Significance
Analysis of protein packing reveals the prevalence of cavities, some of which are evolutionarily conserved. Despite the fact that these packing defects are generally destabilizing, they can play important roles in facilitating functionally important protein motions and in the evolution of protein function. In this study, we used site-directed spin-labeling with electron paramagnetic resonance spectroscopy to investigate the structural and dynamical response of an enzyme to engineered ligand-binding cavities. The results show that the engineered cavities introduce conformational fluctuations between a native-like state and a new state. Ligand binding shifts the equilibrium toward the native-like state, suggesting binding via conformational selection, although additional structural adaptation is observed for one mutant. The results underscore the potential role of cavities in modulating molecular flexibility.
Keywords: EPR, site-directed spin labeling, DEER, benzene, saturation recovery
Abstract
The studies presented here explore the relationship between protein packing and molecular flexibility using ligand-binding cavity mutants of T4 lysozyme. Although previously reported crystal structures of the mutants investigated show single conformations that are similar to the WT protein, site-directed spin labeling in solution reveals additional conformational substates in equilibrium exchange with a WT-like population. Remarkably, binding of ligands, including the general anesthetic halothane shifts the population to the WT-like state, consistent with a conformational selection model of ligand binding, but structural adaptation to the ligand is also apparent in one mutant. Distance mapping with double electron-electron resonance spectroscopy and the absence of ligand binding suggest that the new substates induced by the cavity-creating mutations represent alternate packing modes in which the protein fills or partially fills the cavity with side chains, including the spin label in one case; external ligands compete with the side chains for the cavity space, stabilizing the WT conformation. The results have implications for mechanisms of anesthesia, the response of proteins to hydrostatic pressure, and protein engineering.
Globular proteins have overall packing densities similar to those of crystalline solids (1) but nevertheless contain cavities and pockets that can range from a few to hundreds of cubic angstroms (2, 3). These native packing defects are generally destabilizing (4, 5), and improving the packing of the protein interior may be a general way to increase protein stability. Indeed, small-to-large mutations that fill native cavities can increase stability (6–8), but with a concomitant loss of function (7, 8). Thus, cavities in the protein interior can play a critical role in function. One role of cavities may be to allow alternative packing arrangements of the core. The resulting ensemble of conformational substates could give rise to promiscuity in protein–protein interactions (9) and allosteric behavior (7, 8, 10). In addition, large-to-small mutations that generate cavities may have played an important role in the evolution of protein function (11).
Considering the potentially important role of cavities in sculpting the energy landscape of proteins, the present study was undertaken to investigate, in solution, the structural and dynamical response of a protein to engineered cavities introduced by large-to-small mutations. T4 lysozyme (T4L) was selected for study because of the large database of crystal structures of ligand-binding cavity mutations and the corresponding thermodynamic characterization from Matthews and coworkers (4, 12–15). Remarkably, comparison of the WT and mutant structures showed very little difference or limited local relaxation near the cavity, although the mutations were strongly destabilizing (4, 15).
In the present study, we examined the cavity-creating mutants L121A/L133A, L133G, and W138A in solution, free from the confines of the crystalline lattice, using site-directed spin labeling together with continuous wave (CW), pulsed saturation recovery (SR), and double electron-electron resonance (DEER) methods. The CW spectra and SR reveal the existence of multiple substates in equilibrium as well as the time domain for exchange between them. DEER spectroscopy measures interspin distance distributions between pairs of spin labels, and thereby directly reveals the structural heterogeneity in an equilibrium mixture and provides clues to the structure of the contributing species. Collectively, the CW, SR, and DEER data suggest a model in which the engineered cavities populate new substates of the protein that are in equilibrium with a native-like conformation; the new substates appear to be alternative packing arrangements that fill or partially fill the cavity. Binding of nonendogenous ligands to the engineered cavities shifts the equilibrium toward a native-like state, generally consistent with a conformational selection model of ligand binding; however, for the W138A mutant, the DEER data indicate additional structural adaptation upon ligand binding. Plausible structural models for the new substates, which are not seen in the crystal structures, are discussed.
Experimental Strategy and Results
Three large-to-small mutations were introduced at buried sites in the C-terminal domain of T4L to generate cavities. T4L mutants L121A/L133A and L133G have known crystal structures (14, 15); the engineered cavities are large (>170 Å3) and bind nonpolar ligands such as benzene (14). Although a crystal structure has not been reported for W138A, it apparently does not bind benzene (14). However, modeling shows that W138A should generate a new large cavity (∼233 Å3) capable of ligand binding. A model of the W138A cavity and models of the X-ray structures showing the cavities of the other mutants are provided in Fig. S1.
For each of the cavity mutants, a nitroxide side chain designated R1 (Fig. 1A, Inset) was introduced at solvent-exposed sites to serve as a sensor of local structure. At such sites, R1 typically has weak or no interactions with the protein, as reflected by a single-component electron paramagnetic resonance (EPR) spectrum, and introduces little structural perturbation (16–19). The R1 sensor sites were selected to monitor the helix in which the cavity-creating mutation was introduced, or in a neighboring helix that shares the cavity. Where relevant, the selection was guided by the corresponding crystal structure that revealed subtle changes due to the presence of the cavity. In particular cases, R1 sites distal to the cavity were selected to look for global changes. For simplicity, we will refer to any T4L species without the native cysteines (the so-called “pseudo-WT”) and without a cavity mutation as WT′. This includes species containing spin labels at nonperturbing solvent-exposed sites.
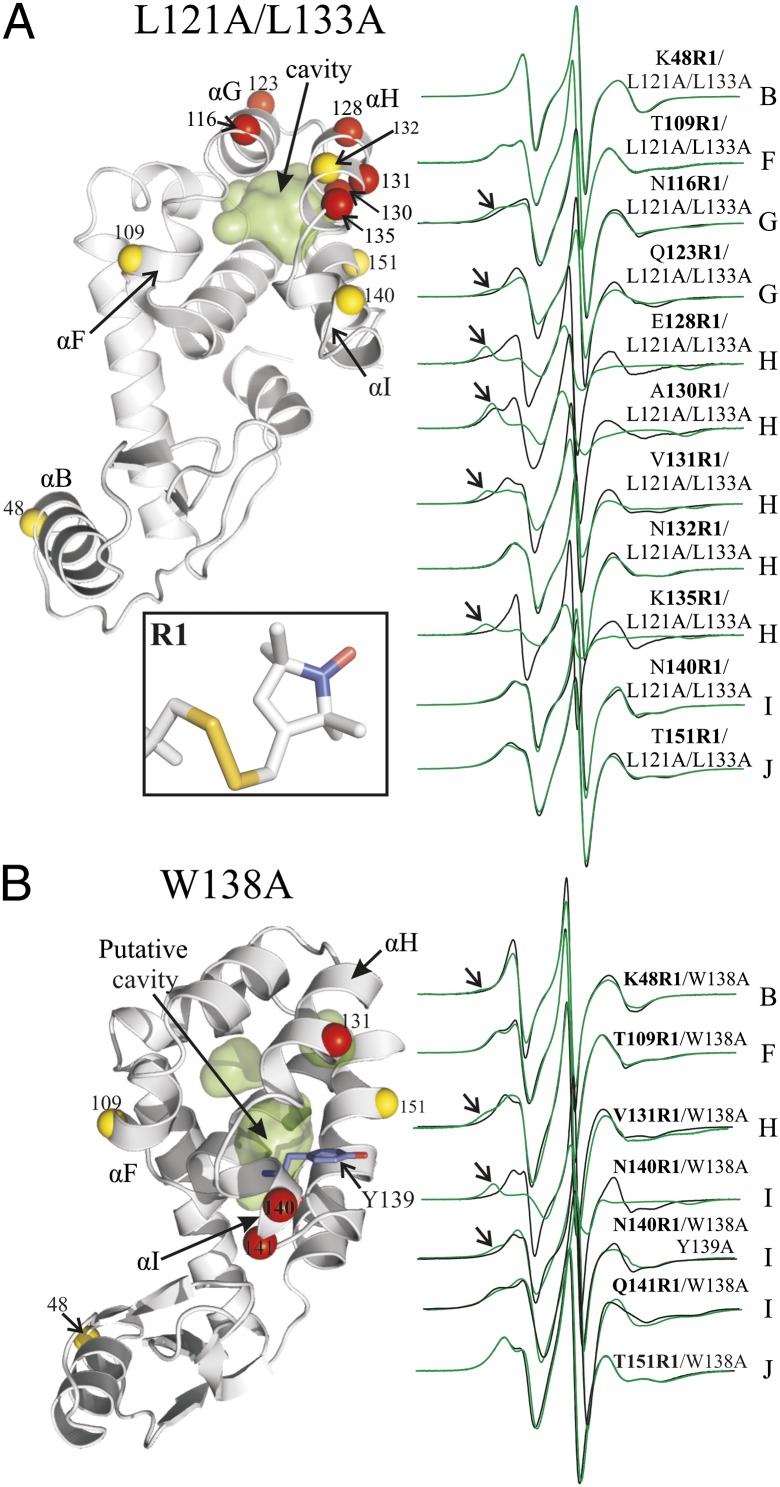
Fig. 1.
