Significance
Epstein–Barr virus nuclear antigen (EBNA) leader protein (LP) and EBNA2 (E2) up-regulation of virus and cell gene expression is important for human B-lymphocyte conversion to continuous, potentially malignant, lymphoblast cell lines. Although the molecular mechanism(s) underlying LP and E2 regulation of cell gene expression have been partially elucidated, LP ChIP-sequencing studies have now revealed that LP and LP/E2 interact, genome-wide, with human B-cell transcription factors, mostly at or near prepatterned promoter sites, to increase cell transcription factor occupancies, increase activation-associated histone marks, and positively affect cell gene transcription.
Keywords: genome-wide ChIP-seq analysis, gene expression
Abstract
Epstein–Barr virus (EBV) nuclear antigens EBNALP (LP) and EBNA2 (E2) are coexpressed in EBV-infected B lymphocytes and are critical for lymphoblastoid cell line outgrowth. LP removes NCOR and RBPJ repressive complexes from promoters, enhancers, and matrix-associated deacetylase bodies, whereas E2 activates transcription from distal enhancers. LP ChIP-seq analyses identified 19,224 LP sites of which ∼50% were ±2 kb of a transcriptional start site. LP sites were enriched for B-cell transcription factors (TFs), YY1, SP1, PAX5, BATF, IRF4, ETS1, RAD21, PU.1, CTCF, RBPJ, ZNF143, SMC3, NFκB, TBLR, and EBF. E2 sites were also highly enriched for LP-associated cell TFs and were more highly occupied by RBPJ and EBF. LP sites were highly marked by H3K4me3, H3K27ac, H2Az, H3K9ac, RNAPII, and P300, indicative of activated transcription. LP sites were 29% colocalized with E2 (LP/E2). LP/E2 sites were more similar to LP than to E2 sites in associated cell TFs, RNAPII, P300, and histone H3K4me3, H3K9ac, H3K27ac, and H2Az occupancy, and were more highly transcribed than LP or E2 sites. Gene affected by CTCF and LP cooccupancy were more highly expressed than genes affected by CTCF alone. LP was at myc enhancers and promoters and of MYC regulated ccnd2, 23 med complex components, and MYC regulated cell survival genes, igf2r and bcl2. These data implicate LP and associated TFs and DNA looping factors CTCF, RAD21, SMC3, and YY1/INO80 chromatin-remodeling complexes in repressor depletion and gene activation necessary for lymphoblastoid cell line growth and survival.
Epstein–Barr virus (EBV) nuclear antigens EBNALP (LP) and EBNA2 (E2) are EBV-encoded transcription factors (TFs) that are coordinately expressed within hours after EBV infection of resting B lymphocytes (RBLs) and are important for B-lymphocyte conversion to lymphoblastoid cell lines (LCLs) (1–7). However, the biochemical mechanisms by which LP and E2 coordinately affect RBL transformation to LCLs are largely unknown. LP coactivates transcription by heterodimerizing with HA95 and Hsp70/72 to relocate HDAC4 from the nucleus to the cytoplasm, displaces Sp100 and Hp1α from ND10 bodies, and disrupts matrix-associated deacetylase (MAD) bodies, broadly affecting repressor localization in cell nuclei (8–16). LP also decreases repressive NCOR and RBPJ occupancy at E2 sites, without altering E2 occupancy (9).
E2 enhances gene expression by localizing to cell TF sites through RBPJ or ZNF143 (17–19). E2/RBPJ sites localize in six clusters of EBF, ETS1, ZNF143, PU.1, NFκB, and RUNX1 sites. Encyclopedia of DNA elements (ENCODE) ChIP-sequencing experiments (ChIP-seqs) indicated high-level cell TF cooccupancy at E2 sites, consistent with these sites being open to cell or virus TF occupancy. Indeed, E2 chromatin sites in LCLs are open chromatin sites in RBLs, before EBV infection, consistent with EBF and RBPJ as pioneering factors that displace nucleosomes (17). E2 increases H3K4me1 signals allowing E2 and cell TF occupancy and transcription activation. The E2 activation domain recruits basal and activation-related cell TFs, including TAF40, TFIIH, TFIIE, and histone acetylases P300, CBP, and PCAF (17–21). Focusing on 88 dynamically E2-regulated genes, using ENCODE chromosome conformation capture (3C) data, the transcriptional start site (TSS) of 50 E2 dynamically regulated genes are in proximity to approximately three E2 enhancers per gene. These enhancers are 61% on the same chromosome and at a median distance of ∼330 kb from their affected genes. The combined effect of three E2 enhancers accounts for E2’s strong up-regulatory effects (17). E2 induces MYC expression within 24 h of RBL infection. MYC then drives RBL cell cycle entry and proliferation (14, 22). However, the nearest RBPJ and E2 sites are >100 kb from myc (17). FISH and 3C assays connect the myc TSS to an E2 site at −428 kb from myc (17).
