Significance
Pheromone recognition by insect olfactory organs is critical for the ability of insects to locate mates. The silkworm moth Bombyx mori has long served as a model organism for studies of this process. Key components in the sensory organs have been identified, including the pheromone bombykol, pheromone-binding protein (BmorPBP), ligand-degrading enzymes, and the pheromone receptor, but many details of the mechanism allowing highly sensitive and selective pheromone detection are still elusive. Here, it is shown that a pH-dependent conformational polymorphism of BmorPBP affords highly selective transport of the pheromone, demonstrating an active role for BmorPBP in ligand discrimination.
Keywords: amide proton exchange, conformational equilibrium, insect odorant-binding protein, NMR structure, selective transport of pheromones
Abstract
The Bombyx mori pheromone-binding protein (BmorPBP) is known to adopt two different conformations. These are BmorPBPA, where a regular helix formed by the C-terminal dodecapeptide segment, α7, occupies the ligand-binding cavity, and BmorPBPB, where the binding site is free to accept ligands. NMR spectra of delipidated BmorPBP solutions at the physiological pH of the bulk sensillum lymph near pH 6.5 show only BmorPBPA, and in mixtures, the two species are in slow exchange on the chemical shift frequency scale. This equilibrium has been monitored at variable pH and ligand concentrations, demonstrating that it is an intrinsic property of BmorPBP that is strongly affected by pH variation and ligand binding. This polymorphism tunes BmorPBP for optimal selective pheromone transport: Competition between α7 and lipophilic ligands for its binding cavity enables selective uptake of bombykol at the pore endings in the sensillum wall, whereas compounds with lower binding affinity can only be bound in the bulk sensillum lymph. After transport across the bulk sensillum lymph into the lower pH area near the dendritic membrane surface, bombykol is ejected near the receptor, whereas compounds with lower binding affinity are ejected before reaching the olfactory receptor, rendering them susceptible to degradation by enzymes present in the sensillum lymph.
Insects can discriminate a myriad of physiologically irrelevant chemical compounds in the environment from essential chemical signals, such as sex pheromones (1), and minimal modifications in pheromone molecules can render them inactive (2). This outstanding selectivity is coupled with high sensitivity provided by thousands of pheromone detectors (i.e., olfactory receptor neurons) housed in the insect antennae, where it has been estimated that a single pheromone molecule may be sufficient to activate an olfactory neuron (3). This paper describes stringent ligand selection by a structural polymorphism of the Bombyx mori pheromone-binding protein (BmorPBP), which appears to represent a critical step in ligand recognition amplifying specificity.
The antennae of the male silkworm moth B. mori are covered with 17,000 trichoid sensilla that are specifically tuned to bombykol (4), which is a sex pheromone emitted by the females of the species. Each sensillum contains one or several dendritic nerve endings from olfactory receptor neurons, and it is filled with a concentrated aqueous solution (≈3 mM) of BmorPBP (5). The pheromone bombykol is highly hydrophobic and gains access to the sensillum through pores penetrating the sensillum wall, from where it is transported across the aqueous solution to the bombykol receptor at the olfactory neuron dendritic membrane by BmorPBP (6) (Discussion).
Two different conformations of BmorPBP, BmorPBPA and BmorPBPB, give rise to distinct NMR spectra (7, 8). Due to slow conformational exchange on the NMR chemical shift time scale in mixtures of the two forms, integration of the NMR signals allows determination of the relative populations of the two forms over a wide range of solution conditions (8). The structure of BmorPBPB was determined in crystals of the bombykol complex obtained at pH 8.2 (9), and an NMR structure of BmorPBPA was obtained in solution at pH 4.5 (10).
The NMR solution structure of BmorPBPA determined at pH 4.5 consists of a cluster of six α-helices surrounding a seventh α-helix formed by the C-terminal dodecapeptide segment (10). In the crystal structure of the BmorPBPB complex with bombykol, the C-terminal dodecapeptide segment is not visible (9). An NMR structure determination at pH 6.5 in a solution that was devoid of bombykol but had not been delipidated after expression of the recombinant BmorPBP in Escherichia coli yielded a similar BmorPBP conformation and showed that the C-terminal dodecapeptide segment is flexibly disordered (11). After delipidating the protein to remove heterologous ligands that might copurify with the protein (12), Lautenschläger et al. (13) found that BmorPBP in crystals obtained at pH 7.5 in the absence of ligands adopts the A-form, with the C-terminal dodecapeptide forming an α-helix in the ligand-binding site of the protein. This structure is nearly identical to the NMR structure of BmorPBPA determined at pH 4.5, with an rmsd value of 0.90 Å calculated for the backbone heavy atoms of residues 10–142, and it was solved by molecular replacement with this NMR structure (13).
The observation of BmorPBPA in crystals grown at pH 7.5 indicated that studies of insect pheromone-binding proteins in solution should also be extended to delipidated protein preparations. An NMR structure of the Antheraea polyphemus pheromone-binding protein 1 (PBP1) determined in a delipidated solution at pH 4.5 showed that this protein also adopts an A-form, with the α-helical, C-terminal dodecapeptide segment in the ligand-binding site (14). Furthermore, the equilibrium between the A- and B-forms of the A. polyphemus PBP1 was shown to be largely affected by addition of ligands (15). In this paper, we follow up on these earlier observations, using NMR experiments with delipidated BmorPBP over a wide pH range in the absence and presence of bombykol and related compounds. In combination with the results of previous investigations of a C-terminally truncated variant of BmorPBP (16), and of amide proton exchange measurements (discussed below), these systematic studies of the dependence of conformation on pH and ligands resulted in unique insights into the role of BmorPBP in achieving outstanding sensitivity and specificity of pheromone signaling in B. mori.
