Significance
Medulloblastoma is a common malignant pediatric brain tumor. Gene expression data have indicated that the tumors fall into four molecular subgroups, “Wnt,” “Hedgehog,” “group 3,” and “group 4.” With the exception of the Hedgehog subgroup, few functional data exist defining key molecular pathways driving tumor growth. Using a transposon mutagenesis approach, we identified genes that functionally cooperate with Hedgehog signalling to promote tumorigenesis in a Ptch1 mouse model of medulloblastoma. Surprisingly, the genes we identified were able to accurately define all four human molecular subtypes, not just Hedgehog, when used to interrogate published expression data. Thus, we have functionally defined key regulatory networks that illustrate both the differences and commonalities between tumor subgroups indicating a number of therapeutic strategies.
Abstract
The Sleeping Beauty (SB) transposon mutagenesis screen is a powerful tool to facilitate the discovery of cancer genes that drive tumorigenesis in mouse models. In this study, we sought to identify genes that functionally cooperate with sonic hedgehog signaling to initiate medulloblastoma (MB), a tumor of the cerebellum. By combining SB mutagenesis with Patched1 heterozygous mice (Ptch1lacZ/+), we observed an increased frequency of MB and decreased tumor-free survival compared with Ptch1lacZ/+ controls. From an analysis of 85 tumors, we identified 77 common insertion sites that map to 56 genes potentially driving increased tumorigenesis. The common insertion site genes identified in the mutagenesis screen were mapped to human orthologs, which were used to select probes and corresponding expression data from an independent set of previously described human MB samples, and surprisingly were capable of accurately clustering known molecular subgroups of MB, thereby defining common regulatory networks underlying all forms of MB irrespective of subgroup. We performed a network analysis to discover the likely mechanisms of action of subnetworks and used an in vivo model to confirm a role for a highly ranked candidate gene, Nfia, in promoting MB formation. Our analysis implicates candidate cancer genes in the deregulation of apoptosis and translational elongation, and reveals a strong signature of transcriptional regulation that will have broad impact on expression programs in MB. These networks provide functional insights into the complex biology of human MB and identify potential avenues for intervention common to all clinical subgroups.
Medulloblastoma (MB) is the most common malignant pediatric brain tumor, comprising 20–25% of all tumors diagnosed in the CNS. Recent advances in therapy have lifted 5-y survival rates for average-risk MB to 80%; however, the outcome for high-risk patients remains poor (1, 2). Additionally, survivors often experience significant neurological, intellectual, and physical disabilities as a consequence of the aggressive nature of existing treatment protocols (3, 4). Clearly, less invasive and more effective therapies are urgently required.
MB is well established as a biologically heterogeneous disease, with several studies identifying at least four subgroups with distinct clinical, biological, and genetic profiles (5–8). Preliminary evidence has pointed to additional intersubgroup heterogeneity (7, 9, 10), with future studies on larger cohorts likely to refine the four major subgroups of MB as consisting of multiple subgroups potentially arising from different cellular origins. Despite these advances, targeted therapies based on the molecular abnormalities of MB subgroups are not currently in use. Although inhibitors of the Sonic Hedgehog (SHH) pathway component Smoothened (SMO) had shown early promise (11), studies have since revealed that resistance has emerged (12). Furthermore, although some molecular abnormalities of each of the four subgroups have been successfully defined, particularly for the SHH and WNT subgroups, the specific molecular drivers required for the initiation and progression of MB are still not well understood.
To discover candidate cancer genes that function as key drivers of MB, we performed an insertional mutagenesis screen using the inducible Sleeping Beauty (SB) transposon system (13). We bred mice containing the SB DNA transposon, T2Onc3, and a Cre-inducible SB transposase to the well-characterized Patched1 (Ptch1)-heterozygous (Ptch1lacZ/+) model of MB (14). We hypothesized that mice harboring the active SB transposon would acquire mutations that accelerate the rate of tumorigenesis in this sensitized MB model, therefore identifying drivers of MB. To date, one study has reported a SB mutagenesis screen in the Ptch1-inactivated mouse model of MB, restricting SB transposase within granule neuron progenitors (GNPs) of the cerebellum via the Math1Cre promoter (15). Cerebellar GNPs derived from the upper rhombic lip were previously identified as one potential cell of origin for MB (16). However, additional studies have shown that the SHH subgroup MB may have multiple cellular origins (17, 18). In light of this, the approach taken in this study is distinct and complementary to the previously published SB screen in MB, whereby we chose to not restrict SB mutagenesis to committed cells of the neuronal lineage but rather drive ubiquitous transposase activity in a broad range of developing tissues. We observed significant acceleration of MB formation in mice with SB insertional mutagenesis compared with Ptch1lacZ/+ controls. Sequencing of SB-derived tumors identified 77 common insertion sites (CISs)/loci associated with accelerated MB development. We subsequently validated that haploinsufficiency of the highly ranked CIS, Nfia, accelerates MB formation in vivo. Furthermore, protein coding genes from the CIS list were capable of accurately clustering the four molecular subgroups of human MB, confirming that the murine SB mutagenesis model recapitulates the genetic complexity of human MB.
Significance
Gene expression and genome organization studies have revealed that MB can be divided into four subgroups Hedgehog (SHH), WNT, group 3, and group 4. Genome sequencing has revealed a number of potential molecular defects in each subgroup with the challenge being to discern the “drivers” from the “passengers.” We used a SB transposon screen in mice prone to SHH MB to identify regulatory networks that function to promote MB initiation and growth. Unexpectedly, the regulatory networks identified discriminated between all four MB subgroups, indicating functional networks that can be used to target tumors based on their commonalities and differences.
Results
SB-Induced Mutagenesis Drives MB Formation in the Ptch1 Heterozygous Mouse Model.
To identify candidate genes involved in driving MB formation, we performed a SB transposon-mediated insertional mutagenesis screen in the genetically predisposed Ptch1lacZ/+ mouse model. Ptch1lacZ/+ mice were bred to mice carrying a β-actin Cre (βCre) transgene, which is ubiquitously expressed beginning early in development (19). βCre; Ptch1lacZ/+ were then crossed to a compound transgenic line containing 30 copies of the mutagenic SB transposon (T2Onc3) and a Lox-STOP-Lox SB-transposase allele knocked into the ubiquitous Rosa26 locus (Rosa26SB11). T2Onc3 has been engineered to carry the ubiquitously expressed CAG promoter to activate expression of protooncogenes upon insertion at appropriate locations. In addition, T2Onc3 carries splice acceptor sites in both orientations and a bidirectional poly(A) signal and can therefore inactivate the expression of tumor suppressor genes when integrated within the coding region in either orientation.
