Significance
Many bacteria respond to environmental cues by producing cyclic diguanosine monophosphate (c-di-GMP), which binds to proteins to modulate transitions between sessile and motile lifestyles important for chronic and acute infections, respectively. The breadth of protein targets and mechanisms of action of c-di-GMP remain unknown. In the opportunistic pathogen Pseudomonas aeruginosa, c-di-GMP binds to FleQ, a transcription factor that is the master regulator of flagella synthesis and a member of the large family of AAA+ ATPase proteins. We found that c-di-GMP competitively inhibits FleQ ATPase activity and interacts with the ATP-binding site on FleQ. We suggest that AAA+ ATPases from other bacteria may respond to c-di-GMP, and that interference with ATPase activity is one mechanism of c-di-GMP action.
Keywords: transcription factor, σ54, AAA+ protein, competitive inhibition
Abstract
The transcription factor FleQ is a bacterial AAA+ ATPase enhancer-binding protein that is the master activator of flagella gene expression in the opportunistic bacterial pathogen Pseudomonas aeruginosa. Homologs of FleQ are present in all Pseudomonas species and in many polarly flagellated gamma proteobacteria. Cyclic diguanosine monophosphate (c-di-GMP) is a second messenger that controls the transition between planktonic and biofilm modes of growth in bacteria in response to diverse environmental signals. C-di-GMP binds to FleQ to dampen its activity, causing down-regulation of flagella gene expression. This action is potentiated in the simultaneous presence of another protein, FleN. We explored the effect of c-di-GMP and FleN on the ATPase activity of FleQ and found that a relatively low concentration of c-di-GMP competitively inhibited FleQ ATPase activity, suggesting that c-di-GMP competes with ATP for binding to the Walker A motif of FleQ. Confirming this, a FleQ Walker A motif mutant failed to bind c-di-GMP. FleN, whose gene is regulated by FleQ, also inhibited FleQ ATPase activity, and FleQ ATPase activity was much more inhibited by c-di-GMP in the presence of FleN than in its absence. These results indicate that FleN and c-di-GMP cooperate to inhibit FleQ activity and, by extension, flagella synthesis in P. aeruginosa. The Walker A motif of FleQ is perfectly conserved, opening up the possibility that other AAA+ ATPases may respond to c-di-GMP.
Bis-(3′-5′)-cyclic diguanosine monophosphate (c-di-GMP) is an ubiquitous second messenger in bacteria, where it affects the transition between a motile planktonic lifestyle and an adhesive biofilm lifestyle by binding to various effector proteins to modulate enzymatic activity, protein–protein interactions, transcription, and translation (1–6). One of these effector proteins is FleQ (PA1097), an enhancer-binding protein (EBP) transcription factor from the opportunistic pathogen Pseudomonas aeruginosa.
FleQ contains an N-terminal FleQ domain, a central AAA+ ATPase domain, and a C-terminal helix-turn-helix DNA-binding domain. It activates expression of biofilm-related genes, including genes for Pel and Psl exopolysaccharide (EPS) in response to binding of c-di-GMP at an unknown site (7, 8). EBPs typically act in conjunction with σ54-RNA polymerase; however, FleQ regulates EPS gene expression independent of σ54, most likely with σ70 (8, 9).
In addition to regulating genes for biofilm formation, FleQ has a second, better-known role as a σ54-dependent master regulator of P. aeruginosa flagella gene expression (9, 10). FleQ activates expression of the two-component regulatory genes fleSR, as well as genes for the assembly of the flagella export apparatus and for the initiation of flagella basal body assembly. FleSR controls genes to complete the basal body-hook structure (9, 11).
The regulation of both flagella and biofilm genes is also under the control of a second protein, FleN. Mutations in fleN (PA1454) lead to an up-regulation of flagella gene expression and a smalldown-regulation of biofilm genes (7, 12). The expression of fleN is under the control of FleQ. FleN is a putative ATPase containing a deviant Walker A motif (13). FleN and FleQ interact with one another in the presence as well as in the absence of ATP or c-di-GMP, and a FleQ/FleN complex binds to pel, fleSR, and flhA promoter DNA (7, 8, 14).
Transcriptome studies have shown that the expression of most FleQ-controlled flagella genes is down-regulated by 1.5- to 2-fold when intracellular c-di-GMP is high (7, 15). To investigate the basis for this effect, we tested the influence of c-di-GMP on FleQ ATPase activity. In general, ATP hydrolysis by EBPs like FleQ provides energy for remodeling the σ54-RNA polymerase closed complex, allowing loading of the template strand of DNA into the active site of the RNA polymerase (16, 17). Energy is transferred by direct interaction of σ54 and the EBP. Our results indicate that FleQ has a strong cooperative ATPase activity, and that FleN and c-di-GMP inhibit FleQ ATPase activity, thereby explaining their effects in depressing the ability of FleQ to activate flagella gene expression. Our results further suggest that an intact Walker A motif is necessary for the binding of c-di-GMP to FleQ.
Results
FleQ Exhibits Cooperative ATPase Activity.