LP and E2 cooperatively activate virus and cell gene transcription following transient or stable B-lymphocyte transfection (9, 23, 24). The experiments described here were undertaken to identify the mechanisms through which LP and E2 affect cell gene transcription in LCLs.
Results and Discussion
LP, LP/E2, and E2 Genome Distributions.
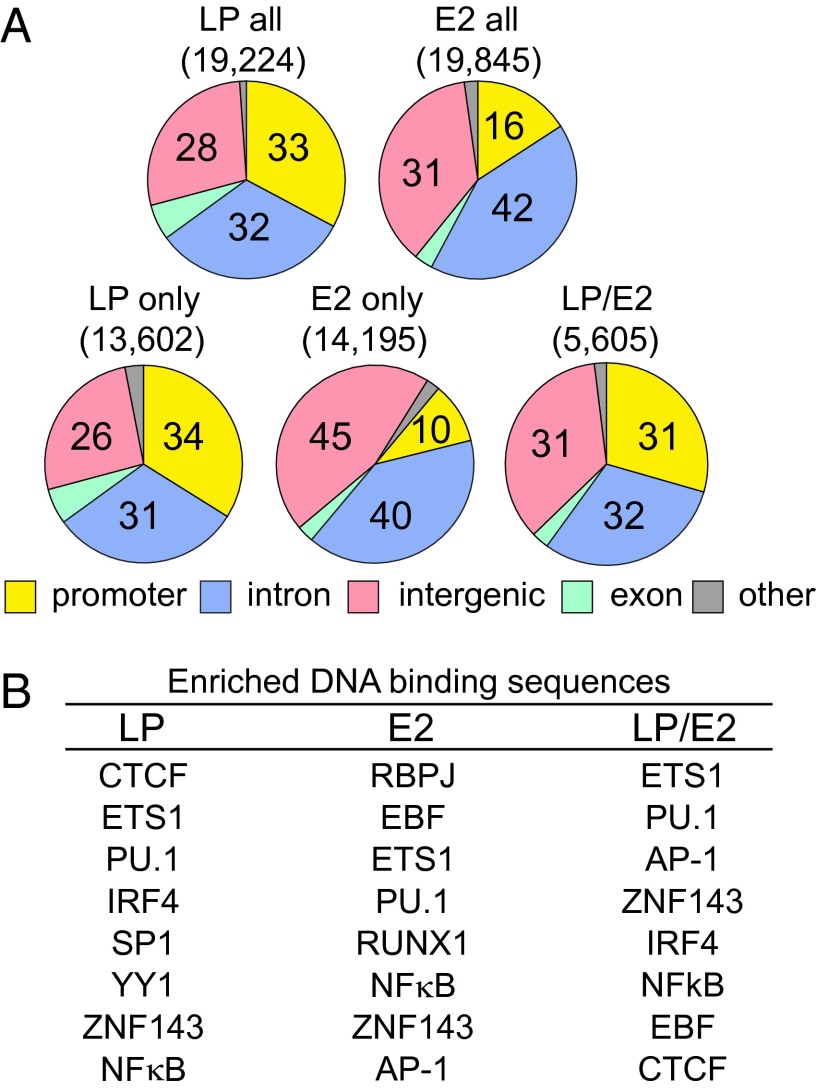
Duplicate LCL LP and an E2 ChIP-seq dataset (17) were analyzed using HOMER, with a false discovery rate of P < 0.001. LP localized to 19,224 sites and E2 localized to 19,845 sites (Fig. 1A) (17). In contrast to E2 sites without LP, which are mostly at enhancers (17) and only 10% at promoters (defined as −1 to +0.1 kb from a TSS), LP sites without E2 were 34% at promoters and more than 50% of LP sites without E2 were within 2 kb of a TSS (Fig. S1), indicating that LP is much more promoter localized than E2 (Fig. S1). The 5,605 LP/E2 sites were also 31% promoter associated, similar to the 13,602 LP-only sites, which were 34% promoter associated (Fig. 1A). These data indicate that LP is substantially promoter localized and dominantly maintains a similar level of promoter localization with E2 cooccupancy.
Fig. 1.