Results
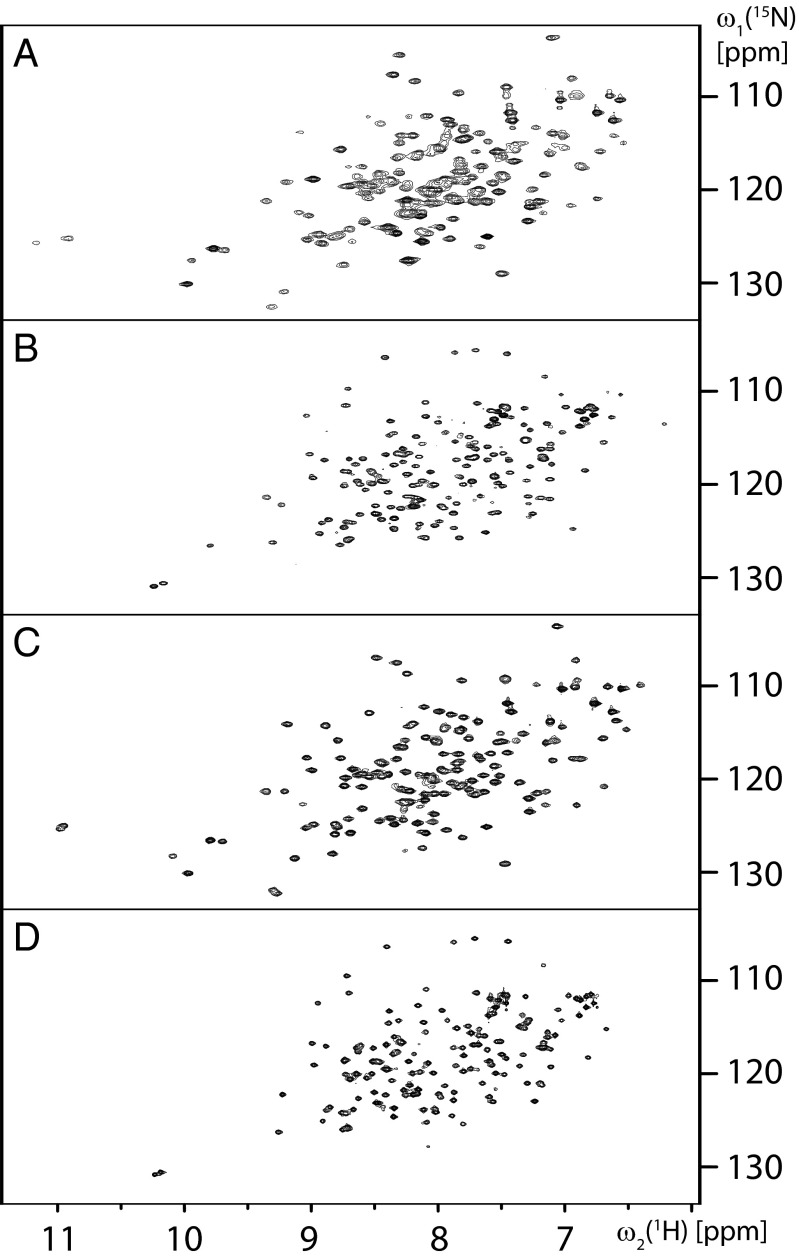
Purified 15N-labeled BmorPBP (8) was delipidated at pH 4.5 (Materials and Methods). To survey the conformational states of the protein in delipidated solutions, 2D [15N,1H]-heteronuclear multiple quantum coherence (HMQC) correlation NMR spectra were recorded in 50 mM potassium phosphate buffer at variable pH values in the range of 8.0–4.0, without and with addition of bombykol. Throughout, we observed only the previously described spectra of BmorPBPA (8, 10) and BmorPBPB (8, 11). At pH 8.0, the protein displayed a complete set of BmorPBPB signals (Fig. 1A), whereby some cross-peaks were slightly shifted relative to the BmorPBPB spectrum obtained previously with nondelipidated samples at pH 6.5 (11), probably due to deprotonation of nearby groups at the higher pH value. At pH 6.5, only the spectrum of BmorPBPA was observed (Fig. 1B). After addition of an equimolar amount of bombykol to the solution at pH 6.5, only the spectrum of BmorPBPB was seen (Fig. 1C). Upon subsequent change of the pH to 4.5 in the solution containing BmorPBP and bombykol, only BmorPBPA was observed (Fig. 1D). The observations at pH 8.0 and 6.5 in delipidated BmorPBP solutions show that the conformational polymorphism with the A- and B-forms is an intrinsic property of BmorPBP, because it is also observed in the absence of ligands. In addition, the data in Fig. 1 B–D document that the dynamic equilibrium between the conformations A and B is strongly affected by ligand binding as well as by variation of the pH value of the BmorPBP solution, which coincides with the behavior of the A. polyphemus PBP1 (14, 15). The data of Fig. 1 further indicate that all published BmorPBPB structures are of BmorPBP bound either to bombykol (9) or to exogenous ligands that had copurified with the protein (11). In the following, we further investigate the structural and functional implications of these observations.
Fig. 1.
Survey of conformational states adopted by the BmorPBP in delipidated aqueous solution. Shown are 2D [15N,1H]-HMQC correlation NMR spectra at temperature (T) = 20 °C of 0.3 mM solutions of uniformly 15N-labeled BmorPBP containing 50 mM potassium phosphate and 1 mM NaN3 in four different conditions. (A) pH = 8.0. (B) pH = 6.5. (C) Same as B after addition of a saturating amount of bombykol, showing the spectrum of the 1:1 complex with this ligand. (D) Same as C after adjusting the pH to 4.5.
Competitive Ligand Binding to BmorPBP.