Cavity mutants introduce new conformational states in solution. (A) L121A/L133A. (B) W138A. (Left) Ribbon models of the T4L cavity mutants (PDB ID code 251L for L121A/L133A; for W138A, the cavity was modeled using PDB ID code 3LZM). The surfaces of the cavities are shown in green. The spheres at Cα identify the sites where R1 was introduced, one at a time. Red spheres identify sites where spectral changes were observed between the WT and the cavity mutants; yellow spheres indicate the sites with no change. Residue Y139 is shown as a stick representation in the W138A mutant. (Right) EPR spectra of the indicated sites in the WT′ (black) and cavity mutants (green). Arrows identify relatively immobile components. (Inset) Structure of R1 side chain.
CW EPR Spectra of R1 Reveal New Conformational Substates in the Cavity-Creating Mutations.
T4L L121A/L133A and L133G.
Crystal structures show that the L121A/L133A and L133G mutations merge two native cavities into a single large cavity with volumes of 180 and 176 Å3, respectively (Fig. S1). Compared with the WT protein, the structural changes due to the L133G and L121A/L133A mutations were similar and subtle (≤0.8 Å), and limited to the helical bundle (helices E–J) around the cavity (14, 15). Both mutants bind nonpolar ligands such as benzene with approximately millimolar affinity (14).
Fig. 1A shows a model of the L121A/L133A mutant based on the crystal structure, along with the location of R1 sensor sites and the spectra compared with those of the WT′. The spectra for the WT′ protein have been published previously and interpreted in terms of the protein structure and dynamics (16–20); the basic principles of spectral analysis are outlined in SI Materials and Methods. The spectra of 131R1, 132R1, 140R1, and 151R1 in the WT′ reflect a simple anisotropic motion characteristic of R1 at solvent-exposed sites in relatively rigid helical segments, whereas the spectra of 48R1, 109R1, 123R1, 128R1, and 135R1 reflect higher mobility of the nitroxide consistent with their location near or at the helix termini (16). The spectra of 109R1 and 116R1 have two components; for 109R1, the two components apparently arise from different rotamers of R1 (21). Residue 130R1 is unique among the sites investigated in that it is located at a contact site between helices H and J, where the nitroxide interacts with the protein, as revealed by the presence of a second spectral component reflecting immobilization (arrows, Fig. 1A). This residue plays a special role in the present study and will be considered separately from the other sensor sites.
In the 121A/133A cavity mutant, new spectral components corresponding to immobilized states of the nitroxide appear for sensor mutants in helices G and H, which surround the cavity (116, 123, 128, 131, 135), but not at sites in helices B, F, I, and J (48, 109, 140, 151) or at site 132R1 in helix H. The absence of a spectral change for 132R1 is of particular interest and shows that introduction of the cavity, although in immediate contact with helix H, does not lead to a change in the secondary structure or activation of fast backbone dynamic modes (20, 22). In each case where an immobilized state appears, it unequivocally identifies a new substate(s) of the protein in solution in which the nitroxide has interactions with the environment. The L133G mutant, monitored by sensors in helices H (131R1), I (151R1), and F (109R1), gave similar results to the L121A/133A mutant, as anticipated from the similarity of the cavities (Fig. S2).
The EPR spectra of R1 in 121A/133A can be simulated by a one- or two-component macroscopic order microscopic disorder (MOMD) model (23) to give the populations, orders, and rates of nitroxide motion for each component (Fig. S3). In the case of two-component spectra, the component corresponding to the highest mobility has the same dynamic parameters as the WT′, suggesting equilibrium between substates, one of which has a WT′-like conformation (Fig. S3). It should be noted that the substate identified by the immobilized nitroxide may, in fact, itself be heterogeneous, because spectral resolution with respect to motion is poor in this slow-motion regime for X-band EPR. However, treating the spectra as two-state (mobile and immobile) will not change the conclusions of this study.
From the simulations, the immobilized components amount to ≈50% of the population for 116R1, 123R1, and 131R1 and to ≈90% for 128 and 135. However, the R1–protein interactions that give rise to the immobilized states in the cavity mutants are weakly attractive, as judged by increases in the melting temperature (Tm) for reversible unfolding due to the introduction of the R1 side chain (Table S1). The attractive interaction presumably biases the population, and spectral analysis likely overestimates the true populations of the state giving rise to the immobilized component (Discussion).
T4L W138A.
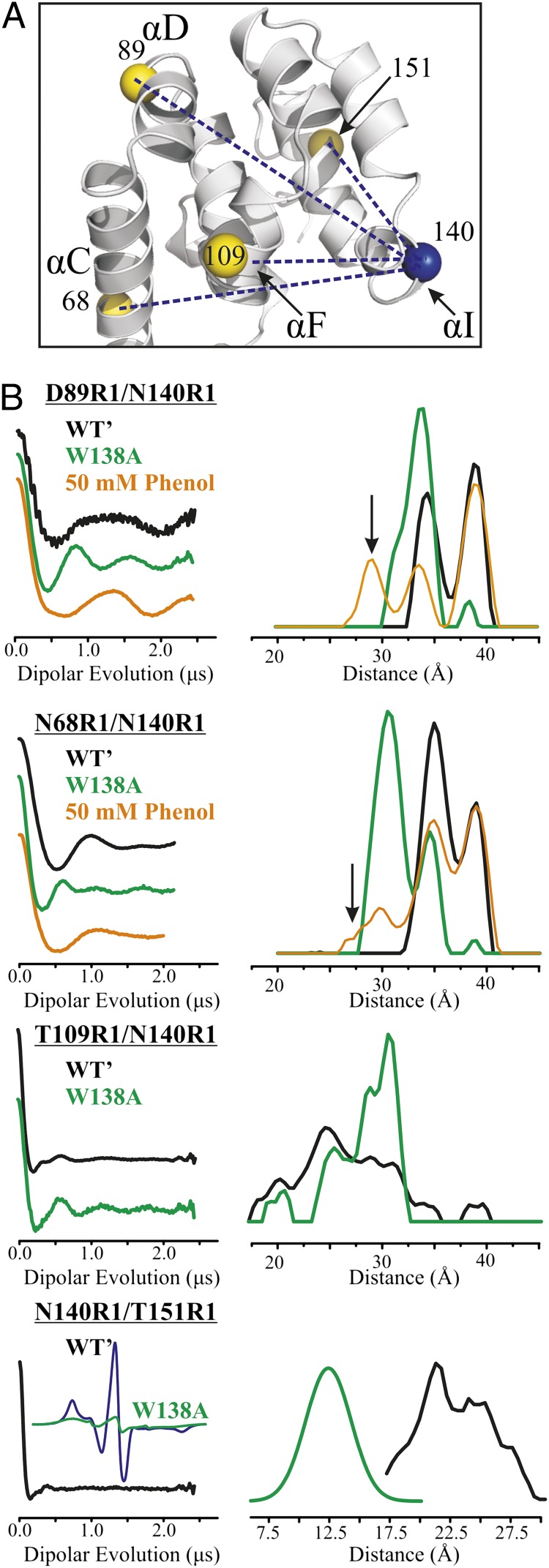
Modeling of T4L W138A suggests that the mutation should create an internal cavity between helices H, I, and J (Fig. S1). Thus, R1 sensors were placed in each of these helices, as well as in remote helices B and F (Fig. 1B). The EPR spectra of 131R1 (helix H) and 140R1 (helix I) show the appearance of a new immobile component in the mutant relative to the WT′, whereas those for 151R1 (helix J) and 109R1 (helix F) are essentially unchanged, suggesting a localized nature of the effect. Spectral simulations show that the component in the EPR spectrum of 140R1/138A corresponding to the more mobile state is essentially identical to that of the WT′ (Fig. S3); the immobile component strongly dominates in the case of 140R1. Although residue 48R1 in helix B is nearly 30 Å from the cavity, a minor population (∼5%) of a relatively immobile component appears in the cavity mutant. Importantly, mutation Y139A in the 138A protein strongly suppresses the formation of the immobilized component in the 140R1 spectrum (Fig. 1B), indicating a key role for this residue in stabilizing the new state, as will be discussed below.
Pulsed SR EPR Confirms Conformational Exchange Between Substates in the 121A/133A Cavity Mutant.
The appearance of two-component spectra in the cavity mutants clearly signals a structural change in the protein, but the origin of the two components could either be due to two rotamers of the R1 side chain in a single new conformation or to two conformations of the protein. These cases can apparently be distinguished by SR EPR methods, as discussed in detail elsewhere (21, 24) and outlined in SI Materials and Methods. Briefly, SR measures the recovery of the EPR signal following a saturating microwave pulse applied to the maximum absorbance of the nitroxide MI = 0 resonance (Fig. 2). For two spectral components arising from rotamer exchange, the recovery has been found to be monoexponential due to fast exchange on the nanosecond time scale. On the other hand, for slow protein conformational exchange, the relaxation is biexponential and SR data in the presence of various concentrations of a relaxation reagent nickel ethylenediaminediacetate (NiEDDA) can be analyzed in terms of the exchange lifetime in the range of 1–70 μs. In addition, the data provide a measure of the residue solvent accessibility (21, 25).