Genomic distribution and TF DNA binding sequences enriched at LP, E2, and LP/E2 sites. (A) Pie charts showing the genome-wide distribution of all LP or E2 sites, as well as the genome-wide distribution of LP-only, E2-only, and LP/E2 sites. (B) List of most highly enriched cell transcription factor DNA binding sequences at LP, E2, and LP/E2 sites (±100 b) (P < 0.01).
Cell TF Sites Associated with LP or E2.
LP sites (±100 bp) were significantly enriched (from P < 0.01 to <1 × 10−958) for cell TF binding sites important in lymphocyte development, including CTCF, ETS1, PU.1, IRF4, SP1, YY1, ZNF143, NFκB, and RUNX1 sites (Fig. 1B). LP-associated cell TF sites were unaffected by increasing the LP site search window to ±250 bp.
The LP site-associated TFs are remarkable for their importance in B-cell development and mature B-cell responses to antigen. CTCF is a transcription insulator, which associates with YY1, RAD21, and SMC3 to mediate long-range chromatin interactions (25–38). YY1-associated INO80 chromatin-remodeling complexes and PU.1 have prominent roles in development, immune responses (39), and chromatin domain transcription (40). ZNF143 and RBPJ mediate Notch or E2 interaction with cognate DNA (41, 42) in tissue development and in EBV RBL conversion to LCLs (42). BATF/JUN/FOS/ETS family proteins heterodimerize with IRF4 or IRF8 and are essential for mature B-lymphocyte immune responses (43).
Most cell TF binding sites at LP sites were also at E2 and LP/E2 sites (Fig. 1B, compare LP, E2, and LP/E2 columns), consistent with the hypothesis that LP and E2 evolved to cooperatively up-regulate transcription through RBL genome-wide sites that are prepatterned for up-regulation of B-cell growth and survival. HOMER did not recognize a de novo LP DNA sequence, which may indicate that LP is not a DNA sequence-specific binding protein.
Cell TF Cooccupancy Levels at LP, E2, and LP/E2 Sites.
ENCODE ChIP-seq data were used to determine cell TF occupancies at LP, LP/E2, and E2 sites. Cell TFs with statistically significant enrichment at LP, E2, or LP/E2 sites were YY1, SP1, PAX5, BATF, IRF4, ETS1, RAD21, PU.1, CTCF, RBPJ, ZNF143, SMC3, NFκB, and TBLR1. LP sites were more occupied with most (9 of 15) of these factors than E2 sites, except for RBPJ and EBF, which were more highly occupied at E2 sites. E2 stabilizes RBPJ interaction with DNA (Table 1) (17). However, 11 of 15 LP/E2 sites were more highly occupied by cell TFs than LP or E2 only sites (Table 1). These data are consistent with LP’s derepressive and cooperative effects with E2 being important in transcription activation.
Table 1.
Cell TF cooccupancy at LP, LP/E2, or E2 sites
| TF | LP (13,602) | LP/E2 (5,605) | E2 (14,195) | |||
| No. sites | % Total | No. sites | % Total | No. sites | % Total | |
| YY1 | 8,059 | 59 | 3,965 | 71 | 4,812 | 34 |
| SP1 | 7,506 | 55 | 4,423 | 79 | 5,305 | 37 |
| PAX5 | 7,055 | 52 | 4,199 | 75 | 5,220 | 37 |
| BATF | 5,618 | 41 | 3,987 | 71 | 5,607 | 39 |
| IRF4 | 5,526 | 41 | 3,870 | 69 | 4,753 | 33 |
| ETS1 | 4,822 | 35 | 2,662 | 47 | 2,172 | 15 |
| RAD21 | 4,529 | 33 | 1,822 | 33 | 2,495 | 18 |
| PU1 | 4,064 | 30 | 2,999 | 54 | 4,276 | 30 |
| CTCF | 3,800 | 28 | 686 | 12 | 822 | 6 |
| RBPJ | 3,737 | 27 | 4,434 | 79 | 11,529 | 81 |
| ZNF143 | 3,417 | 25 | 1,113 | 20 | 916 | 6 |
| SMC3 | 3,127 | 23 | 955 | 17 | 923 | 7 |
| NFκB | 3,074 | 23 | 2,828 | 50 | 3,614 | 25 |
| TBLR1 | 2,594 | 19 | 2,472 | 44 | 2,130 | 15 |
| EBF | 1,285 | 9 | 1,920 | 34 | 5,030 | 35 |
Although LP’s frequent localization to promoters and E2’s frequent localization to enhancers might limit their cooperation in transcription activation, their high occupancies with many of the same cell TFs, including YY1, SP1, PAX5, BATF, IRF4, ETS1, PU1, CTCF, RBPJ, RAD21, SMC3, NFκB, and TBLR1, enable multiple dynamic interactions among E2, LP, and their associated cell TFs (9, 23, 24). E2 interactions at enhancer sites and LP at promoter sites, with the similar interacting cell TFs, positions LP and E2 in proximity to each other to mediate DNA looping and transcription activation (17).
