Significance
The traditional, Edisonian method of research involves taking a system and examining its properties. A preferred alternative is to target a suite of properties and then design the most appropriate system in this context. Here we illustrate such a design approach for the self-assembly of colloids grafted with single-stranded DNA. When two types of colloids grafted with complementary ssDNA sequences are mixed, DNA hybridization can lead to the formation of ordered colloidal crystals. We use the genetic algorithm to design ssDNA-grafted particles that will assemble into a desired structure. Our methodology is easily generalizable, fast and highly selective, and not only accurately reproduces the parameters relevant to four currently realized crystals but also elucidates four currently unobserved structures.
Keywords: DNA-grafted colloids, inverse design, nanostructures, crystal lattice predictions, evolutionary algorithm
Abstract
In conventional research, colloidal particles grafted with single-stranded DNA are allowed to self-assemble, and then the resulting crystal structures are determined. Although this Edisonian approach is useful for a posteriori understanding of the factors governing assembly, it does not allow one to a priori design ssDNA-grafted colloids that will assemble into desired structures. Here we address precisely this design issue, and present an experimentally validated evolutionary optimization methodology that is not only able to reproduce the original phase diagram detailing regions of known crystals, but is also able to elucidate several previously unobserved structures. Although experimental validation of these structures requires further work, our early success encourages us to propose that this genetic algorithm–based methodology is a promising and rational materials-design paradigm with broad potential applications.
A topic of much interest in the current literature is the self-assembly of colloid particles multiply grafted with ssDNA molecules (1–10). The typical experimental system consists of two types of colloids grafted with complementary ssDNA sequences. Upon cooling, hybridization of the DNA occurs, cross-linking the colloids. Under the right conditions this cross-linking can facilitate the ordering of the colloids into crystal structures. The typical dimensions of colloids result in periodicities comparable to the wavelength of visible length, which have made them attractive for various emergent technologies, e.g., photonic bandgap materials. Classes of plasmonic, light-emitting, and catalytic metamaterials can be realized via the self-assembly of ssDNA-grafted colloids into specified 3D arrays.
Although much work has examined the effects of temperature, DNA length, linker DNA groups, size of colloids, etc., on structure formation, it has been largely empirically driven. However, there has been some progress in theory and simulation on understanding this assembly process (5, 6, 11–13). The recent work of Starr and coworkers, for example, has emphasized the complicated phase and assembly behavior of these materials (11, 12, 14). Travesset and coworkers (5) and Olvera de la Cruz and coworkers (15) have used large-scale molecular dynamics simulations to study equilibrium aspects and the kinetics of self-assembly, including kinetic traps like gel formation. Crocker and coworker developed a quantitative model based on experimental studies to predict ssDNA-induced particle interactions, the driving force for self-assembly (16). Similarly, Frenkel and coworkers has also defined a general accurate theory of valence-limited colloidal interactions (17). In a similar vein, Mirkin and coworkers proposed a rule-based complementary contact model (CCM) to predict the formation of crystal structures by ssDNA-grafted colloids (7). This model was used to explain the four crystal structures experimentally observed.
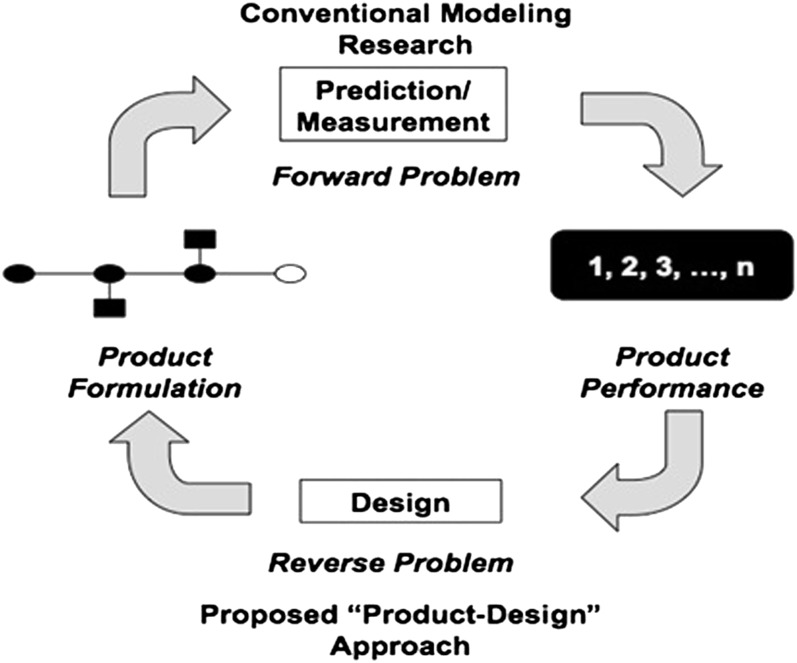
Although the controlled spatial organization of colloidal particles is thus a topic of broad interest, most current work has taken the forward modeling approach: i.e., one starts with a pair of ssDNA colloids with known structures (or product formulation, Fig. 1) and then uses theory and/or simulation to examine the superstructure it assembles into. These predictions are validated against experiments as a means of calibrating the fidelity of the models and the modeling tools. Here we focus on the reverse problem (Fig. 1), where we design ssDNA-grafted colloids that can assemble into desired superstructures.