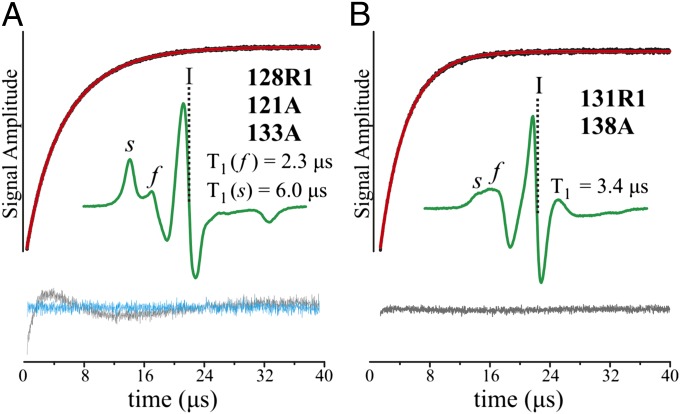
Fig. 2.
SR curves for R1 in T4L cavity mutants. (A) 128R1/121A/133A. The plot shows the signal intensity recorded at position I (vertical dashed lines) in the first derivative spectrum as a function of time following a saturating microwave pulse delivered at the same position (black trace); position I corresponds to the maximum of the absorption spectrum. Also shown is the fit to a double exponential (red trace). The values of T1 for the slowly relaxing [T1(s)] and fast-relaxing [T1(f)] components are given; these correspond to the s and f spectral components. The residuals for a single-component and double-component fit are shown as gray and cyan traces, respectively. (B) Same representation for 131R1/138A. The fit to a single exponential and residuals from the fit are shown.
In the present study, five mutants were selected in the 121A/133A background for SR analysis, namely, 116R1 and 123R1 in helix G and 128R1, 131R1, and 135R1 in helix H. In each case, the SR recovery curves are biexponential, suggesting that the spectra have contributions from conformational exchange; an example for 128R1 is shown in Fig. 2A. For 123R1 and 128R1, analysis of the recovery curves in the presence of various concentrations of the relaxation reagent NiEDDA (21) shows the exchange lifetimes to be ≥70 μs and numerical values cannot be determined. Solvent accessibility determined from SR revealed that the immobilized states are solvent-inaccessible for the five sites investigated (Table S2).
For the T4L 138A cavity mutant, 131R1 and 140R1 were investigated by SR. Despite the two-component nature of the CW spectra, the SR curves are monoexponential, suggesting that the two spectral components arise from R1 rotamer exchange in a new conformational state that is nearly completely populated (≥93%); an example SR relaxation curve is shown for 131R1/138A in Fig. 2B. Solvent accessibility measurements suggest that 140R1 remains solvent-exposed in the cavity mutant, although it is relatively immobilized (Table S2).
Ligand Binding Shifts Conformational Equilibria in the Cavity Mutants Toward the WT′ State.
T4L L121A/L133A.
To investigate the effect of hydrophobic ligand binding on the conformational equilibria in the L121A/L133A mutation, the protein solution was brought into vapor equilibrium with benzene, m-xylene, or anisole at 295K, resulting in a free ligand concentration in solution equal to the corresponding saturated solution value (22 mM, 1.4 mM, and 14.8 mM, respectively). At these concentrations, previous studies show that binding of these ligands to the analogous L133G cavity should be close to saturation (14). Binding of the general anesthetic halothane, not previously reported for T4L cavity mutants, was investigated as a function of concentration.
As examples of the effect of ligand binding, the EPR spectra of the sensors 128R1, 131R1, and 135R1 in the L121A/L133A mutation in equilibrium with benzene vapor (red trace) and 10 mM halothane (blue trace) are shown in Fig. 3 (upper three spectra); the similar effect of other ligands and binding detected by other sensor sites are shown in Fig. S4. It is apparent that ligand binding shifts the population toward the WT′-like state; indeed, the spectra of 128R1, 131R1, and 135R1 in the presence of ligands can be simulated with the same two components as the empty cavity state by variation of only the relative populations (Fig. S3). Similar population shifts upon binding of various ligands using 131R1 as a sensor were obtained for the 133G cavity mutant (Fig. S4A).
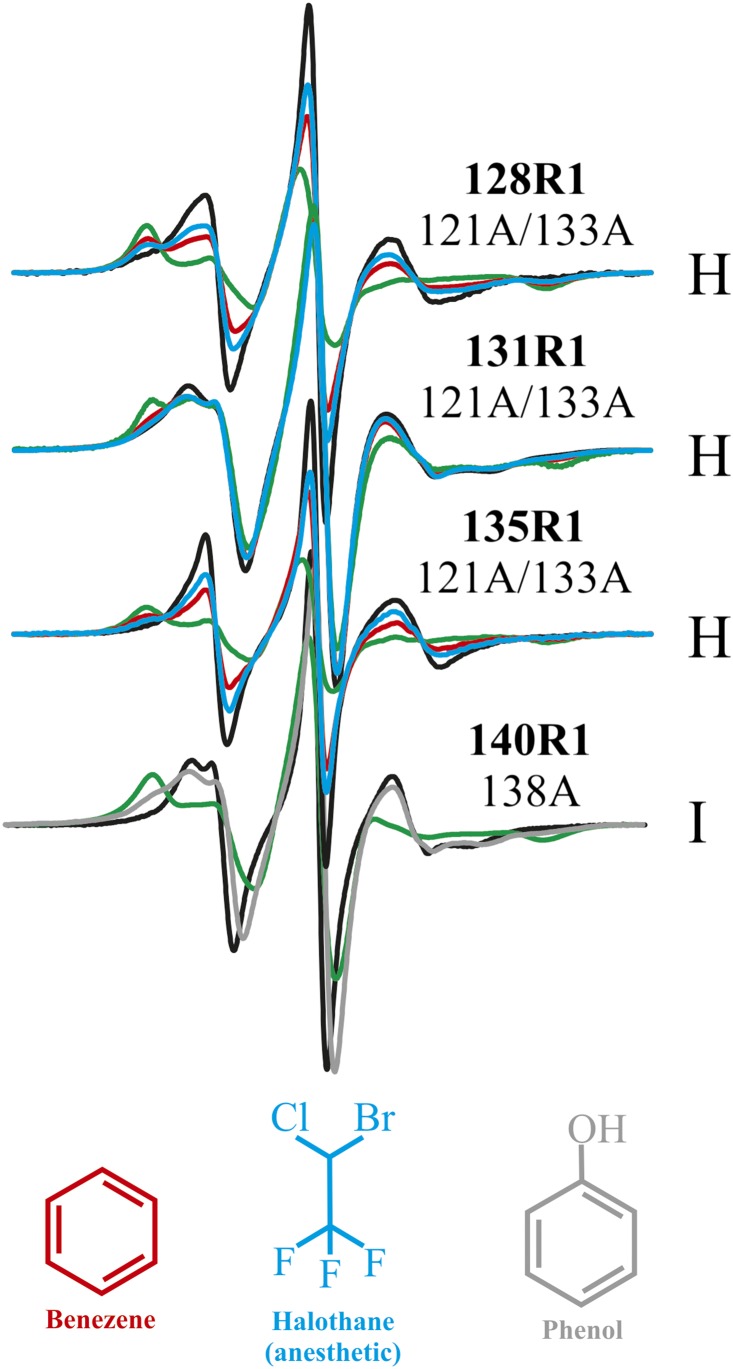
Fig. 3.
Ligand binding to the 121A/133A and 138A cavities shifts the equilibrium toward the WT′-like state. EPR spectra of R1 at the indicated sites in 121A/133A and 138A are shown for the WT′ background (black), the corresponding cavity mutant (green), and in 10 mM benzene (red), with 10 mM halothane (blue) or with 50 mM phenol (gray, for 138A).
An important conclusion from these results is that the ligands bind preferentially to the WT′-like state. Assuming a simple model wherein the ligand binds only to the WT-like state, the equilibria can be represented as
where X is the conformation corresponding to the immobile state of R1, WT′ is the WT′-like state of the cavity mutant, and L is the ligand.
For halothane binding, titrations were done using both 128R1 and 135R1 as sensors to estimate the binding affinity to the 121A/133A cavity. These sites were selected for titration studies because they show the largest spectral differences between the mutant and WT′. Best fits of the data for either sensor to the above model yields K2 = 4 × 105 (K2d ≈ 250 μM) (SI Materials and Methods and Fig. S5).
T4L W138A.
Unlike the 121A/133A mutant, W138A does not bind the hydrophobic ligand benzene (14). However, the polar ligand phenol apparently binds to the cavity, as judged by a shift in the population toward a WT′-like state, as sensed by 140R1 (Fig. 3, lower spectrum). Titration of W138A with phenol using 140R1 as a sensor gives K2 = 2.2 × 102 (K2d ≈ 4.5 mM) for binding to the WT′-like substate in the context of the above model (Fig. S5C). W138A also binds the similar ligand p-cresol with similar affinity (Fig. S4).
DEER Spectroscopy Reveals Structural Differences Between the Cavity Mutants and WT Protein, and Structural Changes upon Ligand Binding to the Cavity.
The magnitude of structural difference between conformational substates can be determined by DEER spectroscopy, which measures interspin distance distributions between two nitroxides in the range of 17–70 Å (26). To explore the structural changes due to the cavity-creating mutants, pairs of R1 side chains were introduced into both 121A/133A and 138A backgrounds. In each case, one member of the pair is a “reference” site, where the EPR spectrum of R1 is not changed by the cavity, or is located at a site distal to the cavity; the other member is located at an “active” site showing spectral changes due to the cavity-creating substitution. Results for the two cavity mutants are presented separately below.