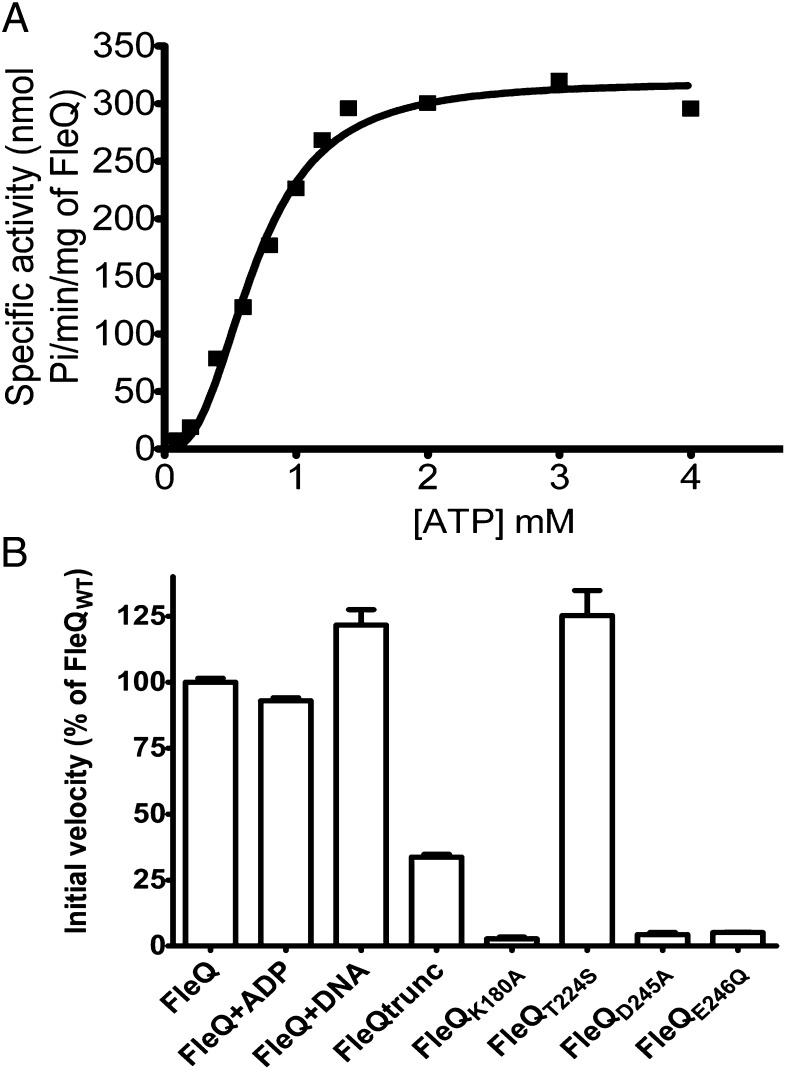
In initial experiments, we found that the ATPase activity of FleQ did not obey classical Michaelis–Menten kinetic relationships (Fig. 1A). Indeed, the enzyme kinetic curves were sigmoidal instead of hyperbolic. The Hill coefficient for ATPase activity of FleQ was approximately 2.5 (Table 1), implying a strong positive cooperativity in sequential binding and hydrolysis of ATP. FleQ ATPase activity was not subject to product inhibition or stimulation (Fig. 1B, FleQ + ADP), and time course experiments showed that FleQ ATPase activity was linear at different ATP concentrations, suggesting that no substrate inhibition occurred at high ATP concentrations. FleQ ATPase activities in the presence or absence of a DNA fragment encompassing the FleQ-binding site of the fleSR promoter (7) were similar, suggesting that FleQ has DNA-independent ATPase activity (Fig. 1B). FleQ has a N-terminal domain of unknown function, known as the FleQ domain. FleQ truncated of this domain had threefold lower ATPase activity relative to full-length FleQ, suggesting that the FleQ domain may positively regulate the activity of the AAA+ domain (Fig. 1B).
Fig. 1.
FleQ has a cooperative ATPase activity. (A) The specific ATPase activity of FleQ (1 µM) as a function of the concentration of ATP. (B) Initial velocity of the ATPase activities of FleQ WT alone or in the presence of 100 μM ADP, 0.25 μM fleS promoter DNA fragment, and ATPase activities of FleQ variant proteins. Expressed as percentage of the activity obtained with FleQ WT. The ATPase activity was assayed with 1 µM protein and 1 mM ATP. The error bars represent SDs.
Table 1.