Fig. 1.
Comparison of conventional and proposed paradigms.
Model
Forward Modeling Using the CCM.
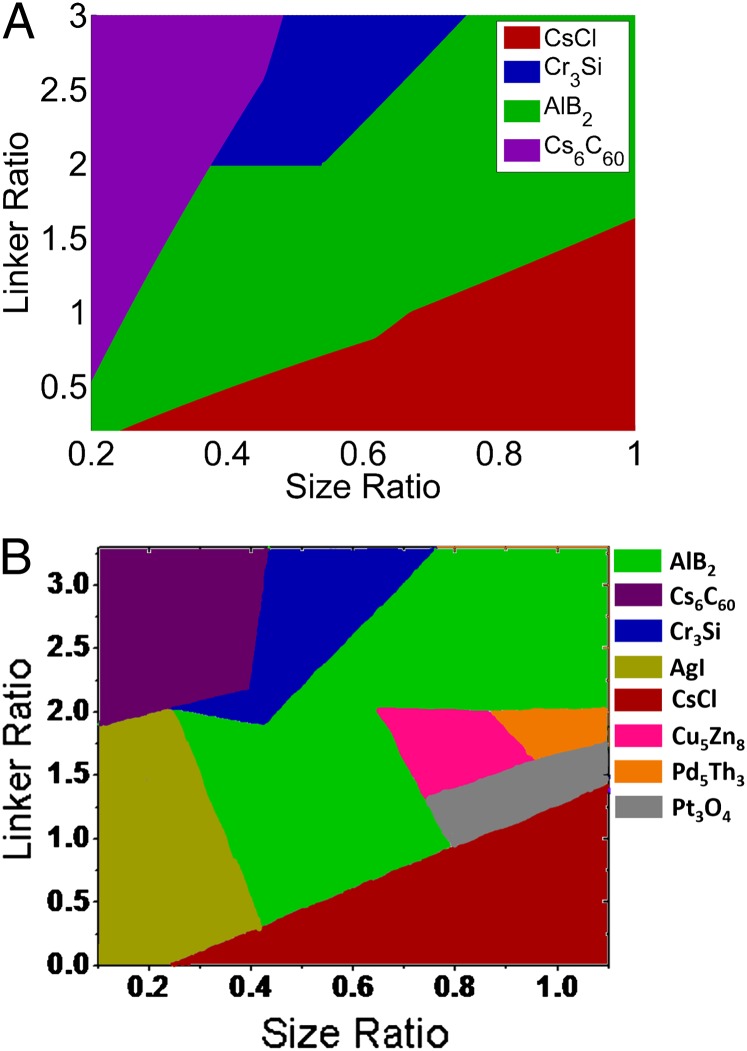
Currently, it is believed that ssDNA-grafted particles form four different crystal structures (7, 8). The primary control variables here are (i) the number of ssDNA grafted to the core, which determines the number of interaction sites, and (ii) the hydrodynamic radius of each colloid, which can be defined as the radius of the colloidal core plus the length of the monodisperse DNA chains grafted onto it. The CCM, based purely on enthalpies, is able to capture these trends (see Fig. 3A) through variations in the hydrodynamic size ratio of the particles including the DNA shells (r), and the ratio of the number of DNA per particle that are able to hybridize, the linker DNA number ratio (l) (5–8). This model calculates the percent of DNA duplexes that could be formed for each particle in a binary crystal structure based on the number of complementary-type nearest neighbors, and number of particles in the unit cell. The structure with the highest predicted percent of DNA duplexes is considered to be the stable structure realized from the building blocks (for further details, see Materials and Methods). It must be stressed here that this model ignores important entropic contributions to the hybridization process as noted by both Frenkel and coworkers and Dreyfus and coworkers (13, 17–19). With this caveat we use this contact model, along with the genetic algorithm (GA) approach, to design the building blocks that will assemble into desired crystal structures selected from the Inorganic Crystal Structure Database.
Fig. 3.
(A) Mirkin predictions using the CCM and (B) predictions from GA using the forward CCM approach.
Reverse Problem Solution Using the Genetic Algorithm.
Reverse or inverse problems, such as the design problem that is the focus of this paper, are usually very difficult to solve because of the following reasons: (i) the complex nonlinear relationships between the parameters of the design problem (such as size ratio of the colloids, the number of DNA bases on each ssDNA-grafted colloid) and the desired crystal structure, and (ii) a large search space of potential solution superstructures that is riddled with local minima. Various approaches have been proposed to address inverse problems, including knowledge-based systems, machine-learning techniques, graph reconstruction methods, enumeration-based algorithms, simulated annealing, and mathematical programming. In general, an inverse solution method should have (i) generality of application; (ii) the ability to handle nonlinear objective functions and the resulting local optima; (iii) ease of implementation and adaptability, particularly in incorporating domain specific constraints; (iv) computational ease in handling large search spaces; and (v) robustness to approximations and uncertainties in the predictions from the forward problem methods. GA are general-purpose, stochastic, evolutionary search-and-optimize strategies based on the Darwinian model of natural selection and evolution, which are particularly appropriate in this context (20–22). However, GAs have certain limitations. Theoretically, there is no guarantee of convergence, although this does not appear to be a serious issue in practice (21). Also, the performance of the algorithm depends on internal parameters such as operator probabilities that may require trial-and-error tuning to determine. In practice, again, this tuning requires only limited effort and the resultant parameters have been found to be robust to model approximations and uncertainties.