T4L L121A/L133A.
For 121A/133A, attention was focused on the motion of helix H, with five R1 pairs involving an active site in this helix (81R1/128R1, 68R1/130R1, 81R1/132R1, 89R1/131R1, and 109R1/131R1) and a sixth pair monitoring helix G (123R1/151R1) (Fig. 4A). The pair that contains residue 130R1 will be considered separately below.
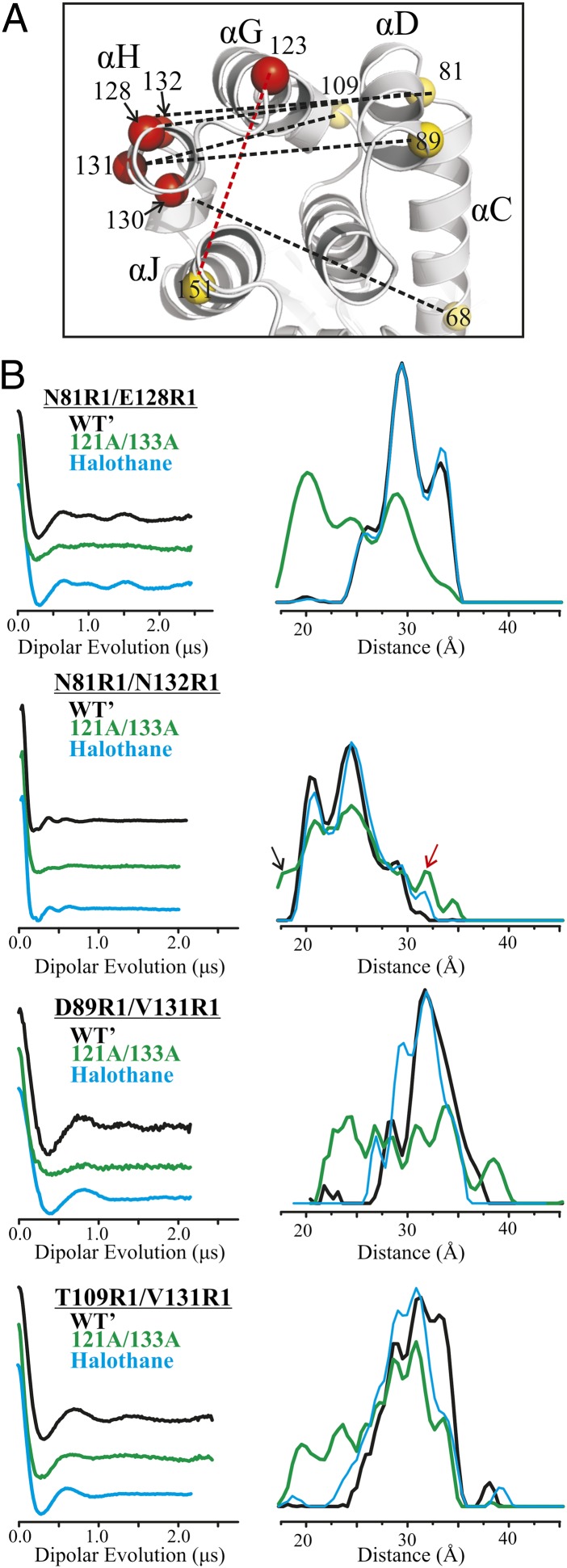
Fig. 4.
Distance measurements of doubly labeled T4L in the WT′ and 121A/133A cavity mutant. (A) Ribbon model of T4L (PDB ID code 251L) shows the sites where R1 was introduced as spheres at the Cα position; yellow and red spheres identify reference and active sites, respectively. Interspin distances measured are shown as dotted lines; black dotted lines identify the interspin distances shown in B and Fig. 5B, whereas the red dotted line identifies an additional distance measured and shown in Fig. S6A. (B) Background-corrected dipolar evolution functions (Left) and corresponding distance distributions (Right) for the indicated doubly labeled protein in the WT′ (black), cavity mutant (green), and cavity mutant with halothane added (light blue).
Fig. 4B shows DEER data in the WT′ and cavity mutant for the four pairs involving 128R1, 131R1, and 132R1, the last two of which monitor the center of helix H. In all cases, the distance distributions in the WT′ protein are multimodal, with modes that could, in principle, be accounted for by rotamers of R1 (27) but could also be due to distinct states of the host helices. Under any condition, the most probable distances are in reasonable agreement with those estimated from modeling the R1 side chain in the crystal structure of the WT protein [Protein Data Bank (PDB) ID code 3LZM] (Table S3).
In general, the distance distributions for the cavity mutant are broader than those for the WT′, and include discrete populations corresponding to the WT′ in addition to new populations shifted by 5–10 Å relative to the mode of the WT′ distribution; these new populations presumably correspond to the immobilized components in the CW spectra of R1 (Fig. 1A). In the interesting case of 132R1 in helix H, the CW line shape does not change due to the cavity, showing that the nitroxide makes no contact with the protein and that the secondary structure of the helix does not change; these conclusions are supported by the fact that the addition of R1 at site 132 does not change the thermal unfolding temperature (Table S1). Nevertheless, the distance distribution between 132R1 and a reference at 81R1 reveals new populations reflecting increased structural heterogeneity in the cavity mutant (arrows, Fig. 4B). Although these populations are small, the data are of sufficiently high quality to detect their presence reliably. In each case, addition of saturating concentrations of ligand results in a shift in the populations toward the WT′ (Fig. 4B). For halothane binding to 81R1/128R1 and 81R1/132R1, the ligand-bound distribution is essentially identical to that for the WT′; for 89R1/131R1 and 109R1/131R1, the distribution is similar but not identical to that for the WT′. Qualitatively similar population shifts were observed upon benzene binding (Fig. S6).
These results strongly support the conclusion based on the CW spectral data, namely, that in solution, the cavity mutants are in equilibrium between a WT-like conformation and another with an alternative packing arrangement, and that ligand binding shifts the equilibrium toward the WT′-like state.
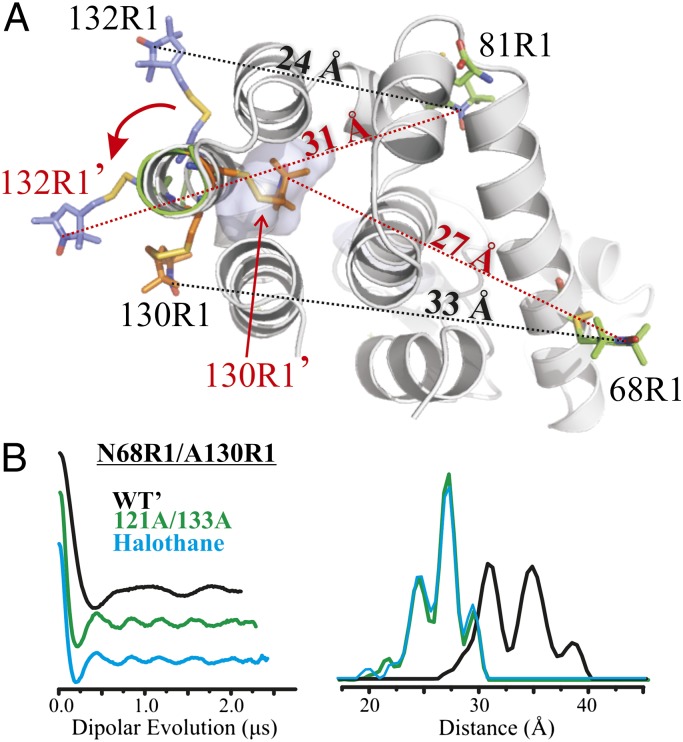
An unusual behavior of residue 130R1 in helix H offers unique insight into the structure of a conformational substate in the cavity mutant. In the WT′ protein, the nitroxide of 130R1 is at a contact site between helices H and J and close to the cavity in the 121A/133A mutation. A stick model of 130R1 based on the known crystal structure (28) in the WT′ is shown in Fig. 5A (gray ribbon), along with the cavity, whose location is shown as a transparent surface. The two-component CW EPR spectrum of 130R1 (Fig. 1A) indicates two discrete states of the nitroxide in solution. Interestingly, SR analysis showed that the two states arise from two conformations of the structure rather than two rotamers of R1 (21), a result that reveals an inherent flexibility in the H helix even in the WT′ structure.
Fig. 5.
Model for the motion of helix H in the 121A/133A mutant compatible with CW data, SR data, and interspin distance measurements. (A) Ribbon model shows an ∼90° rotation of helix H with R1 modeled at reference sites 68R1 and 81R1 and at active sites 130R1 and 132R1 in the WT′ and rotated state. Positions of R1 in the rotated state are indicated with primes, and predicted interspin distances in the rotated states are indicated on the dashed red lines and compared with the corresponding distances in the WT′ (dashed black lines). The models of sites 81R1 and 130R1 in the WT′ state were based on known crystal structures (28). For clarity, only the motion of 132R1 and 130R1 under the rotation is shown; a model illustrating the change in position for 131R1 under the rotation is shown in Fig. S6. (B) Dipolar evolution function and corresponding distance distribution of 68R1/130R1 in the WT′ (black), cavity mutant (green), and cavity mutant in the presence of halothane (cyan). Additional analysis of the model is given in Discussion.