FleQ ATPase kinetic data
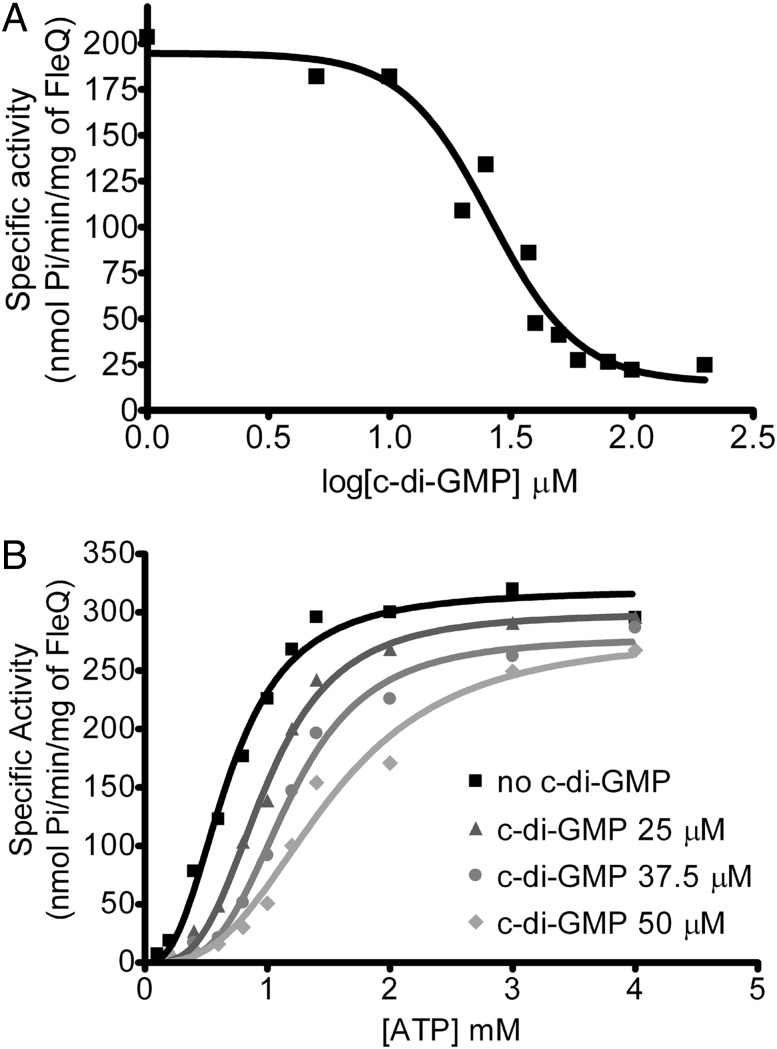
| Vmax* | Khalf† | n‡ | |
| No c-di-GMP | 320 ± 10.6 | 0.68 ± 0.03 | 2.6 ± 0.3 |
| c-di-GMP 25 µM | 300 ± 7.6 | 0.98 ± 0.02 | 3.2 ± 0.3 |
| c-di-GMP 37.5 µM | 280 ± 9.7 | 1.17 ± 0.04 | 3.6 ± 0.4 |
| c-di-GMP 50 µM | 280 ± 23 | 1.50 ± 0.13 | 2.9 ± 0.5 |
Vmax ± SD in nmol Pi/min/mg of FleQ.
Khalf ± SD in mM of ATP.
Hill coefficient ± SD.
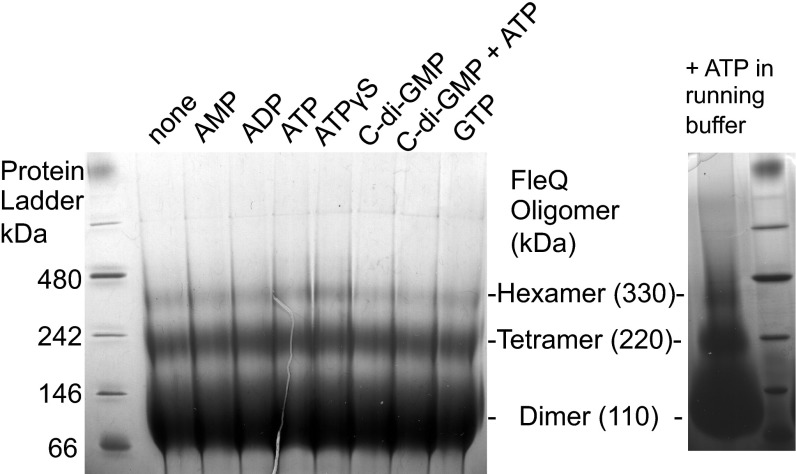
The ATPase activity of EBPs is strongly influenced by their ability to form high-order oligomers (18). EBPs are typically dimeric in their inactive state and must form a higher-order oligomer (mostly hexamers) for high rates of ATP hydrolysis to occur. In most cases, the formation of these active hexamers requires ATP binding, but ligand binding, phosphorylation, or binding to DNA also can influence EBP oligomerization (17). We assessed the oligomeric state of FleQ by running purified protein on a blue native (BN) gel. FleQ dimers were the primary forms detected, but FleQ tetramers and some FleQ hexamers were observed as well (Fig. 2).
Fig. 2.
FleQ forms dimers, tetramers, and hexamers. (Left) BN-PAGE of 50 μg of FleQ mixed with 10 mM AMP, ADP, ATP, ATPγS, or GTP or 1 mM c-di-GMP as indicated at the top of the figure. (Right) The FleQ protein was incubated with 1 mM ATP, and 1 mM ATP was also added to the buffer.
The finding that FleQ forms oligomers is consistent with the cooperative ATPase activity of the enzyme. However, preincubation of FleQ with ATP, ADP, AMP, ATPγS, c-di-GMP, or GTP did not change the oligomeric state of FleQ, as is generally the case for EBPs. To detect possible transient associations, we also performed BN-PAGE in the presence of ATP in the buffer, but found no differences. FleQ dimers, isolated by size-exclusion chromatography, had the same ATPase-specific activity as unfractionated FleQ. FleQ dimers also showed a cooperative ATPase activity, with a Hill coefficient of 2.53 ± 0.03.
AAA+ ATPase domains are characterized by Walker A and Walker B motifs, which are involved in nucleotide binding and hydrolysis, respectively (16, 17). The Walker A motif forms a P-loop with the consensus sequence GxxxxGK(T/S) and interacts with the phosphates of ATP. In the Escherichia coli protein PspF, substitution of the lysine residue of the Walker A motif with an alanine (PspFK42A) abolishes the binding and hydrolysis of ATP (19). The Walker B motif has a consensus sequence of hhhhDE (where h is a hydrophobic amino acid), which may play a role in the coordination of Mg2+, which is necessary for ATP hydrolysis. PspF protein with D107S or E108Q substitutions in the Walker B motif exhibited decreased ATP hydrolysis but increased ATP binding (20).