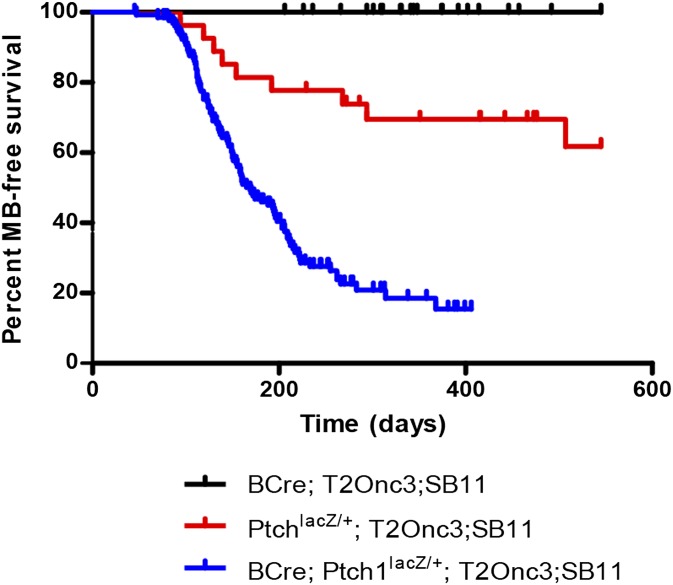
Three cohorts of mice carrying different combinations of alleles were bred and monitored for MB formation (Fig. 1). Ptch1lacZ/+ mice develop MB at a frequency of ∼30% by 1 y of age (14). Consistent with these data, we observed MB formation in 29.6% (8 of 27) of Ptch1lacZ/+; T2Onc3; Rosa26-SB11 controls. By contrast, βCre; Ptch1lacZ/+; T2Onc3; Rosa26-SB11 mice showed increased penetrance of MB at 69.6% (94 of 135) (P < 0.0001, χ2 test). Furthermore, βCre; Ptch1lacZ/+; T2Onc3; Rosa26-SB11 mice presented with a decreased latency of tumor-free survival of 176 d compared with 290 d with Ptch1lacZ/+; T2Onc3; Rosa26-SB11 animals (P < 0.0001, Kaplan–Meier survival analysis) (Fig. 1). Mobilization of the T2Onc3 transposon via βCre-driven expression of SB transposase in Ptch1WT mice (βCre; Ptch1+/+; T2Onc3; Rosa26-SB11) did not develop MB; however, we did observe a background liver and hematological malignancy rate as previously described (13). These data suggest that the SB-insertional mutagenesis identified in this screen will identify those genes involved in driving MB formation in predisposed, hyperproliferative Ptch1lacZ/+ cells.
Fig. 1.
Sleeping Beauty (SB) mutagenesis accelerated tumorigenesis and penetrance of medulloblastoma (MB) in the Ptch1 heterozygous mouse model. Three cohorts of mice were monitored for tumor development, with all three cohorts carrying the SB transposon and SB transposase. Inactivation of tumor suppressor Ptch1 was required for MB development, with β-actin Cre required for expression of SB transposase. Ptch1lacZ/+ SB mutagenized mice succumbed significantly earlier to MB (n = 94 of 135) than animals with inactivated Ptch1lacZ/+ alone (n = 8 of 27) (P < 0.0001, log-rank test).
Sequence Analysis of Transposon Insertion Sites Identifies MB Candidate Cancer Genes.
To identify mutations that cooperated with haploinsufficiency of Ptch1 to increase incidence and accelerate the development of MB, we PCR-amplified and sequenced the transposon insertion sites of 85 MBs from Ptch1lacZ/+ SB-mutagenized mice. Mapping analysis was then performed to identify CISs, defined as genomic loci that contain transposon insertions more frequently than would be predicted by chance. The analysis revealed 77 statistically significant CISs mapping to 56 protein-coding genes, in addition to five predicted or hypothetical genes and a number of noncoding RNAs (P < 0.05) (Table S1). CREB binding protein (Crebbp), mitogen-activated protein kinase kinase kinase 1 (Map3k1), and nuclear factor 1/A (Nfia) were identified as the top three CISs (P < 109), followed by zinc finger DHHC-type containing 9 (Zdhhc9) and tetraspanin 7 (Tspan7) (P < 108). The likelihood of a particular CIS being an “activation” or “inactivation” event is estimated based upon the orientation and location of the transposon with respect to the gene, whereby an appreciable frequency of transposons within the gene in the reverse orientation suggests that the CIS is likely loss of function. Mapping of individual integration events in each tumor for each CIS based on these principles indicated that each represented an “inactivating” event, therefore identifying potential mutations that cooperate with Ptch1 deficiency in driving the onset and/or progression of MB.
Transposon Insertion Sites Identify Cancer Genes That Are Capable of Accurately Clustering Known Molecular Subgroups of Human MB.
Although a number of the CISs have been previously validated in MB, such as Crebbp (20) and Pten (21), the majority have not been previously linked to MB tumorigenesis, and represent potentially novel genetic mechanisms for driving tumor formation. To assess whether the CISs identified from the SB mutagenesis screen capture significant biological features of human MB, a cross-species comparative analysis was performed. We mapped the protein coding genes identified as CISs in the SB mouse model to human orthologs (Table S2) and used these genes to select corresponding probes from a previously published expression array analysis of human MB (8). Remarkably, the human orthologs of the CIS gene set were not only able to cluster the SHH-type human tumors, but accurately defined all four human MB subgroups (Fig. 2A).
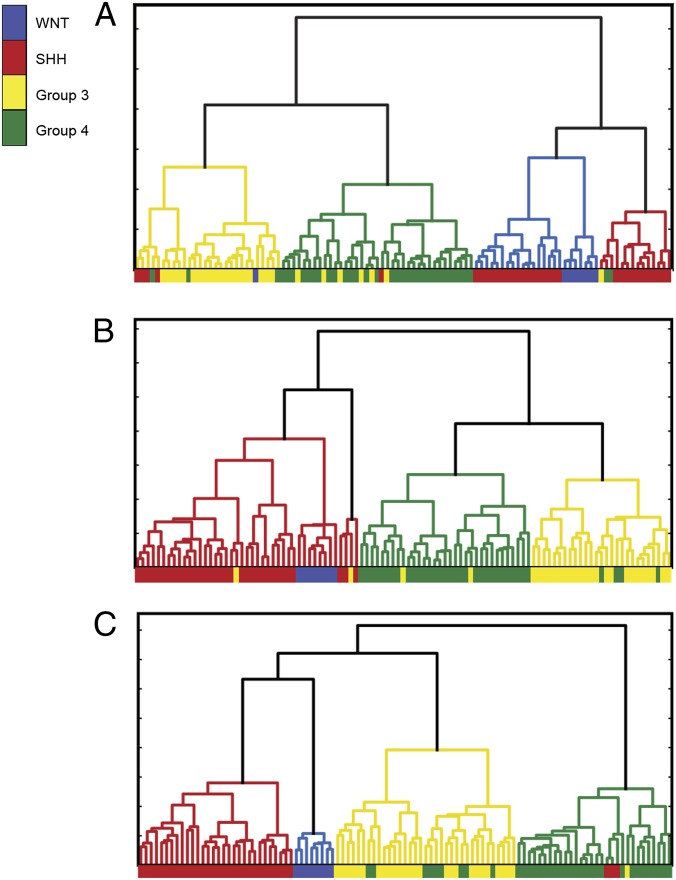
Fig. 2.
