Significance
Transmembrane efflux pumps belonging to the resistance–nodulation–cell division (RND) superfamily are found in all kingdoms of life, and transport substrates out of cells, powered by an electrochemical proton gradient. Here we report two X-ray crystal structures of a Zn(II) efflux pump, ZneA, that capture different intermediate states along the transport cycle. The structures show how passage of substrates through ZneA is regulated by a series of conformational changes in the efflux pump. By comparing the structures of ZneA with other RND efflux pumps, we present a coherent mechanistic model for RND-mediated substrate efflux, which ensures efficient transport of substrates out of the cell.
Abstract
Efflux pumps belonging to the ubiquitous resistance–nodulation–cell division (RND) superfamily transport substrates out of cells by coupling proton conduction across the membrane to a conformationally driven pumping cycle. The heavy metal-resistant bacteria Cupriavidus metallidurans CH34 relies notably on as many as 12 heavy metal efflux pumps of the RND superfamily. Here we show that C. metallidurans CH34 ZneA is a proton driven efflux pump specific for Zn(II), and that transport of substrates through the transmembrane domain may be electrogenic. We report two X-ray crystal structures of ZneA in intermediate transport conformations, at 3.0 and 3.7 Å resolution. The trimeric ZneA structures capture protomer conformations that differ in the spatial arrangement and Zn(II) occupancies at a proximal and a distal substrate binding site. Structural comparison shows that transport of substrates through a tunnel that links the two binding sites, toward an exit portal, is mediated by the conformation of a short 14-aa loop. Taken together, the ZneA structures presented here provide mechanistic insights into the conformational changes required for substrate efflux by RND superfamily transporters.
The resistance–nodulation–cell division (RND) superfamily, named based on the original members’ roles in metal resistance, root nodulation, and cell division (1), is found in all kingdoms of life and is comprised of nine phylogenetically distinct families (2–5). Functional characterization of RND proteins has shown that they are transmembrane efflux pumps that transport a variety of substrates out of cells, powered by an electrochemical proton gradient (6). In Gram-negative bacteria, RND pumps are specific for toxic substrates and largely belong to one of two families; the heavy metal efflux (HME) family and the multidrug hydrophobe/amphiphile efflux-1 (HAE1) family. For the HME- and HAE1-RND–driven efflux systems, a trimer of the RND pump in the plasma membrane is coupled by a hexamer of a periplasmic membrane fusion protein (MFP), also specific for the metal ion substrate in HME-RND–driven efflux systems, to a pore formed by a trimeric outer membrane factor (OMF), forming a continuous conduit that spans the inner and outer membranes (7–10). Each protomer of the RND trimer consists of a transmembrane domain of 12 transmembrane α-helices and two large hydrophilic loops that comprise the substrate-binding porter (or pore) domain and the OMF-coupling docking domain (11).
X-ray crystal structures of only four RND efflux pumps have been described, including just one of the HME family to date, shedding some light on the conformational changes that are necessary for transport (11–20). In two of these structures, those of the HAE1 efflux pumps AcrB from Escherichia coli and of the closely related MexB from Pseudomonas aeruginosa, each protomer of the homotrimer is trapped in a unique conformation, with only a single protomer conducive for substrate binding, suggestive of a functionally rotating mechanism (12, 14, 16). Inactivation of a single protomer thus abolishes transport activity by inhibiting the necessary cycling of structural changes in the trimer (21–23). Earlier crystal structures of AcrB, as well as the more recent structure of the Cu(I) efflux pump CusA from E. coli, do not show conformational differences between the protomers of the threefold symmetric trimer, which crystallizes around a crystallographic threefold axis (11, 18, 19). In the case of CusA, comparison of the structures in the presence and absence of substrate shows that the protomer undergoes a large conformational change, potentially regulating access to the substrate binding site from the periplasm (18).
Here we report X-ray crystal structures of a divalent metal-ion specific RND efflux pump, Zn(II)-specific ZneA from Cupriavidus metallidurans CH34, determined at low vs. high pH. At low pH, the ZneA structure is trapped in an asymmetric conformation with two protomers of the trimer having a single Zn(II) bound. At high pH, the ZneA structure adopts a distinct transport state, showing an additional substrate bound at an interprotomer binding site. Analysis of the two ZneA crystal structures, and comparison with other RND structures, reveals that substrate transport through RND efflux pumps is regulated by distinct conformational changes.
Results
ZneA Is a Proton-Dependent Antiporter Specific for Zn(II).
The Gram-negative bacteria C. metallidurans CH34 grows in millimolar concentrations of metal ions that would normally be toxic to cells (24, 25). As many as 12 HME–RND proteins are carried by C. metallidurans CH34, only four of which are located within their own operon that includes the HME–RND protein, MFP, OMF, as well as a two-component regulatory system or sigma and anti-sigma factors (26–28). The mechanism and binding partners of the remaining eight RND proteins are as yet undefined (27). Of the four that are each located within a full complement operon, the cobalt-zinc-cadmium resistance proteins (CzcCBA) confer resistance to Cd(II), Zn(II), and Co(II), and the cobalt-nickel-resistance proteins (CnrCBA) confer resistance to Co(II) and Ni(II) (29–34). zneBAC is also contained in a full complement operon, and we previously showed that the periplasmic MFP ZneB binds to Zn(II) with a Kd of ∼3 μM (35), but not to Cd(II) or Co(II) (or other divalent metal ions).