As expected, introduction of the corresponding mutations in the putative Walker A motif (FleQK180A) or Walker B motif (FleQD245A and FleQE246Q) of FleQ drastically decreased ATPase activity, whereas replacement of the threonine of the σ54-binding domain with a serine (FleQT224S) had no effect (Fig. 1B). Thus, the properties of the predicted Walker A and B motifs of FleQ are consistent with those of other EBPs.
FleN Exhibits Low ATPase Activity and Inhibits FleQ ATPase Activity.
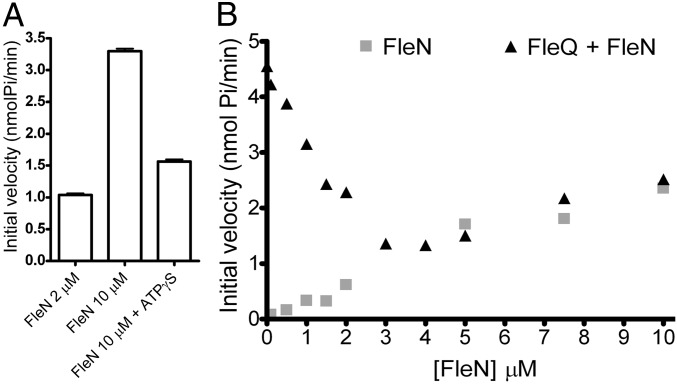
FleN is a putative ATPase with a deviant Walker A motif (xxKGGxxK [T/S]) belonging to a functionally diverse group that includes MinD and ParA (13, 21). These proteins have very low ATPase activity in common. The FleN ATPase activity was also low, and FleN ATPase activity decreased significantly when ATPγS was added as a competitive inhibitor (Fig. 3A). Previously reported data indicate that FleN is a FleQ antiactivator. Flagella gene expression is high in an fleN background (12, 14).
Fig. 3.
FleN inhibits FleQ ATPase activity. (A) The ATPase activity of FleN at different concentrations was assayed in the presence of 1 mM ATP and in the presence or absence of 500 μM ATPγS. (B) The ATPase activity of different concentrations (from 0.1 to 10 μM) of FleN was assayed in the absence or in the presence of 1 μM FleQ and in the presence of 1 mM ATP. The initial velocity of FleN or FleN plus FleQ is plotted against FleN concentration.
To test the hypothesis that FleN inhibits FleQ ATPase activity, we measured the total ATPase activity of a fixed concentration of FleQ (1 μM) incubated with increasing concentrations of FleN (from 0.1 to 10 μM). ATPase activity decreased when the FleN concentration was increased (<3 μM) (Fig. 3B). At higher FleN concentrations (>3 μM), the combined ATPase activity of FleQ and FleN met the curve of FleN activity alone, suggesting that FleQ ATPase activity was completely abolished and that the remaining ATPase activity could be attributed to FleN. This finding indicates that FleN inhibits FleQ ATPase activity and can explain the FleQ-antiactivating role of FleN.
C-di-GMP Competitively Inhibits FleQ ATPase Activity.
C-di-GMP down-regulates flagella gene expression without affecting the binding of FleQ or the binding of FleQ and FleN to the promoter of fleSR (7). Thus, we wondered whether c-di-GMP might inhibit FleQ ATPase activity. Testing this possibility revealed that FleQ ATPase activity decreased with increasing c-di-GMP concentration (Fig. 4A). Kinetic analyses showed that c-di-GMP raised the apparent Khalf (corresponding to the concentration of substrate that produces half-maximal enzyme velocity) values for ATP, but had almost no effect on the apparent Vmax values, indicating that c-di-GMP inhibits FleQ ATPase activity in a competitive manner (Fig. 4B and Table 1).
Fig. 4.
C-di-GMP inhibits FleQ ATPase activity. (A) The ATPase activity of 1 μM FleQ protein, incubated with different c-di-GMP concentrations from 1 to 200 μM, was assayed with 1 mM ATP. (B) The ATPase activity of 1 μM FleQ, preincubated with 25, 37.5, or 50 μM c-di-GMP, and assayed at different ATP concentrations. The activity curves were analyzed according to the Hill equation (Table 1).
An Intact Walker A Motif Is Required for Binding of c-di-GMP to FleQ.
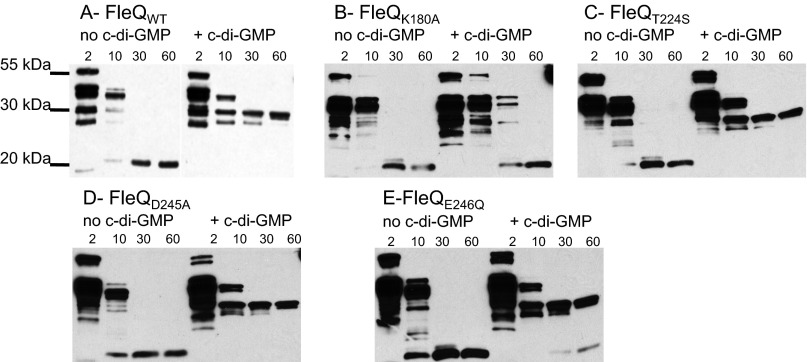
Competitive inhibition of enzyme activity can be most readily explained by binding of substrate and inhibitor to the same site on an enzyme. To test this hypothesis, we purified and subjected to trypsin proteolysis FleQ variants containing amino acid substitutions in the Walker A motif (FleQK180A), the Walker B motif (FleQD245A and FleQE246Q), and the σ54-binding domain (FleQT224S). We have previously reported that WT FleQ undergoes a conformational change on binding of c-di-GMP, which can be detected by a change in its trypsin digestion profile (8). In the absence of c-di-GMP, a major proteolysis fragment of 20 kDa was detected after 30 min of incubation with trypsin, whereas in the presence of c-di-GMP, a different proteolysis-resistant fragment of ∼30 kDa appeared (Fig. 5). This ∼30-kDa fragment did not appear when FleQK180A was subjected to trypsin digestion in the presence of c-di-GMP, suggesting that FleQK180A does not bind c-di-GMP. In comparison, FleQD245A, FleQE246Q, and FleQT224S had the same trypsin digestion profiles as the WT FleQ when incubated with c-di-GMP, indicating that they are able to bind c-di-GMP.