Clustering of human medulloblastoma (MB) samples based on the expression of CIS genes, CIS network, and human gene signatures. Expression of the CISs identified from Sleeping Beauty (SB)-induced MB accurately clustered samples into the four known molecular subgroups of human MB. Unsupervised clustering is based on (A) 49 human protein coding genes that map to the CIS list, with principal component analysis (PCA) reducing these data to the first five principal components; (B) 49 human proteins from A, plus the local interaction network of these proteins; and (C) the human signature gene list from ref. 22, which had been previously optimized to separate the four molecular subtypes of human MB. RI scores for the goodness of clustering of each of these gene sets are (A) 0.469, (B) 0.685, and (C) 0.699. The colored bar below the dendrogram shows the subgroup annotation provided with the expression data (8).
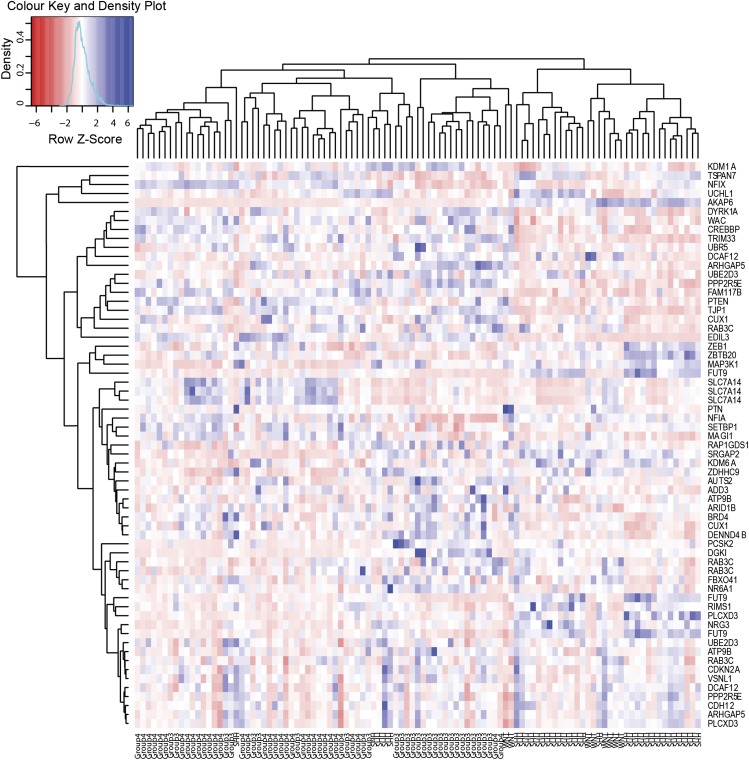
We compared the clustering performance of CIS orthologs to that of well-defined human MB subgroup signatures (8, 22). It is important to note that the subgroup signatures were developed using human MB data and were highly optimized for the task of separating the four subgroups of human MB tumors. To compare our clustering, we used the Rand Index (RI) (23) to evaluate the accuracy of clustering against the known molecular subgroup annotation of the human MB tumors. Using only the genes identified in our mutagenesis screen, unsupervised hierarchical clustering analysis identified clear segregation of MB samples according to molecular subgroup (RI = 0.461, Fig. 2A), with clear clustering of SHH and WNT subtypes both from each other and from the group 3 and 4 tumors, which also separated, although with less accuracy. A similar pattern, including some overlap in group 3 and 4 tumors, is observed when using expression data selected by the human MB subgroup signature (RI = 0.699, Fig. 2C). A heat map of the gene expression values of the CIS-derived genes mapped to human MB dataset shows clear patterns of variability across the molecular subgroups (Fig. 3) and demonstrates the variation in expression underpinning the ability of these genes to successfully cluster the human MB subgroups. To identify which CISs identified in our mutagenesis screen are of particular relevance to specific human molecular subgroups of MB, we identified differentially expressed genes in each of the human subgroups, and then mapped those to the human orthologs of CIS-derived genes (Table 1). A number of the CIS-derived genes were underexpressed across human MB subgroups consistent with their transposon-mediated inactivation in the SB mouse model. Of these, the underexpression of some CIS-derived genes is specific to a certain molecular subgroup. For example, ARID1B is significantly underexpressed in the WNT subgroup alone, with NR6A1 and DENND4B underexpression specific to the SHH subgroup. Specific to group 3, NFIA was the only CIS significantly underexpressed, with underexpressed CIS-derived genes observed in group 4 MB common to one or more of the other subgroups. Although a number of CIS-derived genes were significantly overexpressed in certain human MB subgroups, the majority of these were underexpressed in other subgroups, again reflecting their transposon-mediated inactivation. Taken together, these data represent a truly independent validation that candidate cancer genes identified in this study capture biologically significant features of all molecular subgroups of MB, clearly demonstrating that, although the candidate cancer genes were primarily identified through functional cooperation with Ptch1 to promote MB tumorigenesis, they also underlie all MB subgroups.
Fig. 3.
Heat map and hierarchical clustering of the human MB samples using 49 human protein coding orthologs of the mouse genes from the CIS list.
Table 1.
Differentially expressed human orthologs of CIS-derived genes
| Directionality | WNT | SHH | Group 3 | Group 4 |
| Underexpressed | EDIL3 | EDIL3 | FUT9 | PCSK2 |
| UCHL1 | SLC7A14 | NFIA | AKAP6 | |
| RIMS1 | TJP1 | TSPAN7 | DGKI | |
| ARID1B | ARHGAP5 | AKAP6 | MAP3K1 | |
| PLCXD3 | PTEN | NFIX | NRG3 | |
| TRIM33 | ZBTB20 | SRGAP2 | ||
| NR6A1 | SETBP1 | ZDHHC9 | ||
| PPP2R5E | PTN | TSPAN6 | ||
| CREBBP | MAGI1 | ARHGAP5 | ||
| DENND4B | ||||
| Overexpressed | PTN | PLCXD3 | EDIL3 | CUX1 |
| AKAP6 | ZDHHC9 | PCSK2 | PPP2R5E | |
| KDM6A | WAC | |||
| TSPAN7 | DYRK1A | |||
| ZBTB20 | CREBBP | |||
| ZEB1 | FAM117B | |||
| TSPAN6 | TRIM33 | |||
| DGKI | SETBP1 | |||
| SRGAP2 | PTEN | |||
| UCHL1 | MAGI1 | |||
| NRG3 | RAB3C | |||
| MAP3K1 | TJP1 | |||
| RIMS1 | NFIX | |||
| FUT9 | SLC7A14 | |||
| AKAP6 | EDIL3 |
Significantly underexpressed or overexpressed genes (false discovery rate-adjusted value of P < 0.05) in each subgroup of human MB are listed. CIS-derived genes in bold represent those deregulated in a specific molecular subgroup, whereas those italicized represent the overlap with the Wu et al. (15) dataset. Of the 11 CIS-derived genes that mapped to human orthologs, BRD4 and KDM1a were not significantly differentially expressed.