To investigate the effects of variable pH on the unliganded protein and on BmorPBP–ligand complexes, the NMR signal intensities of BmorPBPA and BmorPBPB were evaluated in 2D [15N,1H]-HMQC spectra measured at different pH values (Materials and Methods). Aqueous solutions of delipidated [u-15N]-BmorPBP showed a B-to-A transition with a midpoint at pH 7.3 (Fig. 2). Addition of each compound in Fig. 2B converted BmorPBPA to BmorPBPB at pH 6.5, but their complexes displayed quite different behavior at variable pH values. In the presence of an equimolar amount of bombykol, the transition midpoint was at pH 5.4, and with hexadecan-1-ol or hexadecan-1-al, it was at pH 5.8 (Fig. 2). These data, and the observation that addition of hexane to delipidated BmorPBP resulted in the formation of BmorPBPB (Materials and Methods), provided additional indications that recombinant BmorPBP expressed in E. coli copurifies with extrinsic ligands, which stabilize the B-form of the protein at pH 6.5 (8, 11).
Fig. 2.
(A) pH-dependent conformational transitions in aqueous solutions of delipidated BmorPBP in the absence and presence of equimolar amounts of bombykol (prepared as in Fig. 1C) or related ligands. The protein concentration was 0.2 mM, T = 20 °C. Plotted vs. pH is the fraction of BmorPBP in the A-form, fA, as derived from the average of the integrals of 20 well-resolved 15N-1H NMR cross-peaks of the conformations BmorPBPA and BmorPBPB in 2D [15N,1H]-HMQC spectra. Experimental data: squares, unliganded BmorPBP; triangles, BmorPBP–hexadecan-1-ol complex (the same result was obtained for the BmorPBP complex with hexadecan-1-al); circles, BmorPBP–bombykol complex. Each dataset was fitted by a sigmoidal curve describing a two-state equilibrium (Eq. 6), and the resulting transition midpoints, pH1/2, are indicated. (B) Chemical formulae of the ligands used.
Amide Proton Exchange Reveals the Presence of BmorPBPB at pH 4.5.
At pH 4.5, only BmorPBPA was seen in the 15N-1H correlation spectra, also after addition of an equimolar amount of bombykol (Fig. 1D). This indicates that the population of the B-form of the protein must be smaller than about 2%. Measurements of amide proton exchange rates in BmorPBP solutions at pH 4.5 in the absence and presence of an equimolar amount of bombykol now provided direct evidence for a small population of BmorPBPB in dynamic equilibrium with the A-form. Exchange rates can largely vary between protons with or without solvent contact, where the observed exchange rates, kobs, are given by Eq. 1 (17–19):
where k1 is the rate of conversion from a solvent-shielded to a solvent-exposed state, k2 is the rate for the reverse process, and kint is the intrinsic exchange rate for solvent-exposed amide protons (18, 19). The ratio of kint to kobs, as expressed by the “protection factor” P, is typically related to the solvent-inaccessible surface area (SISA), with amide protons with large SISA values having high P values. Specifically, amide protons in α-helices are protected by the regular hydrogen-bonding network (18).
In BmorPBP, the helices α3a, α5, and α6 form a cavity that contains either the helix α7 in BmorPBPA (10, 13) or bombykol in the B-form of BmorPBP (9). In nondelipidated BmorPBPB solutions, the amide proton exchange rates in the C-terminal dodecapeptide segment were very close to the intrinsic exchange rates (20), which is consistent with the observation that this polypeptide segment is flexibly disordered on the protein surface (11). In BmorPBPA, the helices 3a, 5, 6, and 7 all have nearly identical SISA values, but exchange from α7 was two orders of magnitude faster than for α3a, α5, and α6 (Fig. 3). Furthermore, the protection factors are quite uniform for all hydrogen-bonded amide protons in α7 (i.e., those of residues 134–141). A single conformational exchange process between the solvent-protected state of α7 and a solvent-exposed state of the corresponding polypeptide segment thus governs the exchange rates in α7, indicating that the increased exchange rate is due to a small admixture of BmorPBPB. The average log P value for residues 134–141 of α7 in unliganded BmorPBP was 3.3 ± 0.2 (Fig. 3A), which corresponds to an equilibrium constant of 2.0 ± 0.1⋅103. At pH 4.5, about 0.05% of the BmorPBP is thus in a state where the polypeptide segment corresponding to helix α7 is solvent-exposed. When combined with the data in Fig. 2, this shows that dynamic equilibrium between the A- and B-forms of the protein is maintained over the entire physiologically relevant pH range.
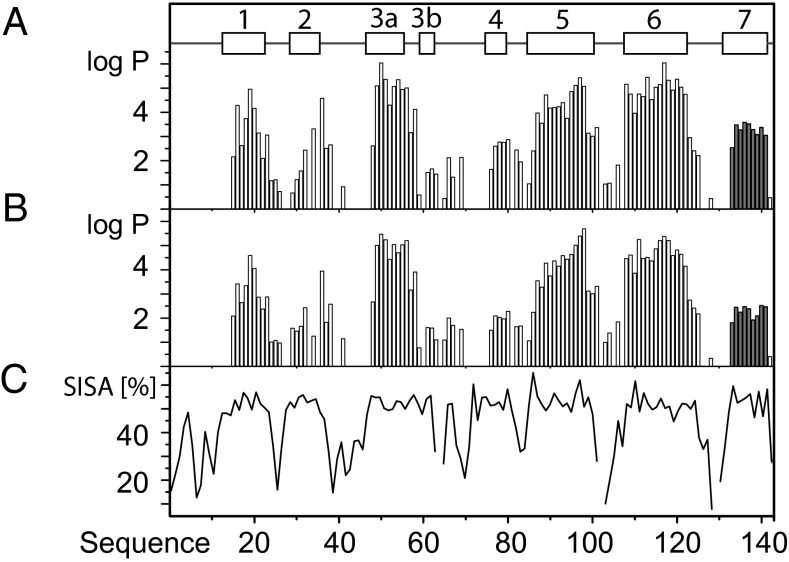
Fig. 3.