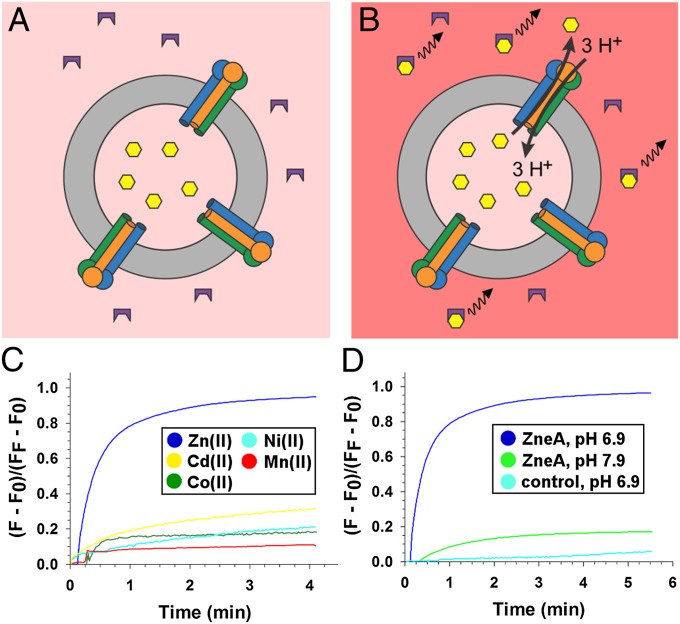
To determine the transport specificity of ZneA, we used a reconstituted proteoliposome assay that measures substrate transport through the transmembrane domain of ZneA (Fig. 1 A and B). ZneA was purified and reconstituted into liposomes containing one of five different divalent metal ions at 1 mM. In the presence of a pH gradient, ΔpH (inside-alkaline), Zn(II) is transported out of the lipid vesicles by ZneA, as monitored by an increase in fluorescence of a membrane-impermeable chelator specific for Zn(II) and other divalent metal ions (FluoZin-3) (36) located outside the proteoliposomes, to ∼95% of the maximal signal generated by subsequently solubilizing the proteoliposomes (Fig. 1C). In the absence of ZneA, liposomes that were treated otherwise identically show essentially no Zn(II) export (<3% of maximal signal upon solubilizing the liposomes; Fig. 1D). With ZneA but in the absence of ΔpH (inside-alkaline), Zn(II) reaches the outside, down the gradient of Zn(II), at a slow rate (∼16% of maximal signal upon solubilizing the proteoliposomes; Fig. 1D). The other cations tested, also at 1 mM [Cd(II), Co(II), Ni(II), Mn(II)], were only slowly released from the ZneA proteoliposomes in the presence of ΔpH (inside-alkaline, ∼10–28% of their maximal signal assessed by solubilizing the proteoliposomes; Fig. 1C). Therefore, we conclude that ZneA is a proton-dependent antiporter that is specific for Zn(II), the same as the Zn(II) specificity shown by ZneB (35).
Fig. 1.
ZneA is a proton-dependent Zn(II) efflux pump. Reconstituted proteoliposome transport assay in the absence (A) and presence (B) of ΔpH (inside-alkaline). ZneA (transmembrane domain as cylinders, porter and docking domains as circles) reconstituted into liposomes (gray ring) were loaded with divalent metal ion (yellow hexagons). The increase in fluorescence of FluoZin-3 (purple) upon divalent metal-ion binding is depicted as a black wavy line. (C) FluoZin-3 fluorescence in the presence of ΔpH (inside-alkaline) across the membrane of ZneA proteoliposomes containing Zn(II) (blue), Cd(II) (yellow), Co(II) (green), Ni(II) (light blue), or Mn(II) (red). (D) FluoZin-3 fluorescence in the presence (blue) or absence (green) of ΔpH (inside-alkaline) across the membrane of ZneA proteoliposomes containing Zn(II) or in the presence of ΔpH (inside-alkaline) across the membrane of control liposomes containing Zn(II) (light blue). A single representative measurement from three independent measurements is shown. Fluorescence (F) was normalized to the maximum fluorescence (FF) measured after disruption of the proteoliposomes with 0.25% (wt/vol) DDM. (F0 represents the fluorescence before addition of the proteoliposomes.)
Efflux of Zn(II) located in the cytoplasm, through the transmembrane domain of ZneA, would be electroneutral at a proton to Zn(II) ratio of 2:1, and electrogenic for all other proton-to-Zn(II) ratios. For electrogenic transport, a membrane potential (ΔΨ) might also serve to energize ZneA mediated efflux of Zn(II). To examine this, Zn(II) efflux out of proteoliposomes was measured in response to ΔΨ generated by a transmembrane gradient of K+ in the presence of valinomycin (Fig. S1). A total of 5 μM of the K+-specific ionophore valinomycin was added to the assay mixture, allowing K+ to diffuse down its concentration gradient, generating ΔΨ. In the presence of ΔΨ (inside-negative), the rate of Zn(II) efflux out of ZneA proteoliposomes is increased, whereas little change is observed in control liposomes (Fig. S1A). In contrast, in the presence of ΔΨ (inside-positive), Zn(II) efflux increases for the ZneA proteoliposomes and the control liposomes, likely because of Zn(II) diffusion out of the liposomes, down its electrochemical potential (Fig. S1B). The stimulation of ZneA-mediated Zn(II) efflux by ΔΨ (inside-negative) suggests that transport through the transmembrane domain is probably electrogenic, with a proton-to-Zn(II) ratio of greater than 2:1.