Local Interaction Networks of Candidate Cancer Genes Are Highly Relevant to Biology Underlying the Molecular Subgroups of MB.
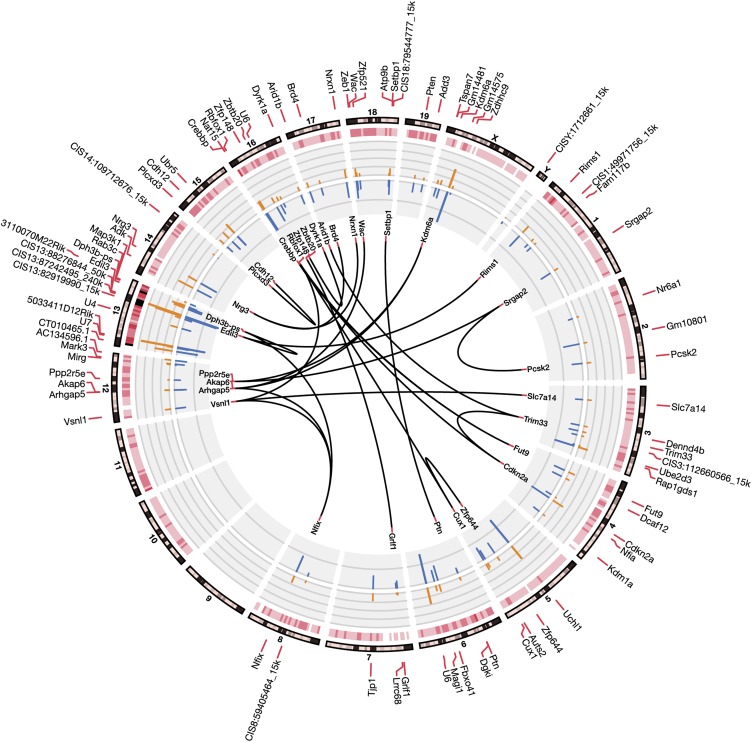
Genes and the proteins they encode do not function in isolation, but operate in networks that are important for the normal function of the cell. To examine potential cooperation between genes identified in our screen, we visualized all 77 CISs on a Circos plot (Fig. 4). Transposon insertions were well distributed across the chromosomes with the exception of chromosome 9. All insertions at chromosome 9 were excluded from statistical analysis for CISs due to the T2Onc3 transposon concatemer being located on this donor chromosome of the transgenic mice and thus commonly coupled to the local hopping phenomena as previously described (13). Interestingly, no comutations were observed for top-ranked CISs, Map3k1 and Nfia (P < 0.01), suggesting that the inactivation of each CIS alone is sufficient to accelerate tumorigenesis on the Ptch1+/− background. In contrast, the majority of CISs were comutated in individual tumors, identified by the links across the interior of the plot, suggesting that some of the candidate cancer genes disrupted by SB mutagenesis may function as components of a broader signaling pathways or networks requiring multiple mutations to drive tumorigenesis.
Fig. 4.
Circos plot of MB common insertion sites (CISs) identified by the Gaussian kernel convolution (GKC) method. Mouse chromosomes are illustrated on the outer perimeter of the plot. GKC CISs are illustrated on the second most outer ring of the plot marking SB insertions in both the forward and reverse orientation. The number of unique insertions at each CIS is represented by orange bars, with the number of tumors containing each CIS represented by blue bars. The black lines in the center connect CISs that significantly co-occur in tumors (Fisher exact test, P < 0.01).
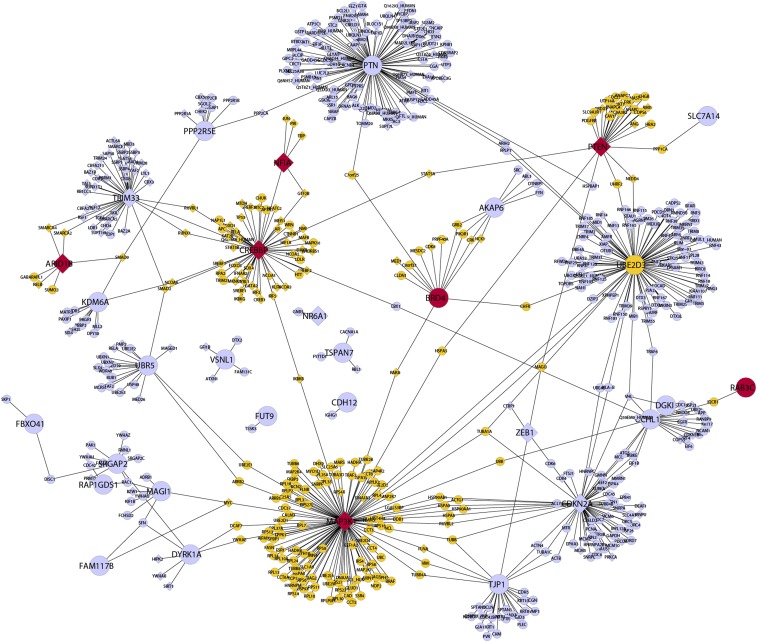
To further explore potential network relationships, the local protein interaction network for each of the protein-coding candidate genes was generated from experimentally determined protein interaction data (Materials and Methods, Bioinformatics), and these local networks were integrated to generate a protein interaction network comprising the CISs and their interacting proteins (Fig. 5). Unsupervised hierarchical clustering of human MB using the network expanded CIS list, coupled with dimensional reduction, greatly improved the accuracy of clustering (RI = 0.685, Fig. 2B) compared with the CIS list alone, and achieved a level equivalent to that obtained using the highly optimized human MB subgroup signatures. Combined, these data indicate that the additional information contained within the expanded CIS–protein interaction network is highly relevant to the molecular changes that characterize the subgroups of human MB.
Fig. 5.
Local protein interaction network for the human orthologs of protein coding genes identified in the SB mutagenesis screen. Local protein interactome hubs for MAP3K1, CREBBP, and NFIA are circled as A, B, and C, respectively. Nodes colored in red represent proteins encoded by CIS-derived candidate genes that overlap with the Wu et al. (15) dataset, whereas yellow nodes represent the proteins that interact with these CIS-derived candidates. Nodes with a diamond shape represent proteins with an annotated role in DNA-dependent transcription.
To elucidate the potential function of each CIS as a tumor suppressor in MB, functional analysis of the local interaction networks of top-ranked CIS genes was performed focusing on Gene Ontology (GO) biological process enrichment. The human ortholog of the Map3k1 protein is known to interact with 115 proteins (Fig. S1A) and is the most highly connected hub in our network. The local network of MAP3K1 shows a strong enrichment for translation elongation genes (GO: 0006414, P = 4.6 × 10−28) and translation (GO: 0006412, P = 2.2 × 10−17). The human ortholog of the next ranked CIS, Crebbp, interacts with 53 proteins, and is the fifth most connected hub in our network (Fig. S1B). The local network of CREBBP shows a strong enrichment for a range of transcription associated processes, including transcription (GO: 0006351, P = 1.3 × 10−13) and positive regulation of DNA-dependent transcription (GO: 0045893, P = 2.2 × 10−16).