There have been several contributions where GA has been used to determine the crystal structures that can be obtained from a given colloid or colloid mixtures, i.e., the forward problem (23–27). In the current work we do not use the GA framework to solve the forward problem of predicting crystal structures formed. Rather, we address the reverse problem of designing ssDNA-grafted colloids that self-assemble into desired superstructures. The proposed methodology is validated against existing experimental data, and, in addition, used to predict several crystal polymorphs that can potentially form within the framework of the forward CCM.
The major steps in the GA framework when applied to this context are: (i) initialization of the DNA-grafted particle population; (ii) prediction of the superstructures formed using these particle building blocks using a forward model (in our case we use the CCM); (iii) fitness (i.e., objective function) evaluation of the superstructures formed; and (iv) reproduction and adaptation of the population building blocks based on their objective function values (new population formation using survival-of-the-fittest criterion; Fig. 2). The reproduction and adaptation step results in an offspring population (new generation) of individuals that contain hybrid traits from the parents in the previous generation. A detailed explanation of the various steps in the GA approach is given in Materials and Methods section.
Fig. 2.
GA approach for the inverse design of ssDNA-grafted colloids. (i) Initialization of the ssDNA-grafted colloid population with defined size and linker ratios (ii) Identification of the stable structure based on the CCM, (iii) Fitness evaluation for the calculated stable structure relative to the desired structure, and (iv) reproduction and adaptation of (r, l) to optimize a particle design.
Results and Discussion
The GA framework, in conjunction with the contact model, correctly predicts the hydrodynamic size ratio and DNA linker ratio for the four currently realized crystal structures, Fig. 3B. More importantly, comparison with the original phase diagram proposed by Mirkin (Fig. 3A) reveals that the GA also predicts the formation of four unique crystal structures—AgI(α−form), Cu5Zn8, Pd5Th3, and Pt3O4—that were previously unobserved in experiments. It should be apparent that these unique structures are formed in the transition region between the domains for the four different experimentally observed structures. This is in agreement with the simulations of Li et al. where different crystal structures were also observed at such boundaries (8).
Population Equilibration Using the GA Framework.
We now comment on the uniqueness of the design procedure based on the GA framework. The mean objective function of each generation gives information about the convergence of the GA toward the final solution, while the average distance between individuals in each generation provides details of the existing diversity in each generation. This latter quantity is a key measure that dictates the exploration of large spaces; in the absence of a broad search space it is likely that the algorithm will converge to local minima, which correspond to suboptimal solutions.
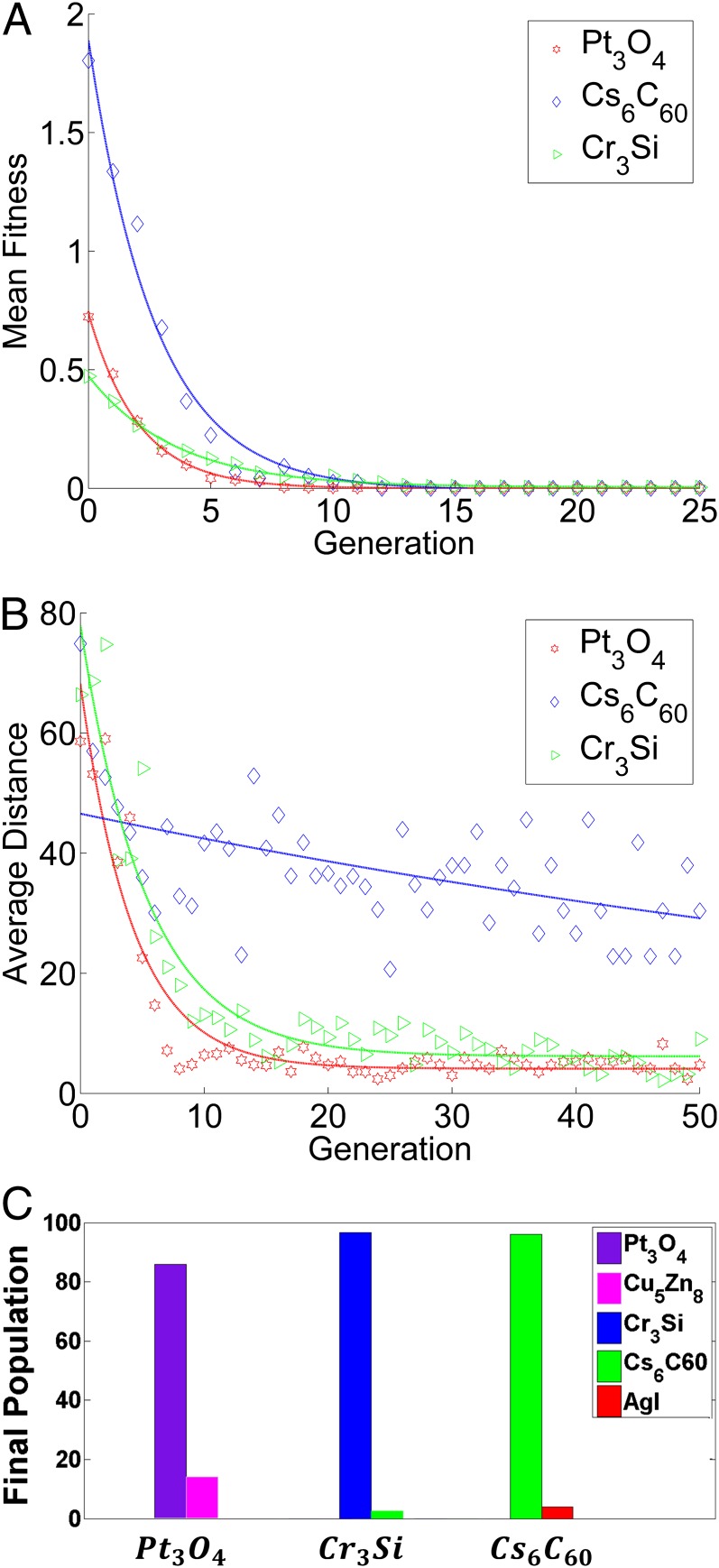
Genetic operations are continued in each generation until the mean objective function value of the generation does not reduce below the threshold value of 10−6. The best objective function values in all of the crystal structures is zero because the objective function, f, has to be minimized and cannot be negative due to its quadratic nature (see Materials and Methods for more details). Fig. 4A shows that the mean objective function for three representative target crystal structures monotonically decay to zero as a function of the number of GA algorithm iterations. This indicates that the entire population moves toward the best objective function (i.e., zero) with an increased number of genetic operations performed as per the rule of the survival of the fittest.