Amide proton exchange measurements revealed the presence of BmorPBPB at concentrations that are too small for direct NMR observation. Data are shown for a delipidated 0.3 mM aqueous solution of BmorPBP in the absence and presence of an equimolar amount of bombykol, pH = 4.5, T = 20 °C. Protection factors (discussed in main text) for individual amide protons of BmorPBP are represented by vertical bars at the respective sequence positions; the data for the helix α7 are highlighted by dark shading. (A) No ligand. (B) With equimolar concentration of bombykol. (C) Plot vs. the sequence of the SISA per residue in BmorPBPA at pH 4.5 (10). The locations of α-helices in BmorPBPA are indicated at the top.
At pH 4.5 in the presence of a stoichiometric amount of bombykol (Materials and Methods), the log P values for the helix α7 were further reduced, with an average value for residues 134–141 of 2.3 ± 0.2 (Fig. 3B). In contrast, the exchange in the helices α3, α5 and α6 is much less affected by the presence of bombykol (Fig. 3 A and B), showing that the ligand-binding cavity is preserved as in BmorPBPB (9, 11). The P value for the helix α7 in the presence of bombykol at pH 4.5 corresponds to an equilibrium constant of 2.0 ± 0.2⋅102, showing that the C-terminal dodecapeptide spends ∼0.5% of the time in a solvent-exposed state.
Reduced Stability of the BmorPBPB–Bombykol Complex at Acidic pH Values.
The equilibrium constant KAB governing the conversion of the B-form to the A-form of BmorPBP can be determined independently either from amide proton exchange experiments (Fig. 3) or from pH titration experiments that monitor the conformational transition (Fig. 2). Assuming that the increased solvent accessibility manifested by the uniformly accelerated HN exchange rates in the C-terminal segment 134–141 was due to transient formation of the conformation BmorPBPB, where the C-terminal polypeptide is flexibly disordered (10, 11, 20), the average protection factor of the polypeptide segment can be related to a good approximation to the equilibrium constant with Eq. 2:
From the populations of BmorPBPA and BmorPBPB, fA and fB, determined by integrating the signals of the two species in 2D [15N,1H]-HMQC spectra recorded at different pH values, the equilibrium constant can be determined with Eq. 3:
The variation of the Gibbs free energy difference between the conformations BmorPBPA and BmorPBPB with pH is given by Eq. 4:
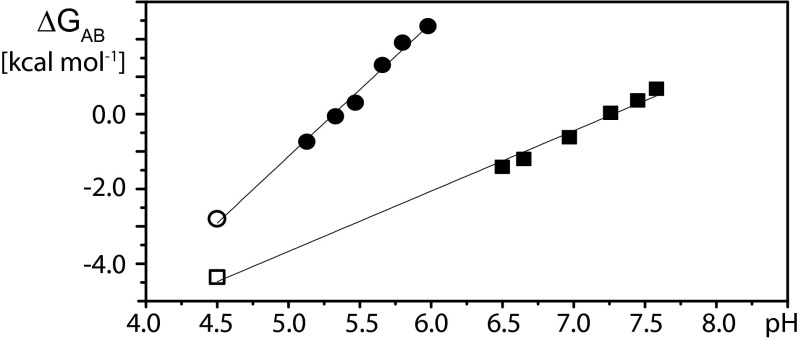
where R is the gas constant and T is the absolute temperature. Combining the data from Figs. 2 and 3 then affords a survey of KAB over the entire physiologically relevant pH range from 4.5 to 7.0 (Fig. 4). An approximately linear relationship is predicted between ΔGAB and pH for a two-state conformational transition driven by the protonation of multiple independently titrating groups with identical pKas in the low-pH regime (21–23):
where ΔG0 is the limiting free energy difference at pH = 0 and n is the number of titratable groups that drive the transition. For BmorPBP, this linear relation is maintained over the entire physiological pH range from 4.5 to 7.0 (Fig. 4). In the presence of ligand, we observe a shift of the transition midpoint, where ΔGAB = 0, to lower pH values, as well as an increase of the slope of the curve near the midpoint, which is indicative of the number of titratable groups that contribute to the conformational transition (21). Table 1 summarizes the results of least squares fits of ΔGAB vs. pH for unliganded BmorPBP and several BmorPBP–ligand complexes. The differences between the ΔGAB values for unliganded BmorPBP and the BmorPBP–bombykol complex at variable pH values can be interpreted as the Gibbs free energy for the binding of BmorPBPB to bombykol. The different slopes of the curves for unliganded BmorPBP and the BmorPBP–bombykol complex then indicate that the binding affinity of bombykol for BmorPBPB, ΔGBB′, as represented by the distance between the two curves, is reduced at lower pH values.
Fig. 4.
Dependence on pH of the Gibbs free energy difference between BmorPBPA and BmorPBPB, ΔGAB. Data derived from experimental measurements with delipidated aqueous solutions of BmorPBP measured at 20 °C are represented by squares for BmorPBP in the absence of bombykol and by circles for BmorPBP in the presence of equimolar bombykol. The values indicated by filled symbols were calculated using equilibrium constants determined with Eq. 3 from the pH titration data in Fig. 2. The values represented by open symbols were obtained with Eq. 2 from the amide proton exchange data in Fig. 3.
Table 1.