The protein encoded by NFIA was not present in the initial interaction network, and consequently, interactions for NFIA were retrieved from the NFI-Regulome database (24) and added into the network. Only five direct interactions are known for NFIA, including TBP, CREBBP, PIR, GTF2B, and JUN, with no significant biological processes enriched for this local interaction network after correction for multiple hypothesis testing.
Loss of Function of Nfia as a Driver of MB Tumorigenesis.
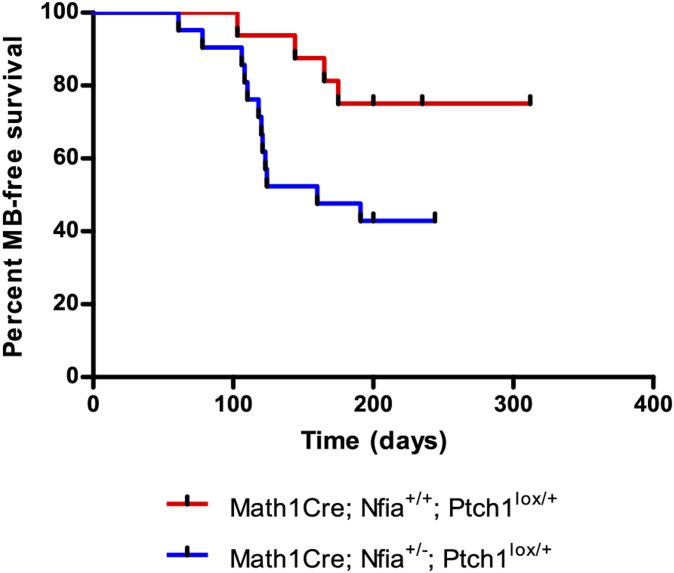
Among other functions, the transcription factor Nfia has previously been demonstrated to play a role in the differentiation of GNPs (25). Given that Nfia was ranked in the top three CISs, we addressed directly whether MB tumorigenicity is influenced by the loss of Nfia. Nfia+/−Ptch1lacZ/+ mice suffer from perinatal lethality due to forebrain defects not evident in control genotypes. Consequently, to examine the role of Nfia in MB formation, we used a Math1Cre transgene to create tissue-specific heterozygosity for Ptch1 given that we have previously demonstrated that conditional inactivation of Ptch1 in GNPs directed via Math1Cre (Math1Cre; Ptch1lox/+) predisposes mice to tumors that resemble human MB (17). We analyzed the onset and frequency of MB formation in Math1Cre; Nfia+/−; Ptch lox/+ mice, compared with Math1Cre; Nfia+/+; Ptch1lox/+ littermate controls. We observed that Math1Cre; Nfia+/+; Ptch1lox/+ mice developed MB at a frequency of 37.5%, similar to that of the Ptch1lacZ/+ model used in the screen (14). In contrast, Math1Cre; Nfia+/−; Ptch1lox/+ presented with a higher incidence of MB (61.90%) (Fig. 6). In addition, Math1Cre; Nfia+/−; Ptch1lox/+ mice demonstrated a significant decrease in tumor latency, with tumors arising at ∼128 d compared with 189 d in Math1Cre; Nfia+/+/Ptch1+/lox mice control mice (P = 0.038, log-rank test) (Fig. 6). Combined, these data confirm that Nfia is a driver of MB tumorigenesis and that reduced Nfia expression cooperates with perturbation in Shh signaling to accelerate MB tumorigenesis in the Ptch1 mouse model.
Fig. 6.
Inactivation of Nfia accelerates MB formation. Kaplan–Meier survival curves comparing Math1Cre; Nfia+/−; Ptch1lox/+ (blue line, n = 12 of 21) experimental mice and control Math1Cre; Nfia+/+; Ptch1lox/+ (red line, n = 6 of 16) mice (P = 0.038, log-rank test).
Comparative Analysis with Previous SB Screen Identifies a Role for Transcription and Translation Elongation in SB-Induced MB.
We compared the list of CIS-derived candidate genes obtained in this study to the previously published SB mutagenesis screen in Ptch1 heterozygous mice (15). Functional analysis of the CIS-derived candidate gene lists from both individual studies revealed a significant enrichment for transcriptional processes (P = 0.045, Wu: P = 8.0 × 10−5), highlighting the presence of many genes with a known role in DNA-dependent transcription in each list. Close comparison of the two CIS lists identified only 14 CIS-derived candidate genes, including top-ranked genes Map3k1, Crebbp, and Nfia that were common to both studies, despite the fact that both screens were performed on the same background model. Although apparently low, this overlap is nonetheless significant (P < 0.001). Interestingly, we note that 9 of the 14 CIS-derived candidate genes common to both SB screens have a known role in DNA-dependent transcription, including Map3k1, Crebbp, Nfia, Nfix, Brd4, Kdm1a, Pten, Wac, and Zbtb20. Enrichment analysis of the local protein interaction network corresponding to the 14 CIS-derived candidate genes in common to both studies once again identified significant enrichment for translation elongation genes (P = 1.2 × 10−20). Combined, the presence of transcription factors in both SB screens suggests the impact of disruption to these genes will be broad and trigger dysregulation of entire programs of gene expression. The enrichment of translation elongation suggests that these transcription factor-mediated deregulated programs of gene expression are potentially linked to this translation elongation and may be therefore implicated in the accelerated MB tumorigenesis observed in both studies.
Discussion
SB mutagenesis is a powerful tool that allows for the unbiased discovery of genes linked to cancer, with previous studies successfully using this approach in several cancer types (13, 26–30). In this study, we used the SB model to identify candidate genes driving the development of MB in conjunction with a mutation in Ptch1. We identified a number of genes with previously recognized roles in MB and/or SHH signaling. One such gene, Crebbp, has been shown to negatively regulate the hedgehog pathway (31, 32), with mutations in Crebbp gene causative of Rubinstein–Taybi syndrome that predisposes to MB (33–35). Whole-genome sequencing of human MB samples recently identified enrichment of mutations in Crebbp in the WNT subgroup of MB, implicating a potential role for this gene in multiple subgroups of MB (20). In addition to Crebbp, haploinsufficiency of Pten was previously shown to promote tumorigenesis in a Shh-activated model of MB (21). The identification of known candidate genes in MB provides proof-in-principle for the validity of the SB mutagenesis model in this disease. The validation of a role for candidate gene Nfia in accelerating MB tumorigenesis provides further support for this, verifying the power of the SB screen as a cancer gene discovery tool for the identification of candidate genes relevant to tumor biology.