Fig. 4.
Dynamic evolution of generations in GA for three desired crystal structures, Pt3O4, Cr3Si, and Cs6C60. (A) Evolution of GA with respect to mean objective function for the three crystal structures. (B) Average distance between individuals in each generation. This quantity is the distance from one individual to all other individuals in the population, averaged across all individuals. (C) Final population for the three crystal structures. Pt3O4 as desired, 86% of the final population are Pt3O4 and 14% of them are Cu5Zn8 structures. Cr3Si as desired, 97% of the final population and 3% of them are Cs6C60 structures. For Cs6C60, 96% of the final population was the desired structure and 4% were the AgI structure.
In these three cases, convergence to a desired crystal structure is achieved. In certain cases, when we assume infeasible crystal structures in the context of CCM model, then the mean objective function does not decay to 0. (All 100 different DNA-NPs in the final GA population yield an objective function greater than zero.) In addition to these nonconverged cases, in most cases, there are important exceptions when we consider converged cases with parameter values that are close to a boundary in Fig. 3. We provide four examples of such special cases (see also Fig. 4C): (i) In one case we found 90% of the desired Cu5Zn8 (f = 0), with 6% AlB2 (f = 0.0128) and 4% Pt3O4 (f = 0.0304). (ii) With Cs6C60 as the desired crystal structure, the final population provided 96% of the desired structure (f = 0), and 4% yielded AgI (α-form) (f = 0.250), which lies at the border of Cs6C60. (iii) With Pt3O4 as the desired crystal structure, the final population provided 86% of the desired structure (f = 0), and 14% yielded Cu5Zn8 (f = 0.011), another border structure; and (iv) with Cr3Si as the desired crystal structure, the final population was comprised of 97% of the desired crystal structure (f = 0) and 3% yielded Cs6C60 (f = 0.023). As it can be seen, the f values of the crystal structures (different from the desired structures) are all close to zero, and in these cases the system might spontaneously yield different crystal polymorphs in coexistence in the framework of CCM model. This likely arises because the contact model is not sensitive enough to distinguish between the different crystal structures, but we suspect that a more detailed free energy calculation could help to clarify this issue.
Analysis of the CCM.
Because the CCM underpins the overall predictive power of the GA framework, it is critical to examine its accuracy. As described earlier, the CCM calculates the percent of DNA linkers duplexed for a given crystal structure, and selects the crystal structure with the highest percent duplexed as the stable phase. This approach is reasonable when the two colloids in the system have approximately the same number of ssDNA chains grafted on them. In situations with high asymmetry in ssDNA strands between the constituents, then, the actual number of ssDNA strands that are hybridized on each particle is the better metric for calculating the system energy. The role of this change in the theory on the predicted phase diagram remains to be explored. Second, we also note that the CCM protocol picks the best crystal for a given combination of linker ratio and DNA size ratio. The stoichiometric amounts of the two constituents are dictated by the equilibrium crystal structure found. Here we emphasize that experiments do not follow this protocol; rather, they select a given stoichiometry and then examine the crystal structures that result. Modifying the theory to account for this real experimental constraint is another aspect that needs to be completed.