Free energy difference between BmorPBPA and BmorPBPB as a function of pH for different BmorPBP–ligand complexes
| Ligand | ΔGAB(pH = 0)* | d(ΔGAB)/d(pH)† | n‡ |
| None§ | −11.1 ± 0.2 | 1.5 ± 0.1 | 1.1 ± 0.1 |
| Hexadecan-1-al | −30.2 ± 2.8 | 5.3 ± 0.5 | 4.0 ± 0.4 |
| Hexadecan-1-ol | −28.7 ± 1.1 | 5.0 ± 0.2 | 3.7 ± 0.2 |
| Bombykol | −18.9 ± 0.7 | 3.6 ± 0.1 | 2.7 ± 0.1 |
ΔGAB(pH = 0) is the free energy difference between BmorPBPA and BmorPBPB extrapolated to pH = 0 in kilocalories per mole.
d(ΔGAB)/d(pH) is the slope obtained from least squares fits of ΔG vs. pH in kilocalories per mole. Least squares r2 values ranged from 0.96 to 0.99.
n is the number of titrating groups participating in the conformational transition.
Delipidated BmorPBP solution.
Discussion and Conclusions
The unparalleled selectivity and sensitivity of bombykol signaling in B. mori, which has long been a paradigm for olfaction in insects (2), is surprising in view of the permissive ways in which BmorPBP binds different lipophilic ligands (24, 25) (Fig. 2; hexane as described in Materials and Methods). Although this apparent promiscuity might appear to indicate that BmorPBP is merely a passive transporter of hydrophobic compounds, with selectivity being entirely dependent on molecular recognition at the receptor (26), the present in-depth characterization of the pH-dependent conformational equilibrium strongly suggests that binding and release of bombykol by BmorPBP is a key component in the mechanism that leads to the high selectivity of the silkworm moth olfactory system. The notion that BmorPBP functions as a nondiscriminating transporter would also hardly be compatible with the observation that this protein is present only in trichoid sensilla (27), which contain dendrites harboring the bombykol odorant receptor 1 (BmorOR1) along with the coreceptor BmorOR2 (now also named BmorOrco) (28–30).
The data in Figs. 1–4 indicate that bombykol is delivered as a free ligand to the BmorOR1/BmorOR2 complex. This would be in line with the observation that sensilla heterologously expressing BmorOR1 and not containing BmorPBP can be activated by bombykol (26, 28, 30). In contrast, our data would seem to be incompatible with the hypothesis that the BmorPBP–bombykol complex is needed to activate the receptor (31, 32). In the following, we propose a mechanism that is compatible with the newly acquired experimental evidence (Figs. 1–4) and can provide for high selectivity of bombykol detection in the B. mori trichoid sensilla.
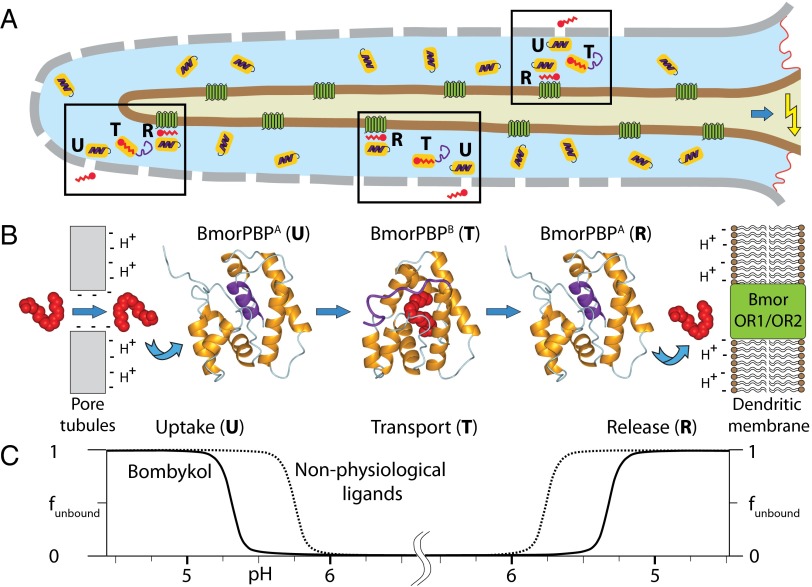
The bulk sensillum lymph has a pH of 6.5 (33), so that BmorPBP is almost exclusively present in the conformation BmorPBPA (Fig. 1B), with the helix α7 in the core of the protein (10, 13). The pore wall has a net negative charge (34) that causes locally a lowering of the pH (35, 36). When bombykol enters through the pore, its affinity for binding to BmorPBP is, in contrast to other compounds in the sensillum lymph, sufficiently high to displace the helix α7 from the hydrophobic cavity and form a complex with BmorPBPB (9, 11). The pheromone-loaded BmorPBPB diffuses to the dendrite of the olfactory neuron that harbors the receptor BmorOR1/BmorOR2 complex (28), where the local pH is reduced by up to 2 pH units due to the negatively charged lipid headgroups of the membrane (35). At this lower pH, the equilibrium between the BmorPBPB–bombykol complex and BmorPBPA is sufficiently shifted toward the conformation BmorPBPA to enable about 99.6% of the bound bombykol to be released to the membrane-standing receptor. It is possible that the residence time near the membrane may be extended by transient interactions of the flexibly unstructured C-terminal dodecapeptide segment of BmorPBPB before the transformation to BmorPBPA. The ligand release may be further assisted by the reduced intrinsic stability of the BmorPBPB–bombykol complex at lower pH values (Fig. 4), but studies of the C-terminally truncated variant protein BmorPBP(1–128), where the helix α7 cannot be formed, show that this is not sufficient to ensure ligand ejection, because the B-form of the complex of BmorPBP(1–128) with bombykol is maintained at pH 4.5 (16). Importantly, our data also indicate that other ligands with lower binding affinity for BmorPBP will be bound further away from the pore wall and released further away from the dendritic membrane surface, where higher pH values prevail and the ligands are susceptible to degradation by aggressive enzymes present in the sensillar lymph (37). Conversely, ligands with higher binding affinity than bombykol would not be released to the receptor. The role of pheromone-binding proteins in optimizing selectivity of the moth sensory system as outlined in Fig. 5 appears to be a general phenomenon. This is supported by the observation that pheromone-binding proteins from Antheraea polyphemus, ApolPBP1 (14), and Amyelois transitella, AtraPBP1 (38, 39), also include a C-terminal dodecapeptide segment that forms a helix in the core of the protein at pH 4.5; by evidence that ApolPBP1 undergoes a conformational transition induced by binding of the physiological ligand (15); and by the high sequence conservation among PBPs of a wide range of moth species (9, 14).