Nfia is a transcription factor that has previously been implicated in the control of the differentiation of GNPs, such that loss of Nfia function leads to an inhibition of GNP maturation and migration (25). Given that GNPs are the cell of origin of SHH MB and possibly some group 3 tumors, we asked whether loss of Nfia function as a single event could lead to an acceleration of MB development in a Ptch1+/− background and indeed this was the case. We modeled this experiment on the global SB experiment such that we combined haploinsufficieny for both Ptch1 and Nfia, although we cannot rule out that in the resultant tumors the wild-type Nfia or Ptch1 allele is not silenced or otherwise inactivated. Mutation or loss of Nfia function per se is not a common feature of human MB (36). Nonetheless, this experiment serves to underline the relationship between mitogenic signals and differentiation in the development of MB. In a series of elegant experiments, Ayrault et al. (37) demonstrated that GNPs transduced with the Shh effector, Gli1, developed MB following orthotopic transplant. However, the additional overexpression of the proneural transcription factor, Math1, significantly increased overall tumor incidence and dramatically reduced tumor latency (37). Because overexpression of Math1 attenuates the differentiation of GNPs to granule neurons, these data clearly demonstrate that MB development is strongly linked to the control of differentiation. Similarly, the data presented here addressing the function of Nfia serve as an additional example of how aberrant differentiation promotes MB formation. Clearly, a greater understanding of the regulator gene network defined by Nfia will provide significant insight into the mechanisms whereby a proliferating GNP driven by a mitogenic signal develops into a MB.
The identification of the SB transposon mutagenesis system has led to the development of novel mouse models of cancer, with a number of the CISs identified in several SB-induced tumors relevant to the equivalent human cancer types (27, 38–40). At the onset of this study, we aimed to solely identify candidate genes that cooperate with the loss of Ptch1 to accelerate MB development, given that the SB screen was performed on a SHH mouse model. However, surprisingly, we observed that the CIS-derived candidate genes and associated protein network were capable of distinguishing the molecular subgroups of human MB, including the difficult-to-distinguish subgroups 3 and 4. These data imply that perturbation of the SHH pathway is implicated in non-SHH molecular subgroups, and indeed, support for this hypothesis is found in a previous study that identified elevated Gli1 expression (a key marker of SHH activation) in 15 of 24 randomly selected MBs (41). Furthermore, the degree of separation of molecular subgroups was equivalent to that obtained using highly optimized subgroup specific signature genes. It is important to note that these signature genes represent the culmination of a series of experimentally derived gene signatures all developed and refined using human MB data (5–8). The predictive power of the CIS-derived candidate genes identified in this study, of an independent mouse model, is particularly surprising and confirmed that the SB mutagenesis model of MB presented here captures critical genetic diversity and common pathways underpinning the distinctive human MB subgroups.
Several transgenic MB mouse models have been constructed through specifically perturbing putative tumor suppressors or oncogenes in the suspected cell of origin for that particular MB subtype (reviewed in ref. 42). As a consequence, these models are generally subgroup specific and do not represent the heterogeneity that exists in the human disease. A comparison of CISs identified in this study to the previously published SB mutagenesis screen in Ptch1 heterozygous mice (15) revealed a small, but significant, overlap. We attribute the observed differences in CIS lists between the two studies as a consequence of targeting SB mutagenesis to distinct cell types. Previously, the targeting of SB mutagenesis to distinct development stages of T lymphopoiesis generated T-cell lymphomas with a significant disparity in CIS profiles (40), suggesting that the degree of cell differentiation influences the genetic events that drive transformation. Based on this, we postulate that Math1Cre-driven SB mutagenesis in the Ptch1 heterozygous mouse model more accurately models SHH-driven MB specifically, whereas the ability of CIS-derived candidate genes identified in this study to classify all of the molecular subgroups of human MB suggests that our SB mutagenesis screen identified driver events of relevance to the underlying biology of each molecular subgroup of human MB. CIS-derived candidate genes identified in this study that overlap with the previously published SB mutagenesis screen are differentially expressed across all molecular subgroups, further supporting that CIS-derived genes identified in this study capture biologically significant features of all molecular subgroups of MB. Although subgroup specific mouse models have proven successful for testing the efficacy of subgroup specific therapies in vivo (43, 44), the model described in this study represents potentially an attractive preclinical model allowing for evaluation of novel approaches to therapy targeting the commonalities of MB pathogenesis, irrespective of molecular subgroup.
Accelerated MB tumorigenesis has been observed in two independent mouse models of SB mutagenesis, irrespective of the distinct cell of origin targeted for mutagenesis. Although a modest degree of overlap was observed between the CIS lists of both studies, the defined overlap was indeed significant; suggesting that the functional impact of the inactivation of the CIS-derived candidate genes is linked to the decreased tumor-free survival or accelerated MB tumorigenesis observed in both screens. Significant enrichment of translation elongation genes from the CIS list common to both studies suggests a role for this process in accelerating MB formation. Recently, Leprivier et al. (45) identified a mechanism whereby tumor cells adapt to metabolic stress by blocking translation elongation. More specifically, nutrient depletion induces the up-regulation of eukaryotic translation elongation factor 2 kinase (eEF2K), which subsequently inhibits eukaryotic translation elongation factor 2 (eEF2) required for translation elongation (45). Elevated eEF2K activity was observed in primary human MB tissue and in both the primary and metastatic components of the previously published SB mutagenesis model of MB (45). Furthermore, high eEF2K expression was correlated with the most aggressive molecular subgroup of MB and decreased overall survival of patients (45). Therefore, these findings implicate a similar mechanism operating in our MB model. Although eEF2K was not identified in our screen, eEF2 was identified as an interacting protein of CIS-derived candidate gene, Cdkn2a, as was eukaryotic translation initiation factor 1β (EIF1B), whereas eukaryotic translation elongation factor 1α (EEF1A2) was identified in our network as an interacting partner of Map3k1. Interestingly, EE1A2 has only recently been reported as an interacting partner of CDKN2A (46), establishing another link between this CIS-derived candidate gene and translation elongation. Given the accelerated MB tumorigenesis noted in both this study and the previously published SB screen (15), these data strongly implicate translation elongation in this process.
SB mutagenesis represents a powerful tool for the unbiased discovery of biology, and here we have demonstrated this through a series of computational and experimental analyses. We have demonstrated a specific role for the transcription factor Nfia in the development of MB derived from Math1-positive GNPs, and additionally have identified networks of interacting proteins that are able to separate human MB subtypes and generate striking insights into the biology of MB. Our analysis implicates candidate cancer genes in the dysregulation of apoptosis and translational elongation, and reveals a strong signature of transcriptional regulation that will have broad impact on expression programs in MB. With the identification of molecular subgroups in MB and many other cancers, many studies concentrate on the development of subgroup-specific treatments. For common cancers, this approach is clearly practicable. However, for the less frequent cancers such as MB, it is debatable whether the clinical resources will be available to trial a series of subgroup-specific approaches as a first-line approach. However, this may not be the case for signaling pathways and molecular networks in common between the subgroups, and the data presented here functionally define a series of pathways held in common between subgroups. Collectively, these results both demonstrate the power of our integrated computational and experimental approach, and open a window into the complex biology of human MB and potential avenues for intervention in all subgroups.