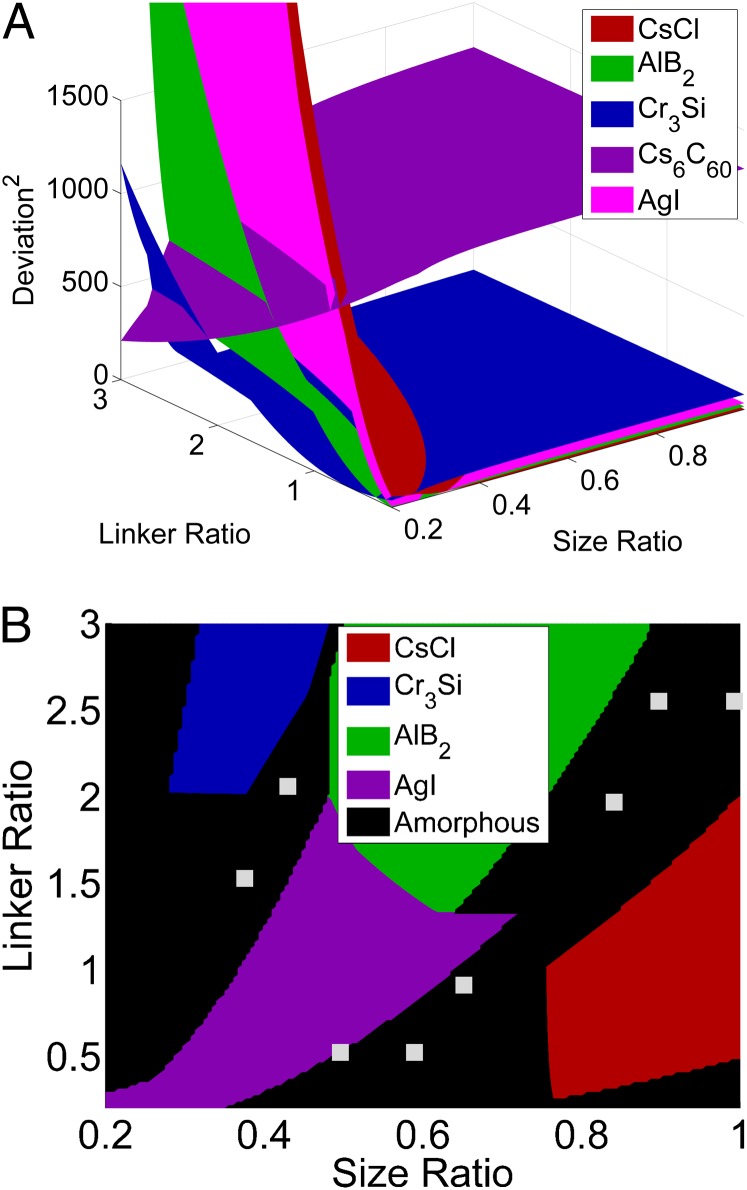
Further, the CCM is, at heart, an equilibrium model that cannot readily incorporate the kinetics of crystal lattice formation. Recent simulations results from Li et al. (8) report the formation of clusters along the boundary between different phases and also in regions of small linker and size ratios. These findings strongly suggest the importance of kinetic constraints on structure formation. In an attempt to study this effect using the CCM, we performed an analysis of the difference between the number of DNA linkers duplexed on each particle type for each equilibrium lattice predicted by the CCM. A plot of the difference squared between these values is shown in Fig. 5A. The results indicate large deviations toward the regions of small linker and size ratios. Therefore, at these locations there is a large fraction of DNA linkers on one particle that have not hybridized and are free to seek out additional interaction. We found that by defining a cutoff of 0.5 for the ratio of the number of duplexed DNA on one particle to the other, we were able to predict the regions of cluster formation (Fig. 5B) that are in agreement with recent experimental and simulation results reported by Li et al. (8). Apparently, such asymmetric binding of DNAs across the two colloids lead to cluster formation, thus frustrating crystal formation. Although this result is interesting, coupling the CCM to a kinetic model (such as a phase field calculation) might allow us to more fully characterize the kinetics of the crystallization events.
Fig. 5.
Symmetry analysis: (A) Deviations away from a balanced number of bound DNAs on the two colloids. Large deviations are observed toward the small size and linker ratio regions. (B) Incorporation of the symmetry analysis into CCM predictions. We predict cluster formation around the phase boundaries, in agreement with simulations from Li et al. (8) (squares).
Additionally, we note that the definition of a crystal in the contact model uses the number of A- and B-type colloids in the unit cell and also the number of complementary neighbors. We propose here that this definition is not unique for identifying a crystal structure. The crystal structure database contains several prototypes that have the same number of nearest complementary neighbors and the same number of atoms in the unit cell. However, they do differ in the total number of nearest neighbors/coordination numbers, which are not parameters in the contact model. This can be seen clearly from Table 1. For example, the contact model does not distinguish between the CsCl and the CuAu structures, which of course are quite different. A model that accounts for distances between different colloid pairs in a crystal and introduces a distance-dependent potential might be of particular use.
Table 1.
Degeneracies from the crystal database
| Crystal | 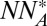 |
NNB | 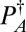 |
PB |  |
CNB |
| CsCl | 8 | 8 | 1 | 1 | 14 | 14 |
| CuAu | 8 | 8 | 1 | 1 | 12 | 12 |
| AlB2 | 6 | 12 | 2 | 1 | 12 | 9 |
| EuGa2 | 6 | 12 | 2 | 1 | 20 | 9 |
| Cr3Si | 4 | 12 | 3 | 1 | 14 | 12 |
| Cu3Au | 4 | 12 | 3 | 1 | 12 | 12 |
NNi nearest neighbor to particle i.