In the cavity mutant, the 130R1 CW spectrum changes to a single component, reflecting a highly ordered state (Fig. 1A and Fig. S3). SR gives a monoexponential recovery curve with a spin lattice relaxation time (T1) for 130R1 of 6.6 μs (Table S2), which is characteristic of immobile nitroxides at buried sites in proteins (21). Moreover, the T1 is insensitive to NiEDDA up to 1.5 mM, which shows that residue 130R1 is solvent-inaccessible (SI Materials and Methods and Table S2). Remarkably, the EPR spectrum and Tm in the 130R1/121A/133A cavity mutant are insensitive to the addition of saturating concentrations of halothane (∼20 mM) (Fig. S4 and Table S1), indicating that halothane does not bind to this mutant. However, the unlabeled 130C/121A/133A variant without R1 binds halothane, as judged by the increase in the Tm in the presence of the ligand (ΔTm ∼ 5.8 °C).
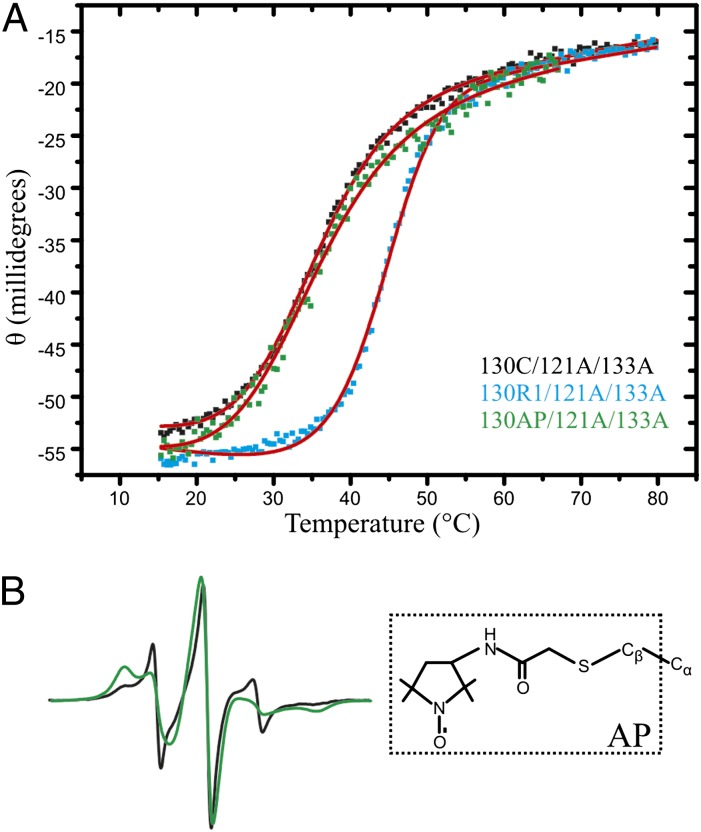
The above data suggest that the nitroxide of 130R1 is sequestered in the engineered cavity of 121A/133A. If R1 is accommodated in the cavity with little strain, the presence of R1 should stabilize the protein due to the overall reduction of cavity volume. Indeed, thermal denaturation studies showed an increase in the melting point (Tm) of ∼8.5 °C for 130R1/121A/133A compared with the same mutant without the R1 side chain (Fig. 6A and Table S1). As a comparison, the maximum increase in melting point due to R1 labeling at the other sites in helices G and H was ΔTm ≈ 3 °C (Table S1).
Fig. 6.
At site 130 in the cavity mutant, the R1 side chain, but not the AP side chain, stabilizes the new state. (A) Thermal denaturation of 130C/121A/133A, 130R1/121A/133A, and 130AP/121A/133A detected by CD. Fits for the unfolding curves are shown in red (SI Materials and Methods); the corresponding Tm values obtained from the fits can be found in Table S1. (B) EPR spectra of 130AP (black) and 130AP/121A/133A (green) are shown, along with the structure of the AP side chain.
One model for a conformational transition that would place 130R1 in the cavity of 121A/133A is a 90° clockwise rotation of helix H, as shown in Fig. 5A (green ribbon), where 130R1′ designates the intracavity position of 130R1. To evaluate this model, DEER distance measurements were made for 130R1/68R1 in both the WT′ and cavity mutant proteins. In the WT′ protein, the distance distribution is strongly bimodal (Fig. 5B), consistent with the two-component CW spectrum (Fig. 1A). The predicted distance based on models of R1 in the WT′ structure is ≈33 Å (Fig. 5A), close to the median of the measured distance distribution. In the cavity mutant, the distance distribution was narrow with a major population at 27.5 Å (Fig. 5B), in good agreement with the predicted distance for 130R1′/68R1 of ≈27 Å (Fig. 5A). As for the CW spectrum, the experimental distance distribution in 130R1/68R1 is insensitive to 20 mM halothane (Fig. 5B).
A question arises as to whether the 130R1 interaction with the protein induces the conformation that allows R1 to reside in the cavity or whether that conformation already exists and the interaction of 130R1 just strongly stabilizes the state due to favorable packing interactions. The distance distributions of 132R1/81R1, 131R1/109R1, and 131R1/89R1 (Fig. 4B) shed light on this issue; the 132R1/81R1 data are of particular interest because 132R1 has no stabilizing interactions with the protein in either the WT or cavity mutant (Fig. 1A). The model of Fig. 5A predicts that the 132R1/81R1 distance in a WT conformation should be ≈24 Å; if a rotated state of helix H already exists in the cavity mutant in the absence of 130R1, it would have a population of around 31 Å (132R1′/81R1; Fig. 5A). Remarkably, the distance distribution in the cavity mutant has a small population at 31 Å (red arrow, Fig. 4B) in addition to those at 21 and 25 Å corresponding to the WT′ conformation, suggesting that a conformation wherein 130R1 would occupy the cavity is already present at equilibrium in the cavity mutant. The distance distributions for both 131R1/109R1 and 131R1/89R1 show new populations at distances compatible with the rotated state of helix H in the cavity mutant (Fig. S6 and Table S3).
The strongly stabilizing interaction of 130R1 is presumably due to the complementary size of the R1 side chain to the hydrophobic cavity and the hydrophobic nature of the disulfide linkage and nitroxide ring of R1. If this is the case, a spin label with a more polar linkage should not enter the cavity. One such spin label is the acetamidoproxyl (AP) side chain shown in Fig. 6B, which lacks the disulfide and contains a polar amide group. The spectrum of 130AP in the WT protein has two components, similar to R1, and there is an increase in a population of a more immobilized state in the 121A/133A background. Unlike R1, the spectrum of 130AP in the cavity mutant shows two components, each of which reflects higher mobility compared with that of 130R1 in the 121A/133A mutant. SR data for T4L 130AP in the presence of NiEDDA show the nitroxide to be solvent-accessible (Table S2), and the thermal stability of the protein due to the introduction of the AP side chain is only slightly decreased (ΔTm ≈ −1°; Fig. 6A). Moreover, the 130AP protein retains the ability to bind halothane, as indicated by an increase in Tm in the presence of the ligand (Table S1). Thus, the AP side chain does not appear to stabilize the new state by occupying the cavity but, nevertheless, detects its presence via changes in the CW spectrum.
T4L W138A.
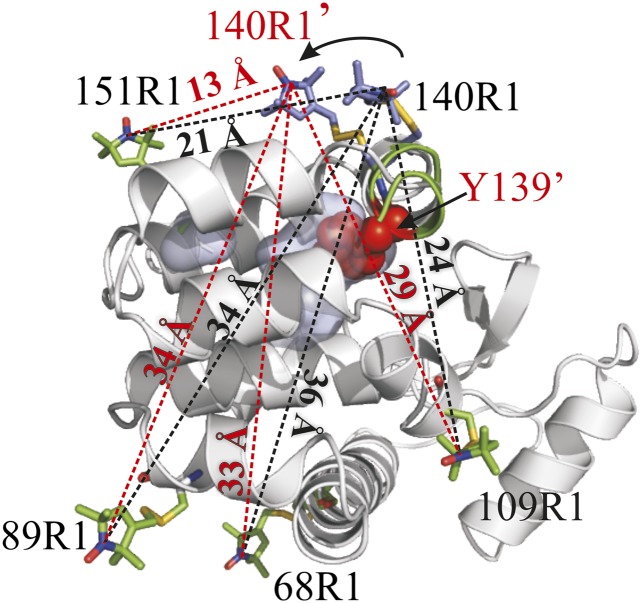
Residue 140R1 becomes predominantly immobilized in W138A (Fig. 1B), signaling the appearance of a new conformation. Nevertheless, it retains relatively high solvent accessibility, as judged by SR in the presence of NiEDDA (Table S2) and is unlikely to be sequestered in the cavity. Mutation of the solvent-exposed Y139 to alanine in the W138A background dramatically reduces the component in the 140R1 spectrum that corresponds to the immobile state (Fig. 1B), indicating an important role for Y139 in stabilizing the new conformation; the mutation Y139A has little effect on the WT protein (17). An attractive model for the new conformation is one in which Y139 confers stability by filling the cavity. This could be achieved by a simple rotation of helix I, reminiscent of the model proposed above for 130R1 filling the 121A/133A cavity via helix rotation. To examine this possibility, four R1 pairs (68R1/140R1, 89R1/140R1, 109R1/140R1, and 151R1/140R1) were constructed for DEER distance mapping (Fig. 7A). Each pair includes 140R1, where the largest spectral difference between the cavity mutant and WT′ was detected (Fig. 1B). For 140R1/151R1, the distance distribution in the cavity mutant was too short for DEER analysis and was measured instead by CW broadening methods (SI Materials and Methods). For all pairs, the WT′ distance distribution is multimodal and the shifts in the distance distributions due to introduction of the cavity (Fig. 7B) are in reasonable agreement with expectations based on a simple ∼90° rotation of helix I that places Y139 in the cavity (Fig. 8). In particular, shifts in the approximate centers of the distributions suggest that 140R1 moves closer to 151R1 (≈−10 Å), closer to 68R1 (≈−5 Å), and away from 109R1 (≈+5 Å), compared with shifts of −8 Å, −3 Å, and +5 Å, respectively, predicted by modeling. The 140R1/89R1 distance distribution shows primarily a shift in population that could reflect changes in rotamers rather than a shift in distance; no change in distance is predicted by the model.