Fig. 5.
Effect of c-di-GMP on the trypsin digestion profile of FleQ WT and mutant proteins. FleQ proteins were incubated in the presence or absence of 500 μM c-di-GMP and subjected to proteolysis with trypsin. Incubation times (min) are indicated. Digestion products were separated by SDS/PAGE, transferred to a nitrocellulose membrane, and visualized with anti-FleQ antiserum.
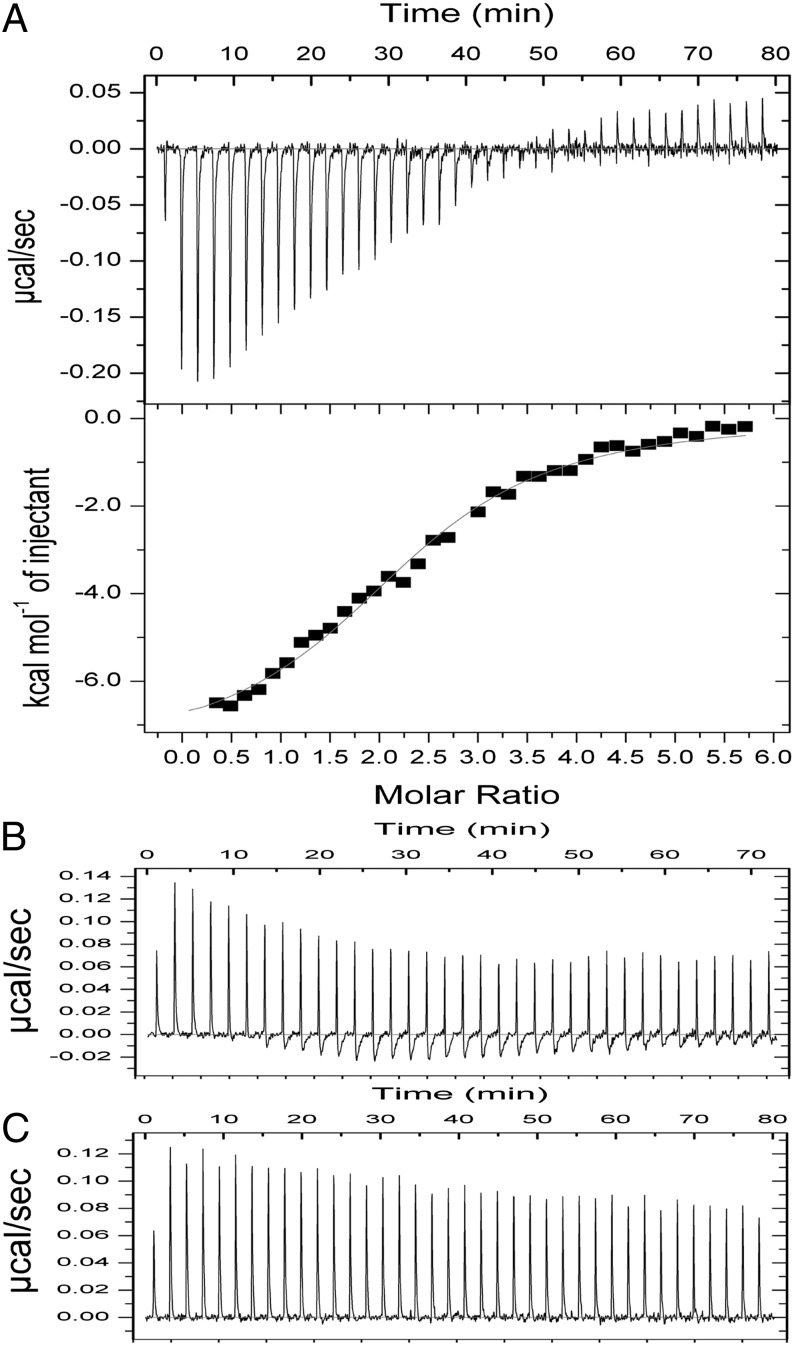
We performed isothermal titration calorimetry (ITC) to further confirm that FleQK180A does not bind c-di-GMP. We found that WT FleQ bound c-di-GMP with a Kd of 7 μM (Fig. 6), comparable to the 15 μM reported by Hickman and Harwood (7). In contrast, when FleQK180A was titrated with c-di-GMP, the amount of heat released was similar to that of buffer titrated with c-di-GMP. These results support the hypothesis that c-di-GMP binds to the ATP-binding site of FleQ, the Walker A motif.
Fig. 6.