Pi number of particle of type i in unit cell (normalized).
CNi coordination number of particle i.
Conclusion
In summary, we show that the GA is a powerful reverse-modeling approach for designing ssDNA-grafted colloids that will assemble into a desired crystal structure. The proposed framework, coupled to the forward CCM, is not only able to successfully reproduce the experimentally validated phase diagram proposed by Mirkin but also elucidates the formation of four previously unobserved crystal structures. The GA framework, however, is limited by the predictive power of the CCM. We believe that systematic improvements to the CCM, which are discussed, are necessary to fully realize the predictive capability of the approach presented here.
Materials and Methods
Genetic Algorithm Framework.
The detailed steps performed by the framework are: (i) Initialization. The initial population consists of 100 entities with variations in the two parameters that define different crystal structures in the CCM: hydrodynamic size ratio varies between 0 and 1, and DNA linker ratio varies between 0 and 3. These parameters are evenly distributed in this phase space to capture the diversity of the original gene pool and to avoid any bias in the setup of the initial population. (ii) Prediction. The second step involves the use of the crystal structure database, along with the contact model to predict the superstructures formed by the population of 100 pairs of particles. In particular, the stable crystal structure Ss (crystal structure corresponding to the maximum percentage of duplexes) for each entity is identified based on the following equation:  , with Ji representing the number of DNA duplexes formed by the ith member of the population. (iii) Fitness evaluation. Let D represent the fraction of DNA duplexed corresponding to the desired crystal structure,
, with Ji representing the number of DNA duplexes formed by the ith member of the population. (iii) Fitness evaluation. Let D represent the fraction of DNA duplexed corresponding to the desired crystal structure,  and
and 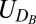 denote the number of atoms of type A and type B in a unit cell corresponding to the desired crystal and
denote the number of atoms of type A and type B in a unit cell corresponding to the desired crystal and  and
and 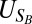 denote the number of atoms of type A and type B in the predicted unit cell. The objective function value for each entity in the population is a sum of squares deviation from this desired structure, namely,
denote the number of atoms of type A and type B in the predicted unit cell. The objective function value for each entity in the population is a sum of squares deviation from this desired structure, namely,  . Thus, when the objective function is zero we have the parameter values that correspond to the desired superstructure, i.e., the fittest superstructure. (iv) Adaptation and optimization. The objective is to obtain the hydrodynamic size ratio and DNA linker ratio for the crystal structure that correspond to an objective function value of zero. To achieve this goal, the initial population is modified using reproduction, cross-over, and/or mutation operators to create a new generation containing children entities. To create the new generation, each entity in the current population is given a rank value r according to their fitness value with highest fit entity having a rank value = 1. The current population is then provided a scaled score equivalent to the inverse of square root of the rank (scaled score
. Thus, when the objective function is zero we have the parameter values that correspond to the desired superstructure, i.e., the fittest superstructure. (iv) Adaptation and optimization. The objective is to obtain the hydrodynamic size ratio and DNA linker ratio for the crystal structure that correspond to an objective function value of zero. To achieve this goal, the initial population is modified using reproduction, cross-over, and/or mutation operators to create a new generation containing children entities. To create the new generation, each entity in the current population is given a rank value r according to their fitness value with highest fit entity having a rank value = 1. The current population is then provided a scaled score equivalent to the inverse of square root of the rank (scaled score  ) (28). This step is followed by the selection procedure where the parents of the new generation are selected using stochastic universal sampling (SUS) algorithm (28). SUS operates by laying out a line in which each entity corresponds to a section of the line of length proportional to the scaled value. This selection algorithm provides a chance for entities with lesser fitness value to be selected, thereby maintaining the population diversity, and selecting higher numbers of parents with high fitness values. Once the parents of the next generation are selected, two of the parents (elite parents) with best fitness functions are retained (reproduced) in the next generation. The fraction of the children (apart from the elite ones) obtained from cross-over operation is set to 0.8. Cross-over between two parents is performed using the scattered cross-over algorithm with a random binary decision vector. Finally, the mutation fraction value of 0.2 is used to obtain the rest of the children in the new generation. These values have been found to be effective based on prior studies (21, 29, 30). Mutation is performed by the adaptive mutation algorithm, which chooses a random step size and direction to modify the individual parents taking into account the bounds on the variables. A description of the adaptive mutation algorithms is provided in (31). The algorithm terminates when there is no further decrease in the objective function value for consecutive generations.
) (28). This step is followed by the selection procedure where the parents of the new generation are selected using stochastic universal sampling (SUS) algorithm (28). SUS operates by laying out a line in which each entity corresponds to a section of the line of length proportional to the scaled value. This selection algorithm provides a chance for entities with lesser fitness value to be selected, thereby maintaining the population diversity, and selecting higher numbers of parents with high fitness values. Once the parents of the next generation are selected, two of the parents (elite parents) with best fitness functions are retained (reproduced) in the next generation. The fraction of the children (apart from the elite ones) obtained from cross-over operation is set to 0.8. Cross-over between two parents is performed using the scattered cross-over algorithm with a random binary decision vector. Finally, the mutation fraction value of 0.2 is used to obtain the rest of the children in the new generation. These values have been found to be effective based on prior studies (21, 29, 30). Mutation is performed by the adaptive mutation algorithm, which chooses a random step size and direction to modify the individual parents taking into account the bounds on the variables. A description of the adaptive mutation algorithms is provided in (31). The algorithm terminates when there is no further decrease in the objective function value for consecutive generations.
