Server issue):W90–W95. [DOI] [PMC free article] [PubMed]
- 25. Nam D, Kim J, Kim SY, Kim S (2010) GSA-SNP: A general approach for gene set analysis of polymorphisms. Nucleic Acids Res 38(Web Server issue):W749–W754. [DOI] [PMC free article] [PubMed]
- 26.Kwon JS, Kim J, Nam D, Kim S. Performance comparison of two gene set analysis methods for genome-wide association study results: GSA-SNP vs i-GSEA4GWAS. Genomics Inform. 2012;10(2):123–127. doi: 10.5808/GI.2012.10.2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiner PB, Kamondi A. Mechanisms of antihistamine-induced sedation in the human brain: H1 receptor activation reduces a background leakage potassium current. Neuroscience. 1994;59(3):579–588. doi: 10.1016/0306-4522(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 28.Curran HV, Pooviboonsuk P, Dalton JA, Lader MH. Differentiating the effects of centrally acting drugs on arousal and memory: An event-related potential study of scopolamine, lorazepam and diphenhydramine. Psychopharmacology (Berl) 1998;135(1):27–36. doi: 10.1007/s002130050482. [DOI] [PubMed] [Google Scholar]
- 29.Verster JC, et al. Acute and subchronic effects of levocetirizine and diphenhydramine on memory functioning, psychomotor performance, and mood. J Allergy Clin Immunol. 2003;111(3):623–627. doi: 10.1067/mai.2003.63. [DOI] [PubMed] [Google Scholar]
- 30.Kay GG, et al. Initial and steady-state effects of diphenhydramine and loratadine on sedation, cognition, mood, and psychomotor performance. Arch Intern Med. 1997;157(20):2350–2356. [PubMed] [Google Scholar]
- 31.Turner C, Handford AD, Nicholson AN. Sedation and memory: Studies with a histamine H-1 receptor antagonist. J Psychopharmacol. 2006;20(4):506–517. doi: 10.1177/0269881106059804. [DOI] [PubMed] [Google Scholar]
- 32.Brignell CM, Rosenthal J, Curran HV. Pharmacological manipulations of arousal and memory for emotional material: Effects of a single dose of methylphenidate or lorazepam. J Psychopharmacol. 2007;21(7):673–683. doi: 10.1177/0269881107077351. [DOI] [PubMed] [Google Scholar]
- 33.Kamboj SK, Curran HV, Curran HV. Neutral and emotional episodic memory: Global impairment after lorazepam or scopolamine. Psychopharmacology (Berl) 2006;188(4):482–488. doi: 10.1007/s00213-006-0552-7. [DOI] [PubMed] [Google Scholar]
- 34.Lieberman HR, Waldhauser F, Garfield G, Lynch HJ, Wurtman RJ. Effects of melatonin on human mood and performance. Brain Res. 1984;323(2):201–207. doi: 10.1016/0006-8993(84)90290-7. [DOI] [PubMed] [Google Scholar]
- 35.Holbrook TL, Galarneau MR, Dye JL, Quinn K, Dougherty AL. Morphine use after combat injury in Iraq and post-traumatic stress disorder. N Engl J Med. 2010;362(2):110–117. doi: 10.1056/NEJMoa0903326. [DOI] [PubMed] [Google Scholar]
- 36.Stahl EA, et al. BIRAC Consortium YEAR Consortium Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pitman RK. Will reconsolidation blockade offer a novel treatment for posttraumatic stress disorder? Front Behav Neurosci. 2011;5:11. doi: 10.3389/fnbeh.2011.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gamache K, Pitman RK, Nader K. Preclinical evaluation of reconsolidation blockade by clonidine as a potential novel treatment for posttraumatic stress disorder. Neuropsychopharmacology. 2012;37(13):2789–2796. doi: 10.1038/npp.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.