Materials and Methods
Transgenic Mice.
To generate the mouse model of MB used in the SB screen, we used the following previously described transgenic mouse lines: T2/Onc3 (13), Rosa-LSL-SB11 (13), Ptch1lacZ/+ (14), and β-Cre mice (19). To generate Math1Cre; Nfia+/−; Ptch1lox/+ mice and appropriate control genotypes, we used the following previously described transgenic mouse lines: Math1Cre (47), Ptch1lox/lox (48), and Nfia+/− mice (49). Mice were monitored daily for tumor development. All experimentation involving animals was reviewed and approved by the Institutional Animal Care and Use Committee of Institute for Molecular and Cell Biology or the University of Queensland animal ethics committee.
Histopathology.
All tumors generated in the SB screen were processed and confirmed MB by the Advance Molecular Pathology Laboratory at the Institute of Molecular and Cell Biology (Singapore).
Linker-Mediated PCR.
Genomic DNA was extracted from tumor tissue using the Puregene Core Kit A (Qiagen), according to the manufacturer’s protocol. Genomic DNA was digested with either BfaI- (left side) or NlaIII (right side) and ligated to BfaI- or NlaIII-specific linkers using T4 ligase. A secondary digest (BamH1, left side; XhoI, right side) was performed to destroy concatemer-generated products. Primary and secondary PCR were performed using primers specific for linker and SB transposon sequences along with Fusion and barcode sequences. PCR amplicons were sequenced using the GS FLX Titanium (Roche).
Sequence Analysis.
Sequences were analyzed for the presence of the barcode, inverted repeat/direct repeat sequences required for transposition, and linker sequences. Genomic sequences were blasted against the mouse genome using BLASTN at 95% stringency and requiring a single match. Sequences were removed if they were on the donor concatamer resident chromosome (Chr 9), in the Engrailed 2 (En2) gene (because the En2 sequence is present within the transposon), and when a single TA dinucleotide contained multiple insertions from several tumors from multiple mice (because of the possibility of a PCR artifact). The remaining 45,481 nonredundant sequences were used to identify a CIS. A CIS was defined as enriched sites of transposon integrations above a background level occurring in at least two independent tumors using modified Gaussian kernel convolution (GKC) method (29, 50) implemented in The Wellcome Trust Sanger Institute bioinformatics pipeline. Multiple kernel scales (widths of 15,000, 30,000, 50,000, 75,000, 120,000, and 240,000 nt) were used to enhance the modified GKC approach. CISs predicted across multiple scales and overlapping in their genomic locations were clustered together such that the CIS with the smallest genomic footprint was reported as the representative CIS. For highly significant CISs with narrow spatial distributions of insertion sites, the 15,000 kernel is typically the scale on which CISs are identified. The P value for each CIS was adjusted by chromosome, and a cutoff of P < 0.05 was used. To control for background integration bias, normal cerebellar DNA from β-cre; Ptch1+/+; T2Onc3; Rosa26-SB11 animals were sequenced. SB common integration sites identified in these animals were excluded from our tumor CIS gene lists.
Bioinformatics.
SB-mutagenesis CISs were mapped to mouse and human protein coding genes using Uniprot release 2013_4 (Tables S1 and S2). Functional analysis was performed using the DAVID web tool (http://david.abcc.ncifcrf.gov/). Gene Ontology FAT categories were selected, and a P value threshold of 0.05 was applied to the Benjamini–Hochberg corrected P value to determine significance. Pathway enrichment was tested against KEGG and Reactome pathways also using DAVID and the threshold described above.
Due to the poor coverage of the mouse interactome, protein interaction networks were built with the human protein set (Table S2) (51). Briefly, an in-house protein interaction database containing protein interactions from six International Molecular Exchange Consortium (52) members (IntACT, MINT, DIP, MatrixDB, BioGrid, and MPIDB) was queried with the Uniprot accession numbers of 49 human proteins of interest to retrieve all direct protein interactions for each listed protein. Thirty-one proteins returned results, whereas the remainder did not have interactions in the dataset. As NFIA was one of the genes lacking protein interaction data in the dataset, protein interactions for NFIA were retrieved from the NFIome database (24). The local interaction networks of each query protein were integrated to generate a protein interaction network with 622 proteins and 666 edges. We used Cytoscape 2.8.2 to analyze and visualize the network. Pathway annotation was retrieved from the KEGG database (www.genome.jp/kegg/). Gene Ontology annotation was retrieved from the Gene Ontology database (www.geneontology.org).
Expression data for a collection of 103 human MB samples was downloaded from the Gene Expression Omnibus (GEO) (GEO accession no. GSE21140). These expression data were generated on the Affymetrix Human Exon 1.0ST Array platform (GEO accession no. GPL5175). Probes corresponding to the human orthologs of CIS genes were selected based on the probe annotation file for GPL5175, and expression data were extracted from the GSE21140 Series Matrix file. No additional probe selection was applied, and if more than one probe corresponded to a listed gene, all probes for that gene were selected. Sample annotation was collected from the GSE21140 series matrix file. The Pei signature used in this study corresponds to the unique, concatenated set of the four gene sets previously published (22). Expression data for the Pei signature were selected in the same way as described above.
Before clustering, all expression datasets were normalized using z-score normalization. Hierarchical clustering was performed with Ward, using the Euclidean distance, average linkage, using the correlation, and Ward using the Euclidean distance and the silhouette index to determine the best number of clusters. K-means clustering was performed with and without the silhouette index, and performance was averaged over 10 repeats. Clustering was performed using Python 2.7 and the Scipy, Numpy, and scikit-lean packages. All clustering results are presented in Table S3. Dimensional reduction was carried out before clustering using principal components analysis, with the number of components determined by inspection of the inflection point of the corresponding plots of the variance against the number of components. To determine which CIS genes were differentially expressed in each subgroup, the human MB dataset (8) was reanalyzed comparing each subtype to the rest in four comparisons. Data from GEO were log2 transformed and z-score normalized using R. Differential expression was calculated using LIMMA. A threshold of 0.05 was applied to the Benjamini–Hochberg corrected P value to identify significantly differentially expressed genes. Once all differentially expressed genes in each group were identified, CIS genes were identified based on their annotated human orthologs (see above).
Supplementary Material
Acknowledgments
We thank Dr. Christelle Adolphe and Dr. Lena Constantin for intellectual input and technical assistance, Dr. Stefan Maetschke for his assistance with the clustering analysis, and Denise Anderson for statistical advice. We also thank Keith and Susan Rodgers for their histopathology support and Christopher Chin for his bioinformatics help. B.J.W. and L.A.G. are supported by Australian National Health and Medical Research Council Grant 1034941 and The John Trivett Foundation Fellowship. M.J.D. is funded by Australian Research Council Grant DP110103384. N.A.J. and N.G.C. were supported by the Agency for Science, Technology and Research, Singapore, and the Cancer Prevention and Research Institute of Texas (CPRIT). D.J.A. and A.G.R. are funded by Cancer Research United Kingdom and The Wellcome Trust. N.A.J. and N.G.C. are both CPRIT Scholars in Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318639110/-/DCSupplemental.