Fig. 5.
Proposed mechanism for selective pheromone transport in B. mori olfaction based on the pH- and ligand-dependent structural polymorphism between the A- and B-forms of BmorPBP. (A) B. mori sensilla with schematic indication in the three boxes of pheromone uptake by BmorPBPA (U), pheromone transport in the complex with BmorPBPB (T), and pheromone release to the OR1/OR2 receptor complex with concomitant conformational transition from BmorPBPB to BmorPBPA (R). The following color code is used: gray, sensillum wall with pores for pheromone entry; light blue, sensillum lymph; brown, neuron dendritic membrane; dark green, odorant receptor complex (BmorOR1/OR2); orange, BmorPBP; purple, polypeptide segment 129–142 of BmorPBP; red, bombykol. On the right, a blue arrow pointing at a yellow arrow indicates signal transfer to the olfactory receptor neuron. (B) Structural basis of the data in A. The locally reduced pH near the pore supports preferential uptake of bombykol over the nonphysiological ligands used in our experiments (Fig. 2B), because it binds with higher affinity to BmorPBP at low pH (Fig. 2A). During the transport phase, ligands in complex with BmorPBPB are protected from degradation by aggressive enzymes present in the sensillum lymph (37). When BmorPBPB–ligand complexes diffuse toward the membrane-standing receptor, their stability is reduced by the acidic milieu near the membrane (7, 35). For bombykol, this results in release to the receptor, whereas less tightly binding ligands are released sooner than bombykol, becoming subject to degradation before reaching the membrane-standing receptor, and more tightly bound ligands would not be released. The images of the BmorPBP structures and bombykol were generated with MOLMOL (43), using the coordinates of BmorPBPA and the BmorPBPB–bombykol complex from the Protein Data Bank depositions 1GM0 and 1DQE. (C) Fraction of ligand not bound to BmorPBP, funbound, indicated along the vertical axis, and pH profile, indicated along the horizontal axis, across the B. mori sensillum cross-section shown in B. Bombykol not bound to BmorPBP may be transiently associated with the pore wall before being taken up by BmorPBP; after release, it would initially be associated with the surface of the neuron dendritic membrane, where it would diffuse to the receptor-binding site.
Finally, the present studies of BmorPBP also provide intriguing observations from the viewpoint of structural biology. Most importantly, the large scale of the conformational change between the A- and B-forms of the protein is quite unique for an otherwise well-structured small globular protein, indicating that there is an unusual structural basis that supports the special physiological role of BmorPBP in ligand discrimination. Furthermore, although only BmorPBPA is observed in crystals grown at pH 7.5 (13), about 80% of the protein in delipidated solution is in the BmorPBPB form at this pH value (Fig. 2). There is thus an indication that the A-form observed in crystals grown at pH 7.5 might have been stabilized relative to the B-form by the crystal packing and/or the reagents used for cryogenic protection.
Materials and Methods
Preparation of NMR Samples.
Uniformly 15N-labeled BmorPBP solutions in 50 mM potassium phosphate buffer at pH 6.5 containing 1 mM NaN3 were prepared as described previously (8). Delipidation was performed by adjusting the pH of the protein solution to 4.5, adding an equal volume of the Lipidex resin from Sigma–Aldrich preequilibrated with 50 mM potassium phosphate buffer at pH 4.5 and 1 mM NaN3, and incubating at 40 °C for 1 h. The delipidated protein was recovered to the extent of 75–90% by centrifuging in a spin column at a low acceleration of gravity and collecting the flow-through. Delipidation was monitored by adjusting the pH to 6.5 and measuring a 2D [15N,1H]-HMQC spectrum, where absence of signals from BmorPBPB indicated complete removal of ligands (Fig. 1B).
Hexadecan-1-ol, hexadecan-1-al, and (10E,12Z)-hexadecadien-1-ol (bombykol) were purchased from Sigma–Aldrich; Bedoukian Research, Inc.; and PHEROBANK (Plant Research International), respectively. Bombykol and hexadecan-1-ol were dissolved in 99.5% atom [2H4]-methanol (Armar Chemicals) at concentrations of 5 and 10 mg/mL, respectively, and hexadecan-1-al was dissolved in >99.9% puriss dichloromethane (Fluka) at 20 mg/mL. Ligands were added to the NMR sample directly on the inside wall of the NMR tube, followed by closing the tube, repeated inversions, and incubation at room temperature for 30 min. To achieve an equimolar ratio of ligand to protein, the ligand was added stepwise and the degree of protein loading was monitored by measuring 2D [15N,1H]-HMQC spectra. Once the signals from BmorPBPA were no longer observed, it was judged that 100% of the protein was bound with ligand, which was typically achieved by addition of ligand in 50% excess of the stoichiometric amount. Initial attempts to dissolve ligands in analytical grade hexane (a common solvent for ligand-binding studies) were abandoned after it was observed that hexane induced the conformational transition to BmorPBPB.
pH Titration Experiments.
pH titrations with 0.15–0.3 mM [u-15N]-BmorPBP solutions were performed in the NMR tube by adding small amounts of 0.1 M HCl to the wall of the NMR tube, followed by repeated inversion of the tube. The resulting pH values were measured in the NMR tube, using a Phillips glass NMR electrode. The relative concentrations of BmorPBPA and BmorPBPB were monitored in 2D [15N,1H]-HMQC spectra measured with water flip-back pulses by integrating the volumes of about 20 well-resolved backbone 15N-HN signals in both conformations and calculating the average peak volumes, VA and VB, and the populations corresponding to the normalized peak volumes, fA = VA/(VA + VB) and fB = VB/(VA + VB). The dependence of fA on pH was fitted with a two-state model, as provided in the software package Origin (OriginLab Corporation):
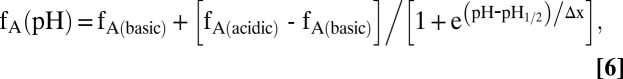 |
where pH1/2 is the pH at the midpoint of the transition; fA(acidic) and fA(basic) are the limiting values of fA at acidic and basic pH, respectively; and Δx represents the width of the transition.