Binding of c-di-GMP to FleQ analyzed by ITC. WT FleQ (A), FleQK180A (B), and buffer (C) were titrated with c-di-GMP. The amount of power (µcal/s) needed to maintain a constant temperature during the titration reflects the amount of heat released on c-di-GMP binding. This is integrated over time to give a specific enthalpy vs. ligand:protein molar ratio plot (A, Lower). Heat from the dilution of c-di-GMP was subtracted before data analysis. The thin line represents the fit of the integrated data to a single-site binding model and gives the following parameters: n = 2.35 ± 0.04, K = 1.39E5 ± 1.32E4 M−1, and ΔH = −7,652 ± 209 cal/mol.
Inhibition of FleQ ATPase Activity by c-di-GMP Is Enhanced by the Presence of FleN.
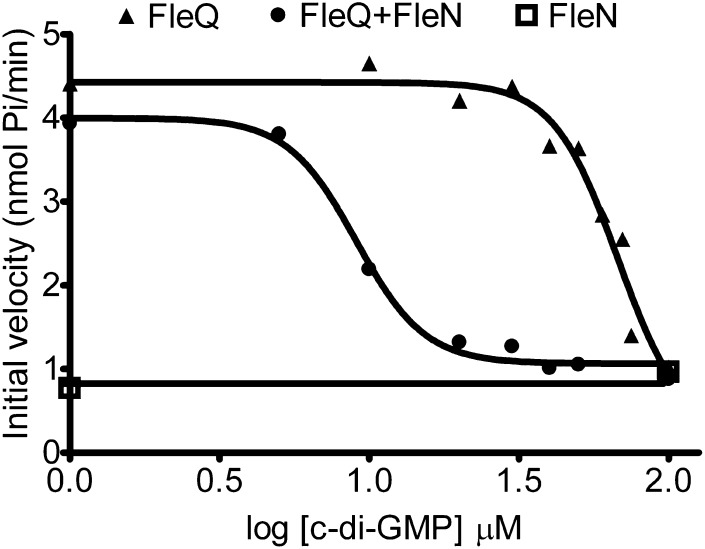
FleQ and FleN interact in the presence as well as in the absence of ATP or c-di-GMP (8). We wondered how c-di-GMP affects FleQ ATPase activity in the presence of FleN. FleQ ATPase activity was much more inhibited by c-di-GMP in the presence of FleN than in the absence of FleN (Fig. 7). The ATPase activity of FleQ alone was inhibited by c-di-GMP with an IC50 value of 67.5 μM (SE, logIC50 of 0.06). This value fell to 9 μM (SE, logIC50 of 0.03) when FleN was present. FleN ATPase activity was not affected by the presence of c-di-GMP. This indicates that inhibition of FleQ ATPase activity by c-di-GMP is enhanced by FleN, and suggests that FleQ and FleN cooperate to detect c-di-GMP.
Fig. 7.
FleQ ATPase activity is inhibited by FleN and c-di-GMP. The ATPase activity of 1 μM FleQ, 1 μM FleN, or 1 μM FleQ plus 1 μM FleN was measured at different concentrations of c-di-GMP (from 1 to 100 μM) and in the presence of 2 mM ATP. The initial velocity of FleN, FleQ, or FleN plus FleQ ATPase activities are plotted against the log of c-di-GMP concentration, allowing determination of the IC50.
Discussion
Here we have presented two lines of evidence suggesting that the Walker A motif of the AAA+ domain of FleQ is involved in the binding of c-di-GMP. We have shown that c-di-GMP competitively inhibits FleQ ATPase activity, with c-di-GMP having a much higher affinity for FleQ compared with ATP (Fig. 4), and that a Walker A mutant of FleQ failed to bind c-di-GMP (Figs. 5 and 6). The most straightforward interpretation of these results is that c-di-GMP binds to the Walker A motif of FleQ; however, in the absence of structural data, we cannot exclude the possibility that c-di-GMP binds elsewhere on FleQ to influence its ATPase activity, and that an intact Walker A motif is required for binding at this alternative site.
The effects of c-di-GMP and FleN on inhibition of FleQ ATPase activity explain their effects on repression of flagella gene expression in vivo. In the absence of c-di-GMP and FleN, FleQ exhibits full ATPase activity (Fig. 1). As with other EBPs, we propose that the hydrolysis of ATP by FleQ remodels σ54-RNA polymerase closed complexes, resulting in the formation of open promoter complexes competent for transcription. Flagella genes under the control of FleQ, including the fleN gene, are expressed (9). The FleN protein interacts with FleQ bound to flagella gene promoters (7, 14) and inhibits FleQ ATPase activity (Fig. 3). This leads to a σ54-RNA polymerase complex in a configuration that is less competent for transcription, resulting in the down-regulation of flagella gene expression such that WT cells synthesize a single polar flagellum.
After surface sensing (22), or depending on the cell cycle (23) or other environmental signals (4, 24, 25), the intracellular concentration of c-di-GMP can increase in P. aeruginosa cells. A recent study showed that the concentration of intracellular c-di-GMP in WT P. aeruginosa cells grown in liquid is approximately 11 µM, but can increase to >30 µM when diguanylate cyclase activity is high (26). C-di-GMP levels can be very low in cells expressing phosphodiesterases (27). C-di-GMP binds to FleQ (7) and further inhibits its ATPase activity (Figs. 4 and 7), leading to further down-regulation of flagella gene expression. Moreover, c-di-GMP inhibition of FleQ ATPase activity is enhanced by the presence of FleN, suggesting that FleQ and FleN cooperate to detect c-di-GMP.