Fig. 7.
Distance measurements of doubly labeled T4L in the WT′ and W138A mutant. (A) Ribbon model of T4L (PDB ID code 3LZM) shows the interspin distances measured as blue dotted lines. Yellow spheres at the Cα identify reference sites. (B) Background-corrected dipolar evolution functions (Left) and corresponding distance distributions (Right) for the indicated doubly labeled protein in the WT′ (black), cavity mutant (green), and cavity mutant with phenol added (orange). Arrows indicate new distances populated with phenol that are not observed in the WT′ or the cavity mutant in the absence of ligand. For W138A/140R1/151R1, the distance was determined by the CW broadening method, as described in SI Materials and Methods. The extent of spectral broadening due to spin–spin interaction is shown by the comparison of the normalized spectra for the double mutant (black trace) and the algebraic sum of the spectra for the individual mutants (green trace), with the latter representing the hypothetical noninteracting spectrum. The broadening, revealed by the reduction in spectral amplitude, is reasonably well accounted for by the Gaussian distribution of distances shown.
Fig. 8.
Motion of helix I in the W138A mutant consistent with CW data and interspin distance measurements. A ribbon model shows an ∼90° rotation of helix I with R1 modeled at reference sites 68, 89, 109, and 151 and at active site 140 in the WT′ (gray ribbon) and rotated state (green ribbon). The position of 140R1 in the rotated state is indicated with a prime, and predicted interspin distances in the rotated state are indicated on the dashed red lines and compared with the corresponding distances in the WT′ (dashed black lines). Models of R1 are based on known rotamers of R1 (17); the model of 151R1 is based on the known crystal structure (17). The model of 140R1 in the mutant required a change in rotamer due to steric clashes in the structure. Native residue Y139 is shown as a sphere representation filling the cavity in the rotated state (Discussion). Experimental distances are tabulated in Table S3.
For the cases investigated (89R1/140R1 and 68R1/140R1), the addition of phenol or p-cresol increases the WT′-like population, but with the appearance of new populations at a shorter distance unique to the ligand-bound state (arrows for phenol, Fig. 7B; similar data for p-cresol are given in Fig. S6), implying a structural adaptation to the bound ligand. Nonspecific binding of phenol or cresol and concomitant change in structure are not likely the origin of the unique distance considering the simple hyperbolic isotherm that describes the binding (Fig. S5). In addition, even at 50 mM, phenol has no effect on the conformation of the WT′ protein, as judged by the lack of spectral changes for R1 residues at sites throughout the protein (Fig. S4C).
Discussion
The data presented above unequivocally identify the presence of new conformational substates in the T4L cavity mutants in equilibrium with a WT′-like conformation. Although the CW spectra of single R1 residues can reveal the existence of conformational equilibria, DEER distance distributions show the full complexity of the conformational space. Multimodal distance distributions are observed even in the WT protein (Figs. 4 and 7). Although the multimodality may be attributed, in part, to rotamers of R1, it is likely that conformational heterogeneity in the WT′ protein makes an important contribution. Evidence for this comes from the narrow and primarily monomodal distance distributions between R1 pairs in rigid proteins such as holomyoglobin [López et al. (22)] and the Rpo4/7 stalk module of the Methanocaldococcus jannaschii RNA polymerase (29).
The width and complexity of the distance distributions increase in the cavity mutants, and the picture that emerges is one of a fluctuating ensemble of conformations. Indeed, some of the broader distance distributions in the mutants resemble those observed by DEER in a molten globule state (22). Based on the CW spectra, the disorder in the protein fold is confined to structures surrounding the cavity.
Ligands apparently bind only to the WT′-like state, displaying conformational selection in ligand binding and, at the same time, indicating that the new states excited by the cavity have alternative packing arrangements that apparently fill or partially fill the cavity, a suggestion that is in line with recent studies in other proteins containing native or engineered cavities (22, 30–33). Precedent for this behavior in T4L is found in the extensively studied cavity mutant L99A, where it has recently been shown that a poorly populated (≈3%) conformational substate in equilibrium with the WT is one in which the cavity is partially filled with a native phenylalanine side chain (31).
The sections below discuss the individual cavity mutants and possible structural models that account for salient features of the collective data.
T4L 121A/133A Mutant.
Although structures corresponding to the alternative packing modes in 121A/133A cannot be determined from the limited data presented here, they allow speculation regarding salient features of the new states. The fact that the line shape of 132R1 is the same in the WT′ and 121A/133A mutant shows that the secondary structure of helix H is retained in the ensemble of states detected by DEER, at least in the central segment sampled by 132R1. The DEER data involving residues 130R1, 131R1, and 132R1 in helix H in the cavity mutant can be accounted for by a new conformation of the protein in which the helix is rotated by ≈90° (Fig. 5A) but may also include an inward displacement and additional distortions of the helix, along with coordinated motions of neighboring structures that would reduce the size of the cavity. Moreover, the pattern of CW spectral changes for these sensors can also be accounted for by this rotation, which moves the Cα of residue i into the position occupied by i-1 in the WT′ structure. Thus, residue 130R1 becomes completely immobilized, consistent with a location in the cavity previously occupied by Ala129; 131R1 adopts a two-component spectrum similar to that of 130R1 in the WT′; and 132R1 is unchanged, with a single component spectrum similar to that of 131R1. Note that the changes in the EPR spectra for residues 128R1 and 135R1, as well as the DEER distance changes for the R1 pair involving residue 128R1, cannot be explained by the simple rigid body helix rotation depicted in Fig. 5. These two residues lie at or near the helix termini; thus, partial unfolding or distortions of the termini that place the nitroxide in a partially buried location may accompany the helical rotation. Of particular significance is that all the buried or partially buried residues of helix H in the 121A/133A mutant are alanines, which may reduce steric constraints and enable the rather large structural changes observed. It should be emphasized that other structural models may account for the data, but the dataset is too sparse to justify modeling of more complex rearrangements involving secondary structural changes.
Little can be said regarding the motion of helix G in the 121A/133A cavity mutant based on the limited data presented, except that the CW EPR spectra of 116R1 and 123R1, and the DEER distance data involving 123R1, are compatible with a rotation of G such as to bring these residues into contact with helix H, giving rise to the immobilized components with low accessibility (Table S2), as observed experimentally. Interestingly, the crystal structure of T4L 121A/133A shows a slight rotation of helix H in the same sense as suggested here (0.6-Å movement of Cα) and a motion of helix G toward H (∼0.6 Å); these small changes are in the direction to reduce the size of the engineered cavity (15). Although such subtle rearrangements cannot explain the magnitude of changes in solution indicated by the EPR data and the lack of ligand binding to the new state, they suggest a trajectory of motion that is consistent with that observed in the absence of crystal lattice constraints.
An interesting question arises when considering the mechanism of ligand binding to engineered cavities in T4L, including the extensively studied L99A. In the WT conformation, which apparently constitutes a major population of the conformational ensemble, there is no access to the cavities. For L99A, the only other detected conformation by NMR dispersion-relaxation was one in which the protein relaxed to fill the cavity partially with a phenylalanine side chain (31). How, then, does the ligand access the cavity? The same question is relevant to the cavity mutants studied here, and a possible answer is provided by the DEER distance distributions, which resolve each state in a heterogeneous mixture. For example, in the 121A/133A cavity mutant, the distance distribution for the pair including 132R1 in helix H shows a small but significant population at shorter (∼18.5 Å) as well as longer distances (arrows, Fig. 4B); the longer distance corresponds to the proposed helix-rotated state. The shorter distance observed exclusively in the cavity mutant could be achieved by a motion in the opposite sense, which could open rather than close the cavity. Thus, this small population in an ensemble could be the “open door” state for ligand entry.
W138A Mutant.
For W138A, CW spectra and DEER distance distributions (Figs. 1B and 7) also reveal at least two conformational substates in solution, one of which is similar to the WT′ protein. The structural difference between the substates appears to be confined to helices H and I. The CW, DEER, and mutagenesis data are all consistent with a model in which Y139 has moved into the cavity in the non-WT conformation via a rotation of helix I (Fig. 8), although other rearrangements involving a change in secondary structure are possible; coordinated motions of helix H would also be required.