Crystallization of Unique Transport States of ZneA.
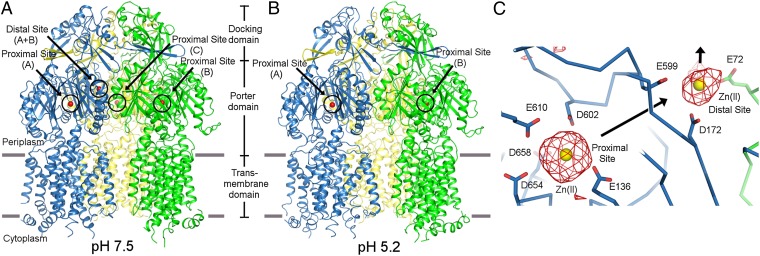
We have determined two independent X-ray crystal structures of ZneA, one at a lower pH (pH 5.2, 3.0 Å resolution) with two biological trimers in the asymmetric unit, and one at a higher pH (pH 7.5, 3.7 Å resolution) with one biological trimer in the asymmetric unit (Fig. 2 A and B and Table S1), from two unrelated C2 crystal forms (Fig. S2 A and B). The weighted 2mFo-DFc electron density maps for both crystal forms were of high quality throughout the trimers (Fig. S2 C and D) except in the regions of the cytoplasmic loops, 4 N-terminal residues, and 20 C-terminal residues. The final refined structures showed good stereochemistry and refinement statistics (Rwork = 27.9% and Rfree = 30.5% at low pH, and Rwork = 22.9% and Rfree = 28.3% at high pH; Table S1).
Fig. 2.
Overall structure of ZneA at high vs. low pH and substrate binding sites. Cartoon representation of the high-pH (A) and low-pH (B) crystal structures of ZneA, viewed parallel to the inner membrane plane. Zn(II) ions are depicted as spheres in red. (C) Zn(II) binding sites of ZneA at high pH. Carbon atoms (stick and ribbon representation) are colored by protomer as defined in A and B. Zn(II) ions are depicted as spheres in yellow. Simulated-annealing omit Fo-Fc electron density is shown in red, contoured at 3σ. The orientation shown is similar to that of A and B.
The ZneA trimer is held together by extensive interprotomer interactions that bury a total surface area of 15,000 Å2. Nearly the entire interface is contributed by the docking domains (57%), which would couple to the hexameric MFP ZneB and the trimeric OMF ZneC to form the complete tripartite assembly, and the large, periplasmic-facing porter domains (33%). The transmembrane domains of ZneA are more disordered (average B values of 100 Å2 at low pH and 134 Å2 at high pH) compared with the porter and docking domains (average B values of 60 Å2 at low pH and 112 Å2 at high pH). Pairwise alignment of each protomer pair of the trimers, using the secondary structure-matching (SSM) algorithm (37), results in ranges of rmsd values of 0.7 to 0.8 Å at low pH, 1.0 to 1.3 Å at high pH, and 0.9 to 1.2 Å between low and high pH protomers, over all common Cα atoms. The differences in structure are spread throughout the entire polypeptide chain, and are more subtle than those of the three protomer conformations [i.e., tight (or binding), loose (or access), and open (or extrusion)] seen in the asymmetric crystal structures of AcrB (12, 14) (rmsd calculated using SSM to be 1.8–2.3 Å). Structural superimposition of AcrB with ZneA using SSM shows that the open (or extrusion) conformation of AcrB is most similar to the protomers of ZneA.
In the high-pH ZneA structure, difference Fourier electron density (>6σ) was observed at two sites, each consistent with a fully occupied Zn(II) binding site, one in the center of the porter domain, termed the proximal site, and one near the exit funnel formed between the three protomers, termed the distal site (Fig. 2C). The proximal site is an anionic pocket for the positively charged Zn(II) ion surrounded by the side chain carboxylates of E136, D602, E610, D654, and D658 (Fig. 2C). The proximal site is conserved in the divalent metal-ion transporters CzcA and CnrA (34% and 35% sequence identity with ZneA, respectively), is less conserved in the monovalent metal-ion transporter CusA (31% sequence identity with ZneA), and is absent entirely from the hydrophobic/amphiphilic drug transporter AcrB (24% sequence identity with ZneA; Fig. S3 A and B). The Zn(II) at the proximal site is ∼5 Å away from, and completely different from, the sole Cu(I) binding site observed for E. coli CusA (18), and is coordinated by different types of residues (Asp/Glu in ZneA vs. Met in CusA) from different parts of the sequence.