The fact that both FleN and c-di-GMP target the ATPase activity of FleQ and that FleQ with or without FleN is maintained bound to the flagella gene promoters in the presence as well as in the absence of c-di-GMP (7), suggests that this organization might be a biological advantage, allowing cells to rapidly respond to changing concentrations of c-di-GMP. Previously reported transcriptome data show only modest decreases in flagella gene expression in response to c-di-GMP (15, 27); however, the equilibrium among all of the components of the flagellum is so important that even a small reduction in gene expression may have a large impact.
FleQ has several features that make it an unusual EBP. Whereas typical EBPs form inactive dimers, which assemble as active hexamers on ATP binding, we found that FleQ forms mostly active dimers and detected few hexamers (Fig. 2). The presence of FleQ dimers with ATPase activity may signify that FleQ binds to flagella gene promoters as a dimer. It may be that the FleQ-σ54-RNA polymerase complex at flagella gene promoters requires less energy than other EBP-σ54-RNA polymerase complexes to form an open complex. The FleQ-binding sites on the flagella promoters have unconventional locations. Most EBPs bind to sites located 100 bp upstream of the transcription start sites. However, FleQ binds downstream of the transcription start site of the flhA, fliE, and fliL promoters; only the fleSR promoter contains a FleQ-binding site located upstream (67 bp upstream) of its transcription start site (11).
C-di-GMP effectors are diverse, especially those linked to transcriptional regulation. Several different transcriptional regulators with different c-di-GMP–binding sites have been identified. Clp from Xanthomonas campestris binds c-di-GMP at its cNMP-binding site (28–30). C-di-GMP binds to the W[F/L/M][T/S]R motif of VpsT from Vibrio cholerae (31). The binding of c-di-GMP to the PilZ domain of MrkH from Klebsiella pneumoniae changes the expression of type 3 fimbriae (32). The c-di-GMP–binding sites in other transcription factors, including Bcam1349 from Burkholderia cenocepacia (33), LtmA from Mycobacterium smegmatis (34), and VpsR from V. cholerae, remain to be identified (35). The X. campestris Clp protein and FleQ belong to well-conserved and well-studied families of transcriptional regulators. In the X. campestris Clp protein, one residue out of the six key residues known to be involved in the binding of cAMP is not conserved, and this particular residue turns out to be important for c-di-GMP binding. In contrast, the Walker A motif of FleQ is perfectly conserved, opening up the possibility that other AAA+ ATPases may bind and respond to c-di-GMP.
Materials and Methods
FleQ Mutant Construction and Protein Purification.
Mutations in the fleQ gene were generated by an overlapping PCR procedure with two sets of divergent and complementary primers, each set containing the expected substitution and the convergent primers FleQ intein up and FleQ intein down previously used to amplify the fleQ gene (7). Primer sequences are available on request. The NdeI-EcoRI fragments thus produced were cloned into pTYB12, generating pFleQK180A-Int1, pFleQT224S-Int1, pFleQD245A-Int1, and pFleQE246Q-Int1. Plasmids were transformed into E. coli ER2566 for expression of fusion protein. FleQ and FleN were purified as described previously (7), and FleQ variants were purified following the same protocol as for WT FleQ. Truncated FleQ (FleQ-trunc) has been described previously (7). The A260/A280 ratio of the purified FleQ proteins and BSA as control were similar, suggesting no contamination of purified protein by nucleotides.
The FleQ protein also was subjected to size-exclusion chromatography. Protein was injected into a Superdex 200 10/300 GL column (GE Healthcare). The buffer was composed of 25 mM Tris (pH 7.5), 100 mM NaCl, 1 mM DTT, and 2 mM MgCl2. Chosen fractions were directly assayed for their ATPase activity immediately after elution from the column.
Limited Proteolysis.
Trypsin digestion was performed as described previously (8). In brief, 8 μg of FleQ WT or variant protein was incubated with 8 μg of BSA and 500 μM c-di-GMP when necessary for 10 min, followed by the addition of 1.6 μg of trypsin. At prespecified intervals, reactions were stopped by plunging 11 μL of the proteolysis mix into cold SDS loading buffer. The samples were heated for 10 min at 95 °C and then analyzed by SDS/PAGE, followed by transfer to nitrocellulose membrane for detection with anti-FleQ antiserum.
ATPase Activity.
ATPase activity was assayed by measuring the production of inorganic phosphate (Pi) using the Enzchek Phosphate Assay Kit (Invitrogen). The reactions were set up as specified by the manufacturer, except that the MgCl2 concentration was increased to 2 mM and 10 mM KCl and 1 mM DTT were added. FleQ WT or variant protein and FleN were added to reaction mixtures as indicated, followed by incubation for 10 min at room temperature before ATP was added to start the reaction. When necessary, c-di-GMP (Axxora; >95% purity), 100 μM ADP, or 0.25 μM DNA fragment encompassing the fleS promoter (corresponding to a ratio of 125 pmol of DNA to 500 pmol of FleQ monomer) was incubated with FleQ with or without FleN for 10 min before the assay. The kinetics of Pi release were monitored by reading at OD360. A standard curve with KH2PO4 as the source of inorganic phosphate was performed. The specific activity (nmol Pi/min/mg of FleQ) versus ATP concentration was used to determine kinetic parameters. These were calculated by the nonlinear regression/allosteric sigmoidal enzyme kinetics model (Hill equation) using GraphPad Prism 4.
In an alternative approach, enzyme velocity was measured at a single concentration of substrate with varying concentrations of a competitive inhibitor (c-di-GMP) to determine the IC50. IC50 values were calculated using the nonlinear regression/sigmoid dose–response (variable slope) kinetics model with GraphPad Prism 4. The same data are presented in Figs. 1A and 4B. Each experiment was performed at least two times, and the maximum error observed was 12%.