The cavity formed by the W138A mutation is polar due to Q105, which forms a hydrogen bond with W138 in the WT protein. Q105 can also act as an H-bonding partner for Y139, providing stability for the new conformation and offering an explanation for the binding selectivity of the mutant for phenol and cresol over the nonpolar benzene. As analogs of the tyrosine side chain, phenol and cresol, but not benzene, could compete with Y139 for H-bonding with Q105 and shift the conformation toward a WT′ state, as observed experimentally. However, the effects of ligand binding to W138A are complex. DEER data show that a WT distance distribution is enhanced upon ligand binding, but with the appearance of new distances not present in either the WT′ or cavity mutant in the absence of ligand (Fig. 7B). Thus, the ligand-bound state has a unique conformation, and it must be assumed that the local structure can relax to adapt to the ligand.
Question of R1 Perturbation in Flexible Proteins.
Introduction of the R1 side chain at surface sites in a protein with a single stable conformation generally leads to decreases, or more rarely, small increases in stability as measured by Tm (16, 34). For R1 at contact sites between helices, such as 130R1 in the WT′ protein, destabilization is observed due to unfavorable steric interactions (16, 28). On the other hand, introduction of R1 into the cavity mutants at surface sites where immobilized states appear results in stabilization of the protein, as judged by an increase in the thermal melting point (Table S1). Thus, the spin label experiences attractive, stabilizing interactions that immobilize the nitroxide in the conformational ensemble of the cavity mutants but not in the WT′. Such interactions will bias the population in favor of the interacting state. For the two-component spectra, an apparent equilibrium constant (Kapp) can be defined as Kapp = X/WT, where X includes the concentration of all immobilized states arising in the cavity mutant; Kapp can be determined directly from the CW spectra but will overestimate the true equilibrium constant (KT) by an amount dependent on the interaction strength, which can vary from site to site where R1 is placed.
The magnitude of the effect can be estimated from the R1–protein interaction energy, which, in principle, can be estimated from the melting curves. However, the existence of two or more conformations in equilibrium (WT, X) belies simple analysis in terms of two-state melting. An order-of-magnitude estimate of the interaction energy can be made assuming that the interaction is hydrophobic adsorption of the nitroxide ring to the surface of the protein surface. The ring is approximately spherical, with a surface area of ≈80 Å2, and adsorption to the protein surface could bury ≈40 Å2. Taking the hydrophobic free energy to be ≈−25 cal/Å2 (35) gives ≈−1 kcal of favorable interaction. Based on the current model for the internal motion of R1 at helix surface sites (17, 20, 36), the adsorption would entail an entropy loss due to restriction of rotamers about the two terminal bonds of R1, each of which is taken to have two most probable configurations (37). This yields an unfavorable contribution to the free energy of −RTln(1/4) ≈ 0.8 kcal/mol, giving a net favorable interaction of ΔGoint ≈ −0.200 kcal/mol. For 116R1, 123R1, and 131R1 in the 121A/133A mutant, spectral simulations give Kapp ≈ 1 (Fig. S3). An attractive interaction of −0.2 kcal/mol would result in Kapp being an overestimation of the true equilibrium constant by ≈40% (KT ≈ 0.7). Thus, the perturbation of the spin label from this source is not large enough to populate strongly a conformation that is not already present in detectable amounts. For sites where the nitroxides are completely buried in the structure, the hydrophobic contribution to the attractive interaction could be ≈ −1.2 kcal/mol. This could apply, for example to, 128R1 and 135R1 in the 121A/133A background, where Kapp ≈ 9. If this were the case, the true equilibrium constant would be KT ≈ 1, the same order of magnitude as that estimated for the 116R1, 123R1, and 131R1 equilibria, but this calculation does not include any unfavorable strain energy that may be involved in reaching the state. Under any condition, in equilibrium ensembles where the free energy difference between conformers is small, the perturbation due to molecular probes in general must be considered; the situation could be considerably larger for very hydrophobic fluorophores.
In principle, the populations in the DEER distance distribution for 132R1 in the 121A/131A mutant can be used to estimate an unperturbed KT assuming that the ≈30% decrease in the WT distance population measures the population of new conformations; with this approximation, KT ≈ 0.3. However, without additional information, a unique correspondence of distances with conformations cannot be guaranteed.
The 130R1 residue in the 121A/133A mutation is a special case, representing an extreme level of population bias due to the side chain interaction. In this case, the entire side chain, including the hydrophobic disulfide, is at least partially removed from solvent and the free energy of transfer to the cavity from hydrophobic sources alone is on the order of −2.5 kcal/mol, as judged by the free energy of transfer of R1 from aqueous to nonpolar media (38); additional contributions from interactions within the cavity could further increase the interaction. In such serendipitous situations, the more polar AP side chain is a better choice and reveals something closer to the true equilibrium populations (Fig. 6B).
Summary and Implications
Collectively, the data presented here show that cavities of a size present in many proteins can allow alternative packing arrangements of the core that are not observed in the crystal structures. Although critical to function, cavities can also lead to anomalous regulation due to the binding of unnatural ligands that shift the populations, as demonstrated here for the T4L cavity mutants. For example, a current theory of general anesthesia, and for the effect of alcohol on protein function, proposes exactly this mechanism; that is, anesthetics and ethyl alcohol function by binding to cavities in ion channels and proteins involved in signal transduction, thus shifting the population of functional conformational substates (39–41). A particularly interesting recent report shows that creation of a cavity mutation due to a phenylalanine-to-alanine substitution bestows alcohol sensitivity to an otherwise insensitive ion channel (39). The data provided here provide direct structural evidence in support of this idea; the general anesthetic halothane binds to the 121A/133A cavity mutant with a Kd close to the clinically effective dose and strongly shifts the populations of substates.
Cavities also play a central role in accounting for the pressure effects on proteins. A protein in solution will respond to application of hydrostatic pressure by changing in a manner to reduce the total volume of the system. This can be accomplished by a shift in conformational equilibria toward substates with the lowest partial molar volume. Extensive NMR studies from Akasaka and coworkers (42, 43) led to the attractive idea that the substates with lowest partial molar volume are those where cavities have been filled with solvent. The results presented here suggest another model that may be appropriate in particular cases; that is, pressure may populate states wherein the protein partial molar volume is reduced by filling the cavity with side chains of the protein. Finally, considering the major effect of cavities on conformational equilibria in solution, large-to-small substitutions in the protein core should be considered in attempting to engineer new functions in proteins.
Materials and Methods
Construction, Expression, Purification, and Spin Labeling of T4L Mutants.
Construction, expression, and purification of T4L mutants was done as previously described (17). Some mutants formed inclusion bodies, and isolation was done as described by Liu et al. (44), with some modifications (SI Materials and Methods). Sample purity of isolated T4L was greater than 95%, as judged by SDS/PAGE electrophoresis. Spin labeling of the T4L mutants was done in buffer consisting of 50 mM 3-(N-morpholino)propanesulfonic acid and 25 mM NaCl at pH 6.8, as described previously (17). The 3-(2-iodoacetamido)-proxyl was obtained from Sigma.
EPR Spectroscopy.
CW EPR spectra were recorded at X-band in a Bruker ELEXYS 580 spectrometer at 295K in buffer containing 30% wt/wt sucrose to eliminate the contribution of the protein rotational diffusion, except for the ligand titration experiment, for which the spectra were recorded in pure buffer. SR experimental procedures are described in detail in SI Materials and Methods. Protein concentrations were in the range of 200–500 μM.
DEER Spectroscopy.
Four-pulse DEER data at 80K were obtained on an ELEXSYS 580 spectrometer operated at Q-band. The protein concentration was at or below 200 μM. Samples of 20 μL in spin-labeling buffer containing 20% vol/vol glycerol in a glass capillary (1.4 i.d. × 1.7 o.d.; VitroCom, Inc.) were flash-frozen in liquid nitrogen. A 36-ns π-pump pulse was set at the maximum absorption spectra, and the observer π/2 (16 ns) and π (32 ns) pulses were positioned 50 MHz (17.8 G) upfield, which corresponds to the absorption maxima of the center-field line. Distance distributions were obtained from the raw dipolar evolution data using the program “LongDistances”. The program is written in LabVIEW (National Instruments) and can be downloaded from www.chemistry.ucla.edu/directory/hubbell-wayne-l.
CD Spectroscopy.
Thermal denaturation studies were performed on a Jasco 810 spectropolarimeter, as described in SI Materials and Methods.
Ligand Binding and Halothane Titration Assay.