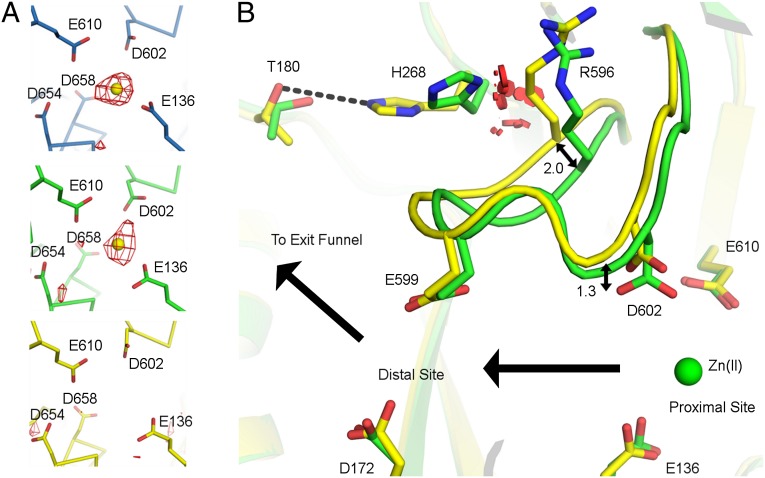
In contrast to the high-pH structure, at low pH, the proximal site is occupied in only two protomers of each trimer and is absent from the third (Fig. 3A). In the unbound protomer, the carboxylate of D602 is displaced ∼1.3 Å away from that observed for the Zn(II) occupied protomers (Fig. 3B). D602 of ZneA is absolutely conserved with both D619 of CzcA and D617 of CusA (Fig. S3B). A D617A CusA mutant exhibits no measurable Cu(I) transport activity (20), despite the observation that D617 is adjacent to the Cu(I) binding site and forms no interactions with Cu(I) (18). In ZneA, the displacement of the carboxylate of D602 away from the Zn(II) ion would reduce affinity for Zn(II), disrupting the proximal site, and likely contributes to the absence of Zn(II) at the proximal site in the unbound protomer.
Fig. 3.
Asymmetry in substrate binding at low pH. (A) The proximal site at low pH is shown, in the same orientation, for each protomer of one ZneA trimer. Carbon atoms (stick-and-ribbon representation) are colored by protomer as defined in Fig. 2. Zn(II) ions are depicted as spheres in yellow. Simulated-annealing omit Fo-Fc electron density is shown in red, contoured at 3σ. (B) Access-loop flexibility defines substrate binding asymmetry at low pH. Zn(II)-bound (green carbon atoms) and apo (yellow carbon atoms) protomers at low pH were aligned by using SSM. Side chains are shown as sticks, and the access loop is shown in cartoon representation. The Zn(II) ion from the Zn(II)-bound protomer is shown as a green sphere. van der Waals overlaps that would occur between the side chains of R596 in the apo conformation and H268 in the Zn(II) bound conformation are shown as red disks. Carbon atoms for the protomers are colored as defined in Fig. 2.
Polar residues in the transmembrane domain of RND efflux pumps have been shown to be critical for transport, and have been suggested to be the sites of proton translocation (29, 38–40). In ZneA, three acidic residues on TM4 (D393, D399, and E406) span a ∼19-Å distance from the center of the transmembrane domain toward the cytoplasm. Structural superposition of all protomer conformations at low and high pH shows that the carboxylate side chains of the three residues all orient in the same direction, between TM4 and TM11, implying a possible role in proton translocation distant from the proximal site. Indeed, mutagenesis of the corresponding conserved residues in CzcA (D402N, D408N, or E415Q) all lead to the loss of proton-dependent transport activity for Co(II), Zn(II), and Cd(II) (29).
Transport of Substrates Through ZneA.
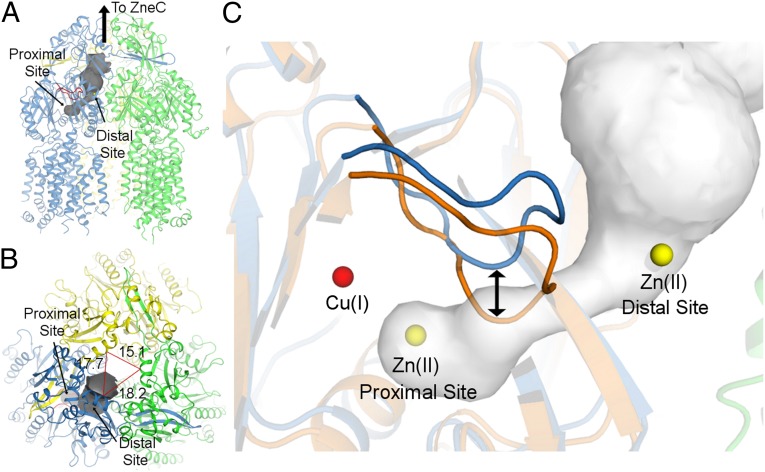
For all protomers at low and high pH, a continuous ∼16-Å-long tunnel extends from the proximal site, through the porter domain, to an opening at the side of the porter domain that is surrounded by the side chains of P129, D172, and E599. From this opening, Zn(II) would have access to the exit funnel, which links through ZneCB to expel Zn(II) out of the cell (Fig. 4 A and B). This tunnel in the porter domain approximately corresponds to that observed for the open (or extrusion) conformation of AcrB (12, 14, 41), except that, in the case of ZneA, the tunnel does not further extend into the docking domain. We do not see a carboxylate-lined tunnel through the transmembrane domain that leads into the proximal site of the porter domain, which would correspond to the methionine-lined transmembrane transport pathway of CusA (18).
Fig. 4.