Separation by BN-PAGE.
In BN-PAGE, Coomassie G-250 binds to proteins and confers a net negative charge while maintaining the proteins in their native state. Purified FleQ proteins (50 μg) were mixed with 10 mM ATP, AMP, ADP, ATPγS, or GTP or 1 mM c-di-GMP in binding buffer containing 10 mM Tris (pH 8), 8 mM magnesium acetate, 50 mM KCl, and 5% glycerol for 10 min at room temperature. The samples were then loaded on a 4–16% NativePAGE Novex Bis-Tris gel (Invitrogen) with dark-blue cathode buffer (0.02% Coomassie G-250), followed by electrophoresis for 2 h at 150 V and room temperature. NativeMarker Unstained (Invitrogen) served as a protein standard.
ITC.
WT FleQ (29 μM), FleQK180A (20 μM), and c-di-GMP (600 μM) were prepared in 25 mM Tris⋅HCl, 300 mM NaCl, and 5 mM MgCl2, and each solution was degassed before use in experiments. ITC measurements were performed with a MicroCal ITC-200 system (GE Healthcare). Except for the first injection, each titration experiment consisted of 37 1-μL injections with a 125-s interval between injections. The ITC data were analyzed with Origin v7.0 software (OriginLab) and fit using a single-site binding model.
Acknowledgments
We thank Seemay Chou and Joseph Mougous for help with size-exclusion chromatography, Marcos Navarro for his ITC protocol and helpful advice, and John Sumida for ITC training and helpful discussions. The ITC experiments were performed at the University of Washington’s Analytical Biopharmacy Core, which is supported by the Washington State Life Sciences Discovery Fund and the Center for the Intracellular Delivery of Biologics. This work was supported by Public Health Service Grant GM56665 from the National Institute of General Medical Sciences.
Footnotes
The authors declare no conflict of interest.
References
- 1.Wolfe AJ, Visick KL. Get the message out: Cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol. 2008;190(2):463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7(4):263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 3.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 4.Mills E, Pultz IS, Kulasekara HD, Miller SI. The bacterial second messenger c-di-GMP: Mechanisms of signalling. Cell Microbiol. 2011;13(8):1122–1129. doi: 10.1111/j.1462-5822.2011.01619.x. [DOI] [PubMed] [Google Scholar]
- 5.Boyd CD, O’Toole GA. Second messenger regulation of biofilm formation: Breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol. 2012;28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP–responsive transcription factor. Mol Microbiol. 2008;69(2):376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baraquet C, Murakami K, Parsek MR, Harwood CS. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012;40(15):7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasgupta N, et al. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(3):809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 10.Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179(17):5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jyot J, Dasgupta N, Ramphal R. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J Bacteriol. 2002;184(19):5251–5260. doi: 10.1128/JB.184.19.5251-5260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasgupta N, Arora SK, Ramphal R. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2000;182(2):357–364. doi: 10.1128/jb.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229(4):1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 14.Dasgupta N, Ramphal R. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2001;183(22):6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Starkey M, et al. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191(11):3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bose D, et al. Dissecting the ATP hydrolysis pathway of bacterial enhancer-binding proteins. Biochem Soc Trans. 2008;36(Pt 1):83–88. doi: 10.1042/BST0360083. [DOI] [PubMed] [Google Scholar]
- 17.Bush M, Dixon R. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev. 2012;76(3):497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyman C, Rombel I, North AK, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275(5306):1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 19.Schumacher J, Zhang X, Jones S, Bordes P, Buck M. ATP-dependent transcriptional activation by bacterial PspF AAA+ protein. J Mol Biol. 2004;338(5):863–875. doi: 10.1016/j.jmb.2004.02.071. [DOI] [PubMed] [Google Scholar]
- 20.Joly N, Rappas M, Wigneshweraraj SR, Zhang X, Buck M. Coupling nucleotide hydrolysis to transcription activation performance in a bacterial enhancer binding protein. Mol Microbiol. 2007;66(3):583–595. doi: 10.1111/j.1365-2958.2007.05901.x. [DOI] [PubMed] [Google Scholar]
- 21.Lutkenhaus J, Sundaramoorthy M. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol Microbiol. 2003;48(2):295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- 22.Güvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66(6):1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christen M, et al. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science. 2010;328(5983):1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Römling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9(2):218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Yan H, Chen W. 3′,5′-Cyclic diguanylic acid: A small nucleotide that makes big impacts. Chem Soc Rev. 2010;39(8):2914–2924. doi: 10.1039/b914942m. [DOI] [PubMed] [Google Scholar]
- 26.Irie Y, et al. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2012;109(50):20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102(40):14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin KH, et al. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell–cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol. 2010;396(3):646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 29.Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191(22):7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao F, He YW, Wu DH, Swarup S, Zhang LH. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J Bacteriol. 2010;192(4):1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krasteva PV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327(5967):866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilksch JJ, et al. MrkH, a novel c-di-GMP–dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011;7(8):e1002204. doi: 10.1371/journal.ppat.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fazli M, et al. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol Microbiol. 2011;82(2):327–341. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- 34.Li W, He ZG. LtmA, a novel cyclic di-GMP–responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res. 2012;40(22):11292–11307. doi: 10.1093/nar/gks923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srivastava D, Harris RC, Waters CM. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol. 2011;193(22):6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]