Addition of benzene, halothane, anisole, and p-xylene to the L133G and L121A/L133A mutants was done by vapor diffusion. Briefly, 5–10 μL of the protein solution was placed on siliconized coverslips suspended in a well containing the ligand (300 μL). The well was sealed immediately and allowed to equilibrate for 1–2 h at room temperature. Incubating for more than 2 h made no difference in the binding of the ligand to the protein, as judged by changes in the EPR spectra. For the halothane titration experiments, a saturated solution of halothane in buffer (20.6 mM) was diluted to the appropriate concentration in the protein solution and the EPR spectra were recorded immediately. Estimation of Kd using EPR data is described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Mark R. Fleissner and Evan K. Brooks for providing the doubly labeled proteins N68R1/N140R1, T109R1/V131R1, D89R1/V131R1, and T109R1/N140R1 in the WT background. We also thank Dr. Joseph Horwitz for providing the facilities for thermal denaturation experiments. All ribbon diagrams of T4L were generated using PyMOL (www.pymol.org/). This work was supported by National Institutes of Health Grant 5R01 EY005216 (to C.J.L.), Jules Stein Eye Institute Training Grant 2T32EY007026-36A1 (to C.J.L.), and the Jules Stein Professor Endowment (to W.L.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318754110/-/DCSupplemental.
References
- 1.Richards FM. The interpretation of protein structures: Total volume, group volume distributions and packing density. J Mol Biol. 1974;82(1):1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- 2.Liang J, Dill KA. Are proteins well-packed? Biophys J. 2001;81(2):751–766. doi: 10.1016/S0006-3495(01)75739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hildebrand PW, Rother K, Goede A, Preissner R, Frömmel C. Molecular packing and packing defects in helical membrane proteins. Biophys J. 2005;88(3):1970–1977. doi: 10.1529/biophysj.104.049585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eriksson AE, et al. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255(5041):178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 5.Vlassi M, Cesareni G, Kokkinidis M. A correlation between the loss of hydrophobic core packing interactions and protein stability. J Mol Biol. 1999;285(2):817–827. doi: 10.1006/jmbi.1998.2342. [DOI] [PubMed] [Google Scholar]
- 6.Munson M, et al. What makes a protein a protein? Hydrophobic core designs that specify stability and structural properties. Protein Sci. 1996;5(8):1584–1593. doi: 10.1002/pro.5560050813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee C, Maeng JS, Kocher JP, Lee B, Yu MH. Cavities of alpha(1)-antitrypsin that play structural and functional roles. Protein Sci. 2001;10(7):1446–1453. doi: 10.1110/ps.840101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogata K, et al. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat Struct Biol. 1996;3(2):178–187. doi: 10.1038/nsb0296-178. [DOI] [PubMed] [Google Scholar]
- 9.Patil A, Kinoshita K, Nakamura H. Hub promiscuity in protein-protein interaction networks. Int J Mol Sci. 2010;11(4):1930–1943. doi: 10.3390/ijms11041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadirvelraj R, Sennett NC, Polizzi SJ, Weitzel S, Wood ZA. Role of packing defects in the evolution of allostery and induced fit in human UDP-glucose dehydrogenase. Biochemistry. 2011;50(25):5780–5789. doi: 10.1021/bi2005637. [DOI] [PubMed] [Google Scholar]
- 11.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324(5924):203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 12.Baase WA, Liu L, Tronrud DE, Matthews BW. Lessons from the lysozyme of phage T4. Protein Sci. 2010;19(4):631–641. doi: 10.1002/pro.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morton A, Baase WA, Matthews BW. Energetic origins of specificity of ligand binding in an interior nonpolar cavity of T4 lysozyme. Biochemistry. 1995;34(27):8564–8575. doi: 10.1021/bi00027a006. [DOI] [PubMed] [Google Scholar]
- 14.Baldwin E, Baase WA, Zhang Xj, Feher V, Matthews BW. Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions. J Mol Biol. 1998;277(2):467–485. doi: 10.1006/jmbi.1997.1606. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, Baase WA, Baldwin E, Matthews BW. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 1998;7(1):158–177. doi: 10.1002/pro.5560070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35(24):7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 17.Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18(5):893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.López CJ, Fleissner MR, Guo Z, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18(8):1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Z, Cascio D, Hideg K, Kálái T, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme. Protein Sci. 2007;16(6):1069–1086. doi: 10.1110/ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Columbus L, Kálai T, Jekö J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in alpha-helices: Analysis by variation of side chain structure. Biochemistry. 2001;40(13):3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 21.Bridges MD, Hideg K, Hubbell WL. Resolving Conformational and Rotameric Exchange in Spin-Labeled Proteins Using Saturation Recovery EPR. Appl Magn Reson. 2010;37(1-4):363. doi: 10.1007/s00723-009-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López CJ, Oga S, Hubbell WL. Mapping molecular flexibility of proteins with site-directed spin labeling: A case study of myoglobin. Biochemistry. 2012;51(33):6568–6583. doi: 10.1021/bi3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg–Marquardt algorithm. J Magn Reson A. 1996;120(2):155–189. [Google Scholar]
- 24.Subczynski WK, et al. Molecular organization and dynamics of 1-palmitoyl-2-oleoylphosphatidylcholine bilayers containing a transmembrane alpha-helical peptide. Biochemistry. 1998;37(9):3156–3164. doi: 10.1021/bi972148+. [DOI] [PubMed] [Google Scholar]
- 25.Pyka J, Ilnicki J, Altenbach C, Hubbell WL, Froncisz W. Accessibility and dynamics of nitroxide side chains in T4 lysozyme measured by saturation recovery EPR. Biophys J. 2005;89(3):2059–2068. doi: 10.1529/biophysj.105.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9(16):1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 27.Fleissner MR, et al. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc Natl Acad Sci USA. 2011;108(39):16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fleissner MR (2007) X-ray structures of nitroxide side chains in proteins: A basis for interpreting distance measurements and dynamic studies by electron paramagnetic resonance. PhD dissertation (University of California, Los Angeles)
- 29.Klose D, et al. Simulation vs. reality: A comparison of in silico distance predictions with DEER and FRET measurements. PLoS ONE. 2012;7(6):e39492. doi: 10.1371/journal.pone.0039492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rick SW, Topol IA, Erickson JW, Burt SK. Molecular mechanisms of resistance: free energy calculations of mutation effects on inhibitor binding to HIV-1 protease. Protein Sci. 1998;7(8):1750–1756. doi: 10.1002/pro.5560070809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouvignies G, et al. Solution structure of a minor and transiently formed state of a T4 lysozyme mutant. Nature. 2011;477(7362):111–114. doi: 10.1038/nature10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meinhold DW, Wright PE. Measurement of protein unfolding/refolding kinetics and structural characterization of hidden intermediates by NMR relaxation dispersion. Proc Natl Acad Sci USA. 2011;108(22):9078–9083. doi: 10.1073/pnas.1105682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roche J, et al. Structural, energetic, and dynamic responses of the native state ensemble of staphylococcal nuclease to cavity-creating mutations. Proteins. 2013;81(6):1069–1080. doi: 10.1002/prot.24231. [DOI] [PubMed] [Google Scholar]
- 34.Fleissner MR, et al. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proc Natl Acad Sci USA. 2009;106(51):21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharp KA, Nicholls A, Friedman R, Honig B. Extracting hydrophobic free energies from experimental data: Relationship to protein folding and theoretical models. Biochemistry. 1991;30(40):9686–9697. doi: 10.1021/bi00104a017. [DOI] [PubMed] [Google Scholar]
- 36.Columbus L, Hubbell WL. Mapping backbone dynamics in solution with site-directed spin labeling: GCN4-58 bZip free and bound to DNA. Biochemistry. 2004;43(23):7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 37.Tombolato F, Ferrarini A, Freed JH. Dynamics of the nitroxide side chain in spin-labeled proteins. J Phys Chem B. 2006;110(51):26248–26259. doi: 10.1021/jp0629487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorgeirsson TE, Russell CJ, King DS, Shin Y-K. Direct determination of the membrane affinities of individual amino acids. Biochemistry. 1996;35(6):1803–1809. doi: 10.1021/bi952300c. [DOI] [PubMed] [Google Scholar]
- 39.Howard RJ, et al. Structural basis for alcohol modulation of a pentameric ligand-gated ion channel. Proc Natl Acad Sci USA. 2011;108(29):12149–12154. doi: 10.1073/pnas.1104480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pentyala SN, Sung K-Y, Chowdhury A, Rebecchi MJ. Volatile anesthetics modulate the binding of guanine nucleotides to the α subunits of heterotrimeric GTP binding proteins. Eur J Pharmacol. 1999;384(2-3):213–222. doi: 10.1016/s0014-2999(99)00625-1. [DOI] [PubMed] [Google Scholar]
- 41.Eckenhoff RG, Johansson JS. Molecular interactions between inhaled anesthetics and proteins. Pharmacol Rev. 1997;49(4):343–367. [PubMed] [Google Scholar]
- 42.Kitahara R, Yamada H, Akasaka K, Wright PE. High pressure NMR reveals that apomyoglobin is an equilibrium mixture from the native to the unfolded. J Mol Biol. 2002;320(2):311–319. doi: 10.1016/S0022-2836(02)00449-7. [DOI] [PubMed] [Google Scholar]
- 43.Kamatari YO, Smith LJ, Dobson CM, Akasaka K. Cavity hydration as a gateway to unfolding: An NMR study of hen lysozyme at high pressure and low temperature. Biophys Chem. 2011;156(1):24–30. doi: 10.1016/j.bpc.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Baase WA, Michael MM, Matthews BW. Use of stabilizing mutations to engineer a charged group within a ligand-binding hydrophobic cavity in T4 lysozyme. Biochemistry. 2009;48(37):8842–8851. doi: 10.1021/bi900685j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.