Transport of substrates between binding sites to the exit funnel. (A) The high-pH structure of ZneA is shown in cartoon representation, with the access loop shown in red. The transport tunnel, shown in dark gray, was calculated with CAVER (51) using the Zn(II) ion at the proximal site as the start point for the calculation. (B) Transport pathway as viewed along the exit funnel of ZneA, 90° relative to that of A. Distances between select Cα atoms near the top of the exit funnel are shown for reference. (C) Transport pathway from the proximal to distal sites in ZneA compared with CusA. ZneA (in blue and green) and CusA (in orange), aligned by using SSM, are shown in cartoon representation. The access loops for ZneA and CusA are shown in darker blue and orange, respectively. The Cu(I) ion from CusA is shown as a red sphere, and Zn(II) ions are shown as yellow spheres. The transport pathway for ZneA is shown in gray. For A and C, the orientation shown is similar to that of Fig. 2 A and B.
For only one protomer at high pH, difference Fourier electron density (>6σ) was seen at the exit site of the tunnel (Fig. 2C), termed the distal site, further supporting this tunnel as a transport pathway in ZneA. Similar to that of the proximal site, the distal site consists of the carboxylates of D172, E599, and an intermolecular E72. For all other protomer conformations at low and high pH, the distal site is absent as a result of displacement of Nα1, on which E72 resides, away from the distal site. Unlike that observed for the proximal site, the distal site of ZneA is not highly conserved in CzcA and CnrA (Fig. S3B). In addition to having fewer carboxylate side chains, the distal site has the side chain amide of K165 within 3.5 Å of the Zn(II) ion, which would electrostatically repel Zn(II) binding. For the proximal and distal binding sites, the distances to the Zn(II) ion (4.1 ± 0.2 Å) are larger than that of a O-Zn(II) coordination bond (∼2.2 Å) (42), implying that Zn(II) carries a hydration shell, and is not tightly constrained at the proximal or distal sites, allowing for movement through the tunnel.
Conformational Regulation of Transport.
In the structures of ZneA, a 14-aa loop lines the transport tunnel, approximately halfway between the proximal and distal sites (Fig. 4C). The loop, which we term the access loop, connects the Cβ2 and Cβ3 strands of the PC1 subdomain and exhibits low sequence conservation with those of CusA or AcrB (Fig. S3A). The corresponding loop of AcrB, alternatively termed the G-loop, switch-loop, or Phe-617 loop, is glycine-rich and has been shown to adopt multiple conformations that are important for regulating substrate binding (13, 15, 43). The access loop of ZneA can also adopt different conformations (Fig. 3B). In the ZneA protomer that lacks Zn(II) at the proximal site, the access loop, on which D602 resides, moves up to 2.0 Å (based on common Cα positions), leading to the disruption of the proximal site, and increasing the diameter of the tunnel in the vicinity of the proximal site by ∼0.5 Å. The space required to accommodate this access-loop conformation is created by a large concomitant movement of the side chain of H268 up to 4.8 Å (based on common side chain atoms), away from the access loop and toward the side chain of T180; in the absence of this H268 movement, a large number of steric clashes would exist between R596 and H268 (Fig. 3B).
The conformations of the access loops of ZneA are very different from those of the corresponding sequence of CusA (Fig. 4C). Structural alignment of ZneA with Cu(I)-bound CusA shows that the access loop of CusA is displaced up to 5.0 Å (based on Cα positions) relative to that of ZneA, and that this difference is larger than that observed for the entire polypeptide chain (rmsd of 2.0 Å, over all common Cα atoms). The conformation of the access loop of CusA would, in ZneA, directly insert into the tunnel between the proximal and distal sites, and thus prevent backflow of substrates from the distal site to the anionic proximal site. Indeed, the transport tunnel observed here for ZneA, through the porter domain and to the exit funnel, is blocked in the structure of CusA by the access loop.
Discussion
An intriguing aspect of HME-RND–driven efflux systems is their apparent ability to recognize and transport metal ions located in the cytoplasm or in the periplasm, termed transenvelope efflux or periplasmic efflux, respectively (44, 45). The low- and high-pH crystal structures of trimeric ZneA reported here show how substrates can be transported through the periplasmic porter domain, from the proximal site in the center of the porter domain to the distal site, through a tunnel seen in all protomers. From the distal site, substrate would be expelled through the nearby exit funnel and out of the cell. Thus, the protomer conformations of ZneA seen here are states that are capable of periplasmic efflux, late in the transport cycle. Indeed, structural comparison of ZneA with CusA shows that the open periplasmic cleft seen in the structure of CusA in complex with Cu(I) (18) is closed in the structures of ZneA (Fig. S4), thus preventing backflow of substrates from the proximal site into the periplasm. The local asymmetries at the proximal site, particularly apparent in the low-pH protomer that lacks substrate bound at the proximal site, are not large, and comparative structural changes in other HME–RND proteins could be accommodated with subtle conformational switches in the vicinity of the proximal site. An electrochemical proton gradient, via proton conduction through the transmembrane domain of ZneA, would power these necessary conformational changes in the porter domain through action at a distance. The side chains of three polar residues on TM4 (D393, D399, and E406), conserved and essential for proton-dependent Zn(II) efflux in CzcA (29), form a network of titratable residues that could serve as the sites of proton translocation, ∼40 to 60 Å from the proximal site of the porter domain. An additional role for the carboxylates of D393, D399, and E406 in binding to Zn(II) during transenvelope efflux is possible. However, the structures of ZneA do not clearly reveal how substrates would be transported through the transmembrane domain, as no carboxylate-lined tunnel, analogous to the methionine-lined tunnel of CusA (18), is present in ZneA.