Amide Proton Exchange Measurements.
For the amide proton exchange measurements, delipidated 0.3–0.5 mM stock solutions of uniformly 15N-labeled BmorPBP in 95% H2O/5% D2O (vol/vol) containing 1 mM NaN3 and 50 mM potassium phosphate buffer were adjusted to pH 4.5 and lyophilized. HN-exchange was initiated by redissolving the lyophilized protein in D2O. For experiments in the presence of bombykol, an equimolar amount of ligand dissolved in [2H4]-MeOH was added to the wall of the NMR tube, and the tube was closed and inverted repeatedly before the NMR measurements. The validity of this method to obtain a 1:1 ratio of BmorPBP to bombykol in the D2O solutions was verified by adding the same amount of bombykol to an aliquot of the same stock solution of delipidated BmorPBP in H2O at pH 6.5, which showed complete conversion of BmorPBPA into BmorPBPB. 2D [15N,1H]-HMQC spectra were recorded at 20 °C with flip-back pulses and the fast Watergate sequence for solvent suppression (40), using a Bruker DRX 500 spectrometer equipped with a triple-resonance cryogenic probehead. The NMR spectra were processed with the program TOPSPIN (Bruker Biospin), and the programs XEASY (41) and Computer Aided Resonance Assignment (CARA; www.nmr.ch) (42) were used for the spectral analysis. Protection factors for individual amino acid residues have been calculated as P = kint/kobs, where kint and kobs are the calculated intrinsic exchange rates (19) and the observed rates of exchange, respectively. Values of kobs were determined by fitting the 15N-HN cross-peak intensities measured at variable exchange times to a single exponential function. For very slowly exchanging amide protons, an upper bound on the exchange rate was estimated from a linear-fit extrapolation to the time point where the intensity was reduced by 1/e.
Note Added in Proof.
Refs. 44 and 45, which propose a model involving recognition of the PBP-pheromone complex by the receptor, should have been cited immediately following references 31 and 32.
Acknowledgments
We thank Dr. R. Horst and Dr. D. H. Lee for exploratory NMR measurements with nondelipidated BmorPBP solutions. Financial support was provided by the Schweizerischer Nationalfonds and the Eidgenössische Technische Hochschule Zürich through the National Centre of Competence in Research “Structural Biology” and by the Agriculture and Food Research Initiative Competitive Grant 2010-65105-20582 from the US Department of Agriculture-National Institute of Food and Agriculture.
Footnotes
The authors declare no conflict of interest.
References
- 1.Leal WS. Proteins that make sense. In: Blomquist GJ, Vogt RG, editors. Insect Pheromone Biochemistry and Molecular Biology: The Biosynthesis and Detection of Pheromones and Plant Volatiles. London: Elsevier; 2003. pp. 447–476. [Google Scholar]
- 2.Kaissling KE. R. H. Wright Lectures on Insect Olfaction. Burnaby, Canada: Simon Fraser Univ Press; 1987. [Google Scholar]
- 3.Kaissling KE, Priesner E. Die Riechschwelle des Seidenspinners. Naturwissenschaften. 1970;57(1):23–28. doi: 10.1007/BF00593550. [DOI] [PubMed] [Google Scholar]
- 4.Steinbrecht RA. Zur Morphometrie der Antenne des Seidenspinners, Bombyx mori L.: Zahl und Verteilung der Riechsensillen. Zeitschrift für Morphologie der Tiere. 1970;68(2):93–126. [Google Scholar]
- 5.Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS. Pheromone reception in fruit flies expressing a moth’s odorant receptor. Proc Natl Acad Sci USA. 2006;103(44):16538–16543. doi: 10.1073/pnas.0607874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leal WS. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 7.Wojtasek H, Leal WS. Conformational change in the pheromone-binding protein from Bombyx mori induced by pH and by interaction with membranes. J Biol Chem. 1999;274(43):30950–30956. doi: 10.1074/jbc.274.43.30950. [DOI] [PubMed] [Google Scholar]
- 8.Damberger F, et al. NMR characterization of a pH-dependent equilibrium between two folded solution conformations of the pheromone-binding protein from Bombyx mori. Protein Sci. 2000;9(5):1038–1041. doi: 10.1110/ps.9.5.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandler BH, Nikonova L, Leal WS, Clardy J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding-protein-bombykol complex. Chem Biol. 2000;7(2):143–151. doi: 10.1016/s1074-5521(00)00078-8. [DOI] [PubMed] [Google Scholar]
- 10.Horst R, et al. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci USA. 2001;98(25):14374–14379. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee D, et al. NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH. FEBS Lett. 2002;531(2):314–318. doi: 10.1016/s0014-5793(02)03548-2. [DOI] [PubMed] [Google Scholar]
- 12.Oldham NJ, et al. Analysis of the silkworm moth pheromone binding protein-pheromone complex by electrospray-ionization mass spectrometry. Angew Chem Int Ed. 2000;39(23):4341–4343. doi: 10.1002/1521-3773(20001201)39:23<4341::AID-ANIE4341>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 13.Lautenschläger C, Leal WS, Clardy J. Coil-to-helix transition and ligand release of Bombyx mori pheromone-binding protein. Biochem Biophys Res Commun. 2005;335(4):1044–1050. doi: 10.1016/j.bbrc.2005.07.176. [DOI] [PubMed] [Google Scholar]
- 14.Damberger FF, Ishida Y, Leal WS, Wüthrich K. Structural basis of ligand binding and release in insect pheromone-binding proteins: NMR structure of Antheraea polyphemus PBP1 at pH 4.5. J Mol Biol. 2007;373(4):811–819. doi: 10.1016/j.jmb.2007.07.078. [DOI] [PubMed] [Google Scholar]