Complementary Contact Model.
Original model.
A brief synopsis of the original description of the CCM calculations is provided (for further details see ref. 7). Given the number of DNA bases on each DNA linker, the maximum hydrodynamic radius of the ssDNA colloid is Rmax = rNP + 0.34x + 0.4, where rNP is the colloid radius, x is the number of DNA bases in the dsDNA regime of the linker, and 0.4 is the length of the short ssDNA strand between the colloid surface and the start of the dsDNA on the DNA linker. An interparticle distance is calculated of the form dAB = rNP,A + rNP,B + 0.255x + 0.8. Here, the value of 0.255 is used for the relaxed length of the dsDNA. With these values, the overlap surface area between the two ssDNA colloid fuzzy spheres and the number of DNA linkers that facilitate the interactions between the two colloids are determined. The fraction of duplexed DNA linkers is then calculated as
 |
where Aoverlap is the overlap area of particle i to the complementary particle, NNi is the number of nearest neighbors of particle i in the unit cell, LT,i is the total number of DNA linkers grafted onto particle i, and NPi is the number of particles of type i in the unit cell. The smaller of the two values calculated serves as the limiting factor and is used to determine the percent of DNA duplexes formed for each particle within the unit cell. This minimal selection emulates the idea that not all of the grafted DNA linkers will eventually hybridize. The process is repeated for all crystal lattices defined within the database, and the structure with the highest percent of DNA linkers duplexed is deemed most stable. This structure is selected by the CCM as the resulting superlattice for the given design parameters.
Symmetry analysis.
To define the locations of amorphous structures (cluster formation), we perform a symmetry analysis in the following manner. First, we take the square of the difference between the number of DNA linkers duplexed, di, calculated for the most stable crystal lattice as defined by the CCM. For the case of the binary systems of A and B particles in this study, this becomes d2 = (dA − dB)2. As shown in Fig. 5A, the deviations in duplexed linkers between the two particles tends to dramatically increase for regions of low linker or size ratios. Using this inherent asymmetry within the system, we define a ratio of the form:
 |
Thus, as R decreases the asymmetry within the system will increase. Defining a value of R = 0.5 as the cutoff for crystal formation, we are now able to quantitatively define a relative energy barrier the system needs to overcome to avoid being trapped in local energy minima that result in kinetic traps and cluster formation (i.e., R < 0.5 → clusters). Note that we use the number rather than percent of DNA linkers duplexed as the criterion for the most stable structure for the results shown in Fig. 5B.
Acknowledgments
Research at Columbia University (B.S., T.V., S.K., and V.V.) was supported by the US Department of Energy (DOE), Office of Basic Energy Sciences (BES), Division of Materials Sciences and Engineering under Award DE-FG02-12ER46909. Y.Z. and O.G. carried out the experimental component of this research at the Center for Functional Nanomaterials, Brookhaven National Laboratory, which is supported by the DOE, BES under Contract DE-AC02-98CH10886.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.J.d.P. is a guest editor invited by the Editorial Board.
References
- 1.Nykypanchuk D, Maye MM, van der Lelie D, Gang O. DNA-based approach for interparticle interaction control. Langmuir. 2007;23(11):6305–6314. doi: 10.1021/la0637566. [DOI] [PubMed] [Google Scholar]
- 2.Nykypanchuk D, Maye MM, van der Lelie D, Gang O. DNA-guided crystallization of colloidal nanoparticles. Nature. 2008;451(7178):549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 3.Xiong HM, van der Lelie D, Gang O. Phase behavior of nanoparticles assembled by DNA linkers. Phys Rev Lett. 2009;102(1):015504. doi: 10.1103/PhysRevLett.102.015504. [DOI] [PubMed] [Google Scholar]
- 4.Xiong HM, van der Lelie D, Gang O. DNA linker-mediated crystallization of nanocolloids. J Am Chem Soc. 2008;130(8):2442–2443. doi: 10.1021/ja710710j. [DOI] [PubMed] [Google Scholar]
- 5.Knorowski C, Burleigh S, Travesset A. Dynamics and statics of DNA-programmable nanoparticle self-assembly and crystallization. Phys Rev Lett. 2011;106(21):215501. doi: 10.1103/PhysRevLett.106.215501. [DOI] [PubMed] [Google Scholar]
- 6.Kim AJ, Biancaniello PL, Crocker JC. Engineering DNA-mediated colloidal crystallization. Langmuir. 2006;22(5):1991–2001. doi: 10.1021/la0528955. [DOI] [PubMed] [Google Scholar]