The access-loop of ZneA, situated between the proximal and distal substrate binding sites, is well positioned to regulate transport through the porter domain. The structures of ZneA, CusA, and AcrB show that the access loops can sample a large conformational space, fitting with a role in regulating substrate transport. Nevertheless, their proposed modes of action are different. The access-loop conformations adopted in the structures of ZneA and CusA would alternately permit and gate substrate extrusion through the tunnel in the porter domain, ensuring that transport is made unidirectional by preventing substrate backflow from the distal site to the proximal site. In AcrB, the corresponding loop would also regulate transit of substrates between binding sites, albeit by alternately occluding substrate binding at one of two sites (13). These different modes of action may be reflective of the sizes of substrates used by HME–RND vs. HAE1–RND efflux pumps, as the smaller binding site footprints of HME–RND efflux pumps would reduce the available volume that could be occluded by conformational changes in the access loop.
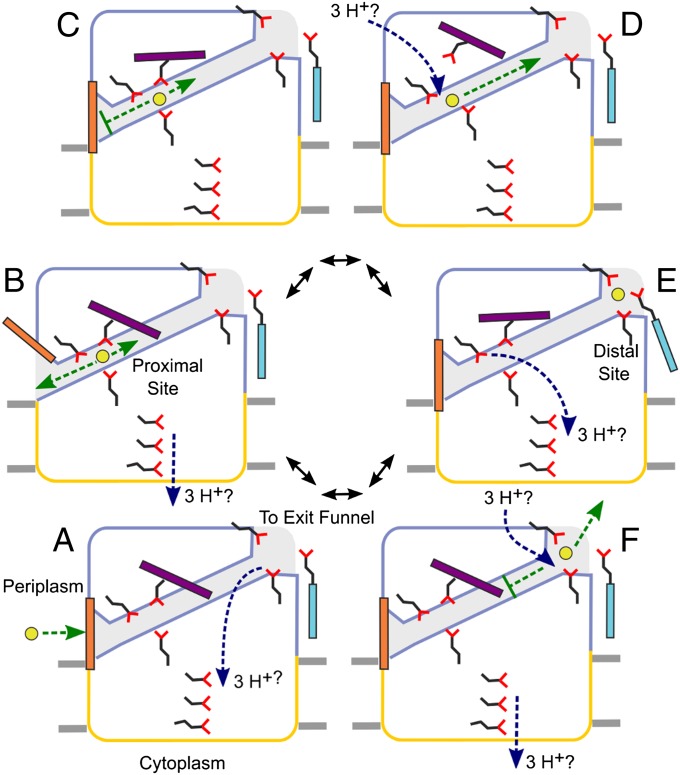
The structures of ZneA, combined with those of CusA and AcrB, suggest a coherent model for the efflux of substrates that are located in the periplasm (Fig. 5). Efflux is made unidirectional by a series of switches that alternately regulate the entry, passage, and exit of substrates through the porter domain, powered indirectly by conformational changes induced by the electrochemical proton gradient across the inner membrane. Initial access to the porter domain is gated (Fig. 5A), with a large conformational change being required to open a periplasmic cleft (Fig. 5B), as is observed in the apo and Cu(I)-bound structures of CusA, respectively (Fig. S4). Opening of the periplasmic cleft may be coupled to proton translocation through the transmembrane domain, or to conformational changes in the MFP (35, 46, 47) if, indeed, the MFP acts as an allosteric activator of the RND pump (45). Closure of the periplasmic cleft in the presence of substrate at the proximal site (Fig. S4) prevents backflow of substrate to the periplasm (Fig. 5C). Alternatively, substrates might also enter the proximal site from the cytoplasm at this point, through the transmembrane domain, if transenvelope substrate efflux is significant in vivo (45). In addition to gate closure, the access loop adopts a conformation that no longer blocks passage through the transport tunnel between the proximal and distal sites (Fig. 5C), reminiscent of a peristaltic mechanism as proposed for AcrB. Dissolution of the proximal site (Fig. 5D), in ZneA caused by local displacements of carboxylate side chains away from Zn(II) and a further conformational change of the access loop, and formation of the distal site (Fig. 5E) permits forward transport of substrate through the porter domain to the distal site. Direct protonation of the carboxylates of the proximal and/or the distal site, before the protons are translocated into the cytoplasm, remains a possibility and would also lead to substrate release by changing the charge of the carboxylates from negative to neutral (44). Dissolution of the distal site and the return of the access loop to the ground state conformation (Fig. 5F) would prevent backflow of substrate to the more anionic proximal site, thereby ensuring efficient transport of substrates out of the cell.
Fig. 5.
Schematic representation of a proposed periplasmic substrate efflux pumping cycle. The periplasmic porter domain is outlined in blue, the transmembrane domain is outlined in yellow, the transport tunnel through the porter domain is colored in light blue, and the membrane bilayer is depicted as gray rectangles. Zn(II) is depicted as a yellow circle, and carboxylate side chains are shown as sticks in black and red. Colored rectangles represent different portions of polypeptide that undergo conformational change during the pumping cycle: periplasmic gate (orange), access loop (purple), and distal site Nα1 helix (cyan). See text for a detailed description of panels A–F.