- 15.Katre UV, Mazumder S, Prusti RK, Mohanty S. Ligand binding turns moth pheromone-binding protein into a pH sensor: Effect on the Antheraea polyphemus PBP1 conformation. J Biol Chem. 2009;284(46):32167–32177. doi: 10.1074/jbc.M109.013383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel E, et al. Dynamic conformational equilibria in the physiological function of the Bombyx mori pheromone-binding protein. J Mol Biol. 2011;408(5):922–931. doi: 10.1016/j.jmb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Hvidt A, Nielsen SO. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 18.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 19.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee D (2003) NMR studies on the structure and function of the Bombyx mori pheromone-binding protein. PhD thesis (Eidgenössiche Technische Hochschule Zürich, Zurich, Switzerland)
- 21.Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 22.Barrick D, Baldwin RL. Three-state analysis of sperm whale apomyoglobin folding. Biochemistry. 1993;32(14):3790–3796. doi: 10.1021/bi00065a035. [DOI] [PubMed] [Google Scholar]
- 23.Ionescu RM, Eftink MR. Global analysis of the acid-induced and urea-induced unfolding of staphylococcal nuclease and two of its variants. Biochemistry. 1997;36(5):1129–1140. doi: 10.1021/bi9609681. [DOI] [PubMed] [Google Scholar]
- 24.Lautenschläger C, Leal WS, Clardy J. Bombyx mori pheromone-binding protein binding nonpheromone ligands: Implications for pheromone recognition. Structure. 2007;15(9):1148–1154. doi: 10.1016/j.str.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou JJ, et al. Characterisation of Bombyx mori Odorant-binding proteins reveals that a general odorant-binding protein discriminates between sex pheromone components. J Mol Biol. 2009;389(3):529–545. doi: 10.1016/j.jmb.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Xu P, Hooper AM, Pickett JA, Leal WS. Specificity determinants of the silkworm moth sex pheromone. PLoS ONE. 2012;7(9):e44190. doi: 10.1371/journal.pone.0044190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinbrecht RA, Ozaki M, Ziegelberger G. Immunocytochemical localization of pheromone-binding protein in moth antennae. Cell Tissue Res. 1992;270(2):287–302. [Google Scholar]
- 28.Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307(5715):1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- 29.Sakurai T, et al. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci USA. 2004;101(47):16653–16658. doi: 10.1073/pnas.0407596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato K, et al. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452(7190):1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 31.Pophof B. Moth pheromone binding proteins contribute to the excitation of olfactory receptor cells. Naturwissenschaften. 2002;89(11):515–518. doi: 10.1007/s00114-002-0364-5. [DOI] [PubMed] [Google Scholar]
- 32.Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133(7):1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaissling KE, Thorson J. In: Hormones and Pheromones in Insects. Receptors for Neurotransmitters. Sattelle DB, Hall LM, Hildebrand JG, editors. Amsterdam: Elsevier; 1980. pp. 261–282. [Google Scholar]
- 34.Keil TA. Surface coats of pore tubules and olfactory sensory dendrites of a silkmoth revealed by cationic markers. Tissue Cell. 1984;16(5):705–717. doi: 10.1016/0040-8166(84)90004-1. [DOI] [PubMed] [Google Scholar]
- 35.van der Goot FG, González-Mañas JM, Lakey JH, Pattus F. A ‘molten-globule’ membrane-insertion intermediate of the pore-forming domain of colicin A. Nature. 1991;354(6352):408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 36.Leal WS. Duality monomer-dimer of the pheromone-binding protein from Bombyx mori. Biochem Biophys Res Commun. 2000;268(2):521–529. doi: 10.1006/bbrc.2000.2158. [DOI] [PubMed] [Google Scholar]
- 37.Ishida Y, Leal WS. Rapid inactivation of a moth pheromone. Proc Natl Acad Sci USA. 2005;102(39):14075–14079. doi: 10.1073/pnas.0505340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu X, et al. NMR structure of navel orangeworm moth pheromone-binding protein (AtraPBP1): Implications for pH-sensitive pheromone detection. Biochemistry. 2010;49(7):1469–1476. doi: 10.1021/bi9020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.di Luccio E, Ishida Y, Leal WS, Wilson DK. Crystallographic observation of pH-induced conformational changes in the Amyelois transitella pheromone-binding protein AtraPBP1. PLoS ONE. 2013;8(2):e53840. doi: 10.1371/journal.pone.0053840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B. 1995;108(1):94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 41.Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6(1):1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 42.Keller R. The Computer Aided Resonance Assignment Tutorial. Goldau, Switzerland: Cantina Verlag; 2004. [Google Scholar]
- 43.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(1):51–55, 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 44.Kaissling KE. Olfactory perireceptor and receptor events in moths: a kinetic model revisited. J Comp Physiol A. 2009;195:895–922. doi: 10.1007/s00359-009-0461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaissling KE. Kinetics of olfactory responses might largely depend on the odorant-receptor interaction and the odorant deactivation postulated for flux detectors. J Comp A. 2013 doi: 10.1007/s00359-013-0812-z. 10.1007/s00359-013-0812-z. [DOI] [PubMed] [Google Scholar]