- 7.Macfarlane RJ, et al. Nanoparticle superlattice engineering with DNA. Science. 2011;334(6053):204–208. doi: 10.1126/science.1210493. [DOI] [PubMed] [Google Scholar]
- 8.Li TI, Sknepnek R, Macfarlane RJ, Mirkin CA, de la Cruz MO. Modeling the crystallization of spherical nucleic acid nanoparticle conjugates with molecular dynamics simulations. Nano Lett. 2012;12(5):2509–2514. doi: 10.1021/nl300679e. [DOI] [PubMed] [Google Scholar]
- 9.Park SY, et al. DNA-programmable nanoparticle crystallization. Nature. 2008;451(7178):553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 10.Leunissen ME, et al. Switchable self-protected attractions in DNA-functionalized colloids. Nat Mater. 2009;8(7):590–595. doi: 10.1038/nmat2471. [DOI] [PubMed] [Google Scholar]
- 11.Hsu CW, Largo J, Sciortino F, Starr FW. Hierarchies of networked phases induced by multiple liquid-liquid critical points. Proc Natl Acad Sci USA. 2008;105(37):13711–13715. doi: 10.1073/pnas.0804854105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu CW, Sciortino F, Starr FW. Theoretical description of a DNA-linked nanoparticle self-assembly. Phys Rev Lett. 2010;105(5):055502. doi: 10.1103/PhysRevLett.105.055502. [DOI] [PubMed] [Google Scholar]
- 13.Mognetti BM, Leunissen ME, Frenkel D. Controlling the temperature sensitivity of DNA-mediated colloidal interactions through competing linkages. Soft Matter. 2011;8:2213–2221. [Google Scholar]
- 14.Dai W, Kumar SK, Starr FW. Universal two-step crystallization of DNA-functionalized nanoparticles. Soft Matter. 2010;6:6130–6135. [Google Scholar]
- 15.Li TI, Sknepnek R, Olvera de la Cruz M. Thermally active hybridization drives the crystallization of DNA-functionalized nanoparticles. J Am Chem Soc. 2013;135(23):8535–8541. doi: 10.1021/ja312644h. [DOI] [PubMed] [Google Scholar]
- 16.Rogers WB, Crocker JC. Direct measurements of DNA-mediated colloidal interactions and their quantitative modeling. Proc Natl Acad Sci USA. 2011;108(38):15687–15692. doi: 10.1073/pnas.1109853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varilly P, Angioletti-Uberti S, Mognetti BM, Frenkel D. A general theory of DNA-mediated and other valence-limited colloidal interactions. J Chem Phys. 2012;137(9):094108. doi: 10.1063/1.4748100. [DOI] [PubMed] [Google Scholar]
- 18.Leunissen ME, Frenkel D. Numerical study of DNA-functionalized microparticles and nanoparticles: Explicit pair potentials and their implications for phase behavior. J Chem Phys. 2011;134(8):084702. doi: 10.1063/1.3557794. [DOI] [PubMed] [Google Scholar]
- 19.Dreyfus R, et al. Simple quantitative model for the reversible association of DNA coated colloids. Phys Rev Lett. 2009;102(4):048301. doi: 10.1103/PhysRevLett.102.048301. [DOI] [PubMed] [Google Scholar]
- 20.Sundaram A, Ghosh P, Caruthers J, Venkatasubramanian V. Design of fuel additives using neural networks and evolutionary algorithms. AIChE J. 2001;47(6):1387–1406. [Google Scholar]
- 21.Sundaram A, Venkatasubramanian V. Parametric sensitivity and search-space characterization studies of genetic algorithms for computer-aided polymer design. J Chem Inf Comp Sci. 1998;38(6):1177–1191. [Google Scholar]
- 22.Katare S, Venkatasubramanian V, Caruthers J, Delgass W. A hybrid genetic algorithm for efficient parameter estimation of large kinetic models. Comput Chem Eng. 2004;28(12):2569–2581. [Google Scholar]
- 23.Stucke D, Crespi V. Predictions of new crystalline states for assemblies of nanoparticles: Perovskite analogues and 3-d arrays of self-assembled nanowires. Nano Lett. 2003;3:1183–1186. [Google Scholar]
- 24. Doppelbauer G, Bianchi E, Kahl G (2010) Self-assembly scenarios of patchy colloidal particles in two dimensions. J Phys Condens Mat 22(10):104105. [DOI] [PubMed]
- 25. Nikoubashman A, Likos CN (2010) Self-assembled structures of Gaussian nematic particles. J Phys Condens Mat 22(10):104107. [DOI] [PubMed]
- 26.Filion L, Dijkstra M. Prediction of binary hard-sphere crystal structures. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;79(4 Pt 2):046714. doi: 10.1103/PhysRevE.79.046714. [DOI] [PubMed] [Google Scholar]
- 27. Fornleitner J, Kahl G (2008) Lane formation vs. cluster formation in two-dimensional square-shoulder systems—a genetic algorithm approach. EPL 82(1):18001.
- 28.Gen M, Cheng R. Genetic Algorithm and Optimization. John Wiley and Sons; 2000. [Google Scholar]
- 29.Venkatasubramanian V, Chan V, Caruthers J. Computer-aided molecular design using genetic algorithms. Comput Chem Eng. 1994;18(9):833–844. [Google Scholar]
- 30. Venkatasubramanian V, Chan V, Caruthers J (1995) Evolutionary design of molecules with desired properties using the genetic algorithm. J Chem Inf Comp Sci 35:188–195.
- 31.Thierens D. Adaptive mutation rate control schemes in genetic algorithms. P Evol Comput. 2002;1:980–985. [Google Scholar]