References
- 1.Gajjar A, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): Long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7(10):813–820. doi: 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 2.Rossi A, Caracciolo V, Russo G, Reiss K, Giordano A. Medulloblastoma: From molecular pathology to therapy. Clin Cancer Res. 2008;14(4):971–976. doi: 10.1158/1078-0432.CCR-07-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat SR, et al. Profile of daily life in children with brain tumors: An assessment of health-related quality of life. J Clin Oncol. 2005;23(24):5493–5500. doi: 10.1200/JCO.2005.10.190. [DOI] [PubMed] [Google Scholar]
- 4.Palmer SL, Reddick WE, Gajjar A. Understanding the cognitive impact on children who are treated for medulloblastoma. J Pediatr Psychol. 2007;32(9):1040–1049. doi: 10.1093/jpepsy/jsl056. [DOI] [PubMed] [Google Scholar]
- 5.Thompson MC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24(12):1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 6.Kool M, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3(8):e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho YJ, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29(11):1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Northcott PA, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Northcott PA, et al. Pediatric and adult sonic hedgehog medulloblastomas are clinically and molecularly distinct. Acta Neuropathol. 2011;122(2):231–240. doi: 10.1007/s00401-011-0846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remke M, et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J Clin Oncol. 2011;29(29):3852–3861. doi: 10.1200/JCO.2011.36.2798. [DOI] [PubMed] [Google Scholar]
- 11.Rudin CM, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361(12):1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yauch RL, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326(5952):572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupuy AJ, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69(20):8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrich LV, Milenković L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277(5329):1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482(7386):529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schüller U, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14(2):123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang ZJ, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14(2):135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grammel D, et al. Sonic hedgehog-associated medulloblastoma arising from the cochlear nuclei of the brainstem. Acta Neuropathol. 2012;123(4):601–614. doi: 10.1007/s00401-012-0961-0. [DOI] [PubMed] [Google Scholar]
- 19.Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- 20.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488(7409):43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellino RC, et al. Heterozygosity for Pten promotes tumorigenesis in a mouse model of medulloblastoma. PLoS One. 2010;5(5):e10849. doi: 10.1371/journal.pone.0010849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei Y, et al. An animal model of MYC-driven medulloblastoma. Cancer Cell. 2012;21(2):155–167. doi: 10.1016/j.ccr.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rand WM. Objective criteria for the evaluation of clustering methods. J Am Stat Assoc. 1971;66(336):846–850. [Google Scholar]
- 24.Gronostajski RM, Guaneri J, Lee DH, Gallo SM. The NFI-Regulome Database: A tool for annotation and analysis of control regions of genes regulated by nuclear factor I transcription factors. J Clin Bioinforma. 2011;1(1):4. doi: 10.1186/2043-9113-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W, et al. Nuclear factor I coordinates multiple phases of cerebellar granule cell development via regulation of cell adhesion molecules. J Neurosci. 2007;27(23):6115–6127. doi: 10.1523/JNEUROSCI.0180-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27(3):264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323(5922):1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starr TK, et al. A Sleeping Beauty transposon-mediated screen identifies murine susceptibility genes for adenomatous polyposis coli (Apc)-dependent intestinal tumorigenesis. Proc Natl Acad Sci USA. 2011;108(14):5765–5770. doi: 10.1073/pnas.1018012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann KM, et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 2012;109(16):5934–5941. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Donnell KA, et al. A Sleeping Beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer. Proc Natl Acad Sci USA. 2012;109(21):E1377–E1386. doi: 10.1073/pnas.1115433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akimaru H, et al. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 1997;386(6626):735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 32.Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog-patched-gli pathway in human development and disease. Am J Hum Genet. 2000;67(5):1047–1054. doi: 10.1016/s0002-9297(07)62934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrij F, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376(6538):348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 34.Skousen GJ, Wardinsky T, Chenaille P. Medulloblastoma in patient with Rubinstein-Taybi syndrome. Am J Med Genet. 1996;66(3):367. doi: 10.1002/(SICI)1096-8628(19961218)66:3<367::AID-AJMG27>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 35.Taylor MD, et al. Medulloblastoma in a child with Rubenstein-Taybi Syndrome: Case report and review of the literature. Pediatr Neurosurg. 2001;35(5):235–238. doi: 10.1159/000050428. [DOI] [PubMed] [Google Scholar]
- 36.Northcott PA, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488(7409):49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayrault O, et al. Atoh1 inhibits neuronal differentiation and collaborates with Gli1 to generate medulloblastoma-initiating cells. Cancer Res. 2010;70(13):5618–5627. doi: 10.1158/0008-5472.CAN-09-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436(7048):272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 39.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436(7048):221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 40.Berquam-Vrieze KE, et al. Cell of origin strongly influences genetic selection in a mouse model of T-ALL. Blood. 2011;118(17):4646–4656. doi: 10.1182/blood-2011-03-343947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hallahan AR, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64(21):7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 42.Markant SL, Wechsler-Reya RJ. Review: Personalized mice: Modelling the molecular heterogeneity of medulloblastoma. Neuropathol Appl Neurobiol. 2012;38(3):228–240. doi: 10.1111/j.1365-2990.2011.01235.x. [DOI] [PubMed] [Google Scholar]
- 43.Romer JT, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/−)p53(−/−) mice. Cancer Cell. 2004;6(3):229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 44.Lee MJ, et al. Hedgehog pathway inhibitor saridegib (IPI-926) increases lifespan in a mouse medulloblastoma model. Proc Natl Acad Sci USA. 2012;109(20):7859–7864. doi: 10.1073/pnas.1114718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leprivier G, et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153(5):1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee MH, et al. Tumor suppressor p16(INK4a) inhibits cancer cell growth by downregulating eEF1A2 through a direct interaction. J Cell Sci. 2013;126(Pt 8):1744–1752. doi: 10.1242/jcs.113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schüller U, et al. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol Cell Biol. 2007;27(23):8259–8270. doi: 10.1128/MCB.00707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellis T, et al. Patched 1 conditional null allele in mice. Genesis. 2003;36(3):158–161. doi: 10.1002/gene.10208. [DOI] [PubMed] [Google Scholar]
- 49.das Neves L, et al. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci USA. 1999;96(21):11946–11951. doi: 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Ridder J, Uren A, Kool J, Reinders M, Wessels L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol. 2006;2(12):e166. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin CJ, Davis MJ, Ragan MA. Towards the mammalian interactome: Inference of a core mammalian interaction set in mouse. Proteomics. 2009;9(23):5256–5266. doi: 10.1002/pmic.200900262. [DOI] [PubMed] [Google Scholar]
- 52.Orchard S, et al. Protein interaction data curation: The International Molecular Exchange (IMEx) Consortium. Nat Methods. 2012;9(4):345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.