Methods
Details of the methods used for protein expression, purification, reconstitution, and structure determination are provided in SI Methods.
Protein Expression, Purification and Crystallization.
The gene encoding ZneA was amplified by PCR from C. metallidurans CH34 genomic DNA and subcloned into pET30b (Novagen). Full-length ZneA, with a C-terminal His6 affinity tag, was expressed in E. coli C43 (DE3) cells (Lucigen), and purified by Ni-NTA affinity chromatography and size-exclusion chromatography.
Crystals of ZneA were grown by hanging-drop vapor diffusion. ZneA was concentrated to 4.5 to 7 mg/mL in buffer containing 20 mM MES, pH 6, 100 mM NaCl, 10% (vol/vol) glycerol, and 0.05% n-dodecyl-α-D-maltoside (α-DDM) or n-dodecyl-β-D-maltoside (β-DDM). After concentration, ZnCl2 was added at a final concentration of 50 μM. For the low-pH crystal form, 0.3 μL of ZneA in β-DDM was mixed with 0.3 μL of well solution consisting of 26% (vol/vol) PEG 400, 100 mM N-(2-Acetamido)iminodiacetic acid, pH 5.2, and 100 mM LiSO4. For the high-pH crystal form, 0.1 μL of ZneA in α-DDM was mixed with 0.1 μL of well solution consisting of 30% (vol/vol) PEG 400, 100 mM Na Hepes, pH 7.5, and 100 mM MgCl2. All crystals were frozen in liquid nitrogen, with no additional cryoprotection being added.
Data Collection, Structure Determination and Refinement.
Diffraction images were collected at beam line 23-ID-B of the Advanced Photon Source (for the low-pH crystal form) and beam line 8.3.1 of the Advanced Light Source (for the high-pH crystal form). The low-pH crystal form belongs to space group C2 with unit cell parameters of a = 223.7, b = 129.0, c = 391.9, and β = 94.6, whereas the high-pH crystal form belongs to C2 with unit cell parameters of a = 215.4, b = 127.1, c = 163.3, and β = 93.1.
The low-pH crystal form was solved by molecular replacement using PHASER (49), using a modified CusA [Protein Data Bank (PDB) ID code 3NE5] search molecule. Six monomers (two biological trimers) are found in the asymmetric unit (∼70% solvent content). The high-pH crystal form was solved by molecular replacement, by using a partially refined low-pH ZneA monomer as the search molecule. Three monomers (one biological trimer) are found in the asymmetric unit (∼60% solvent content). Both structures were refined in PHENIX (50) with several cycles of simulated annealing, followed by coordinate and individual B-factor refinement and translation/libration/screw (TLS) refinement. The low- and high-pH crystal structures were refined at 3.0 Å (R = 27.9% and Rfree = 30.5%) and 3.7 Å (R = 22.9% and Rfree = 28.3%), respectively, and had good overall stereochemistry. Coordinates and structure factors for the high-pH (PDB ID code 4K0E) and low-pH (PDB ID code 4K0J) crystal structures of ZneA have been deposited in PDB.
Supplementary Material
Acknowledgments
We thank the Advanced Photon Source and the Advanced Light Source for synchrotron access; Dr. Robert H. Edwards, Dr. Christopher M. Koth, and Dr. John K. Lee for critical reading of the manuscript; and Dr. H. Ronald Kaback for insight into the energetics of transporters and for suggesting the experiments showing that transport is electrogenic. This work is supported by National Institutes of Health/National Institute of General Medical Sciences Grants P50 GM73210, U54 GM094625, and R37 GM24485, and by National Fund for Scientific Research (Fonds de la Recherche Fondamentale Collective 2.4577.12 and 2.4628.12).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4K0E and 4K0J).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318705110/-/DCSupplemental.
References
- 1.Saier MH, Jr, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11(5):841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 2.Tseng TT, et al. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1(1):107–125. [PubMed] [Google Scholar]
- 3.Saier MH, Jr, Paulsen IT. Phylogeny of multidrug transporters. Semin Cell Dev Biol. 2001;12(3):205–213. doi: 10.1006/scdb.2000.0246. [DOI] [PubMed] [Google Scholar]
- 4.Goel AK, Rajagopal L, Nagesh N, Sonti RV. Genetic locus encoding functions involved in biosynthesis and outer membrane localization of xanthomonadin in Xanthomonas oryzae pv. oryzae. J Bacteriol. 2002;184(13):3539–3548. doi: 10.1128/JB.184.13.3539-3548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, et al. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell. 2002;111(1):63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178(20):5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulsen IT, Park JH, Choi PS, Saier MH., Jr A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol Lett. 1997;156(1):1–8. doi: 10.1111/j.1574-6968.1997.tb12697.x. [DOI] [PubMed] [Google Scholar]
- 8.Dinh T, Paulsen IT, Saier MH., Jr A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol. 1994;176(13):3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta. 2009;1794(5):769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794(5):782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 11.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419(6907):587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 12.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443(7108):173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 13.Eicher T, et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci USA. 2012;109(15):5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeger MA, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313(5791):1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 15.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480(7378):565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 16.Sennhauser G, Bukowska MA, Briand C, Grütter MG. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J Mol Biol. 2009;389(1):134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Tsukazaki T, et al. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474(7350):235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long F, et al. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467(7314):484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su C-C, et al. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature. 2011;470(7335):558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su C-C, et al. Charged amino acids (R83, E567, D617, E625, R669, and K678) of CusA are required for metal ion transport in the Cus efflux system. J Mol Biol. 2012;422(3):429–441. doi: 10.1016/j.jmb.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takatsuka Y, Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J Bacteriol. 2009;191(6):1729–1737. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seeger MA, et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15(2):199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 23.Takatsuka Y, Nikaido H. Site-directed disulfide cross-linking shows that cleft flexibility in the periplasmic domain is needed for the multidrug efflux pump AcrB of Escherichia coli. J Bacteriol. 2007;189(23):8677–8684. doi: 10.1128/JB.01127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mergeay M, Houba C, Gerits J. Extrachromosomal inheritance controlling resistance to cadmium, cobalt, copper and zinc ions: Evidence from curing in a Pseudomonas [proceedings] Arch Int Physiol Biochim. 1978;86(2):440–442. [PubMed] [Google Scholar]
- 25.Mergeay M, et al. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162(1):328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mergeay M, et al. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: Towards a catalogue of metal-responsive genes. FEMS Microbiol Rev. 2003;27(2-3):385–410. doi: 10.1016/S0168-6445(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 27.Janssen PJ, et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE. 2010;5(5):e10433. doi: 10.1371/journal.pone.0010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Lelie D, et al. Two-component regulatory system involved in transcriptional control of heavy-metal homoeostasis in Alcaligenes eutrophus. Mol Microbiol. 1997;23(3):493–503. doi: 10.1046/j.1365-2958.1997.d01-1866.x. [DOI] [PubMed] [Google Scholar]
- 29.Goldberg M, Pribyl T, Juhnke S, Nies DH. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J Biol Chem. 1999;274(37):26065–26070. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 30.Grosse C, et al. Transcriptional organization of the czc heavy-metal homeostasis determinant from Alcaligenes eutrophus. J Bacteriol. 1999;181(8):2385–2393. doi: 10.1128/jb.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grass G, Grosse C, Nies DH. Regulation of the cnr cobalt and nickel resistance determinant from Ralstonia sp. strain CH34. J Bacteriol. 2000;182(5):1390–1398. doi: 10.1128/jb.182.5.1390-1398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tibazarwa C, Wuertz S, Mergeay M, Wyns L, van Der Lelie D. Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J Bacteriol. 2000;182(5):1399–1409. doi: 10.1128/jb.182.5.1399-1409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grass G, Fricke B, Nies DH. Control of expression of a periplasmic nickel efflux pump by periplasmic nickel concentrations. Biometals. 2005;18(4):437–448. doi: 10.1007/s10534-005-3718-6. [DOI] [PubMed] [Google Scholar]
- 34.Nies DH, Rehbein G, Hoffmann T, Baumann C, Grosse C. Paralogs of genes encoding metal resistance proteins in Cupriavidus metallidurans strain CH34. J Mol Microbiol Biotechnol. 2006;11(1-2):82–93. doi: 10.1159/000092820. [DOI] [PubMed] [Google Scholar]
- 35.De Angelis F, et al. Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems. Proc Natl Acad Sci USA. 2010;107(24):11038–11043. doi: 10.1073/pnas.1003908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Bertoglio BA, Devinney MJ, Jr, Dineley KE, Kay AR. The interaction of biological and noxious transition metals with the zinc probes FluoZin-3 and Newport Green. Anal Biochem. 2009;384(1):34–41. doi: 10.1016/j.ab.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(pt 12 pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 38.Murakami S, Yamaguchi A. Multidrug-exporting secondary transporters. Curr Opin Struct Biol. 2003;13(4):443–452. doi: 10.1016/s0959-440x(03)00109-x. [DOI] [PubMed] [Google Scholar]
- 39.Takatsuka Y, Nikaido H. Threonine-978 in the transmembrane segment of the multidrug efflux pump AcrB of Escherichia coli is crucial for drug transport as a probable component of the proton relay network. J Bacteriol. 2006;188(20):7284–7289. doi: 10.1128/JB.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seeger MA, von Ballmoos C, Verrey F, Pos KM. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: Evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry. 2009;48(25):5801–5812. doi: 10.1021/bi900446j. [DOI] [PubMed] [Google Scholar]
- 41.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5(1):e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel K, Kumar A, Durani S. Analysis of the structural consensus of the zinc coordination centers of metalloprotein structures. Biochim Biophys Acta. 2007;1774(10):1247–1253. doi: 10.1016/j.bbapap.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 43.Vargiu AV, Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci USA. 2012;109(50):20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27(2-3):313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 45.Kim E-H, Nies DH, McEvoy MM, Rensing C. Switch or funnel: How RND-type transport systems control periplasmic metal homeostasis. J Bacteriol. 2011;193(10):2381–2387. doi: 10.1128/JB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su C-C, et al. Crystal structure of the membrane fusion protein CusB from Escherichia coli. J Mol Biol. 2009;393(2):342–355. doi: 10.1016/j.jmb.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14(3):577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura T, Hsu C, Rosen BP. Cation/proton antiport systems in Escherichia coli. Solubilization and reconstitution of delta pH-driven sodium/proton and calcium/proton antiporters. J Biol Chem. 1986;261(2):678–683. [PubMed] [Google Scholar]
- 49.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chovancova E, et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput Biol. 2012;8(10):e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.