LP, E2, and LP/E2 sites are differentially occupied by cell TFs.
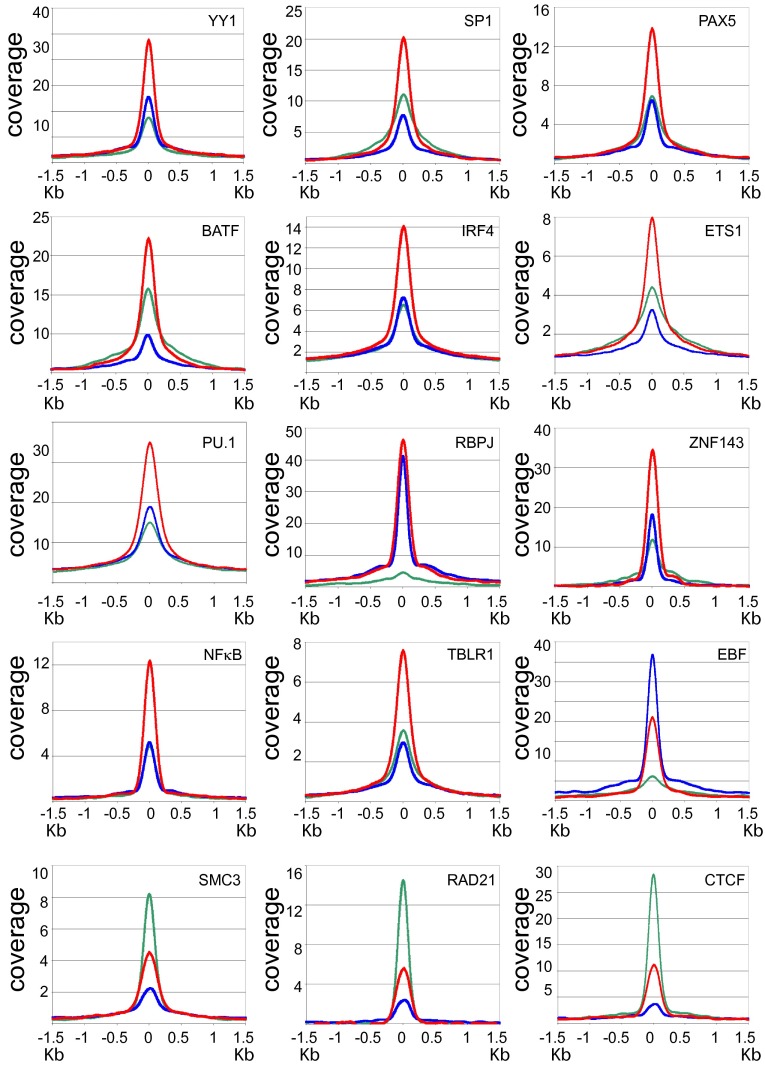
To assess cell TF occupancies at LP, LP/E2, and E2 sites, ENCODE LCL ChIP-seq data were used to quantify average occupancy or coverage of cell TFs at LP, E2, and LP/E2 sites (Fig. 2). The most frequent pattern applied to YY1, SP1, PAX5, BATF, IRF4, ETS1, PU.1, ZNF143, NFκB, and TBLR1 sites, where LP/E2 coverages (red lines) were significantly higher than LP coverages (green lines) or E2 coverages (Fig. 2, blue lines) indicating that LP/E2 sites are more highly occupied by these factors than LP or E2 sites (Fig. 2). However, EBF occupancy was higher at E2 than at LP or LP/E2 sites, consistent with E2 and RBPJ or ZNF143 stabilization of EBF coverage at promoter sites, as previously observed for E2. A third pattern was represented by CTCF, SMC3, and RAD21, whose coverages were highest at LP sites, consistent with their important role in promoter derepression, enhancer-mediated chromatin looping, and INO80/YY1 transcription boundary effects at CTCF sites (31–33, 36–38, 44). At the myc promoter, high CTCF and LP cooccupancies likely prime these sites for distal E2/RBPJ enhancer looping to myc (17) (Fig. S2).
Fig. 2.
Anchor plots of cell TF coverages at LP, LP/E2, and E2 sites. The upper 12 panels show higher cell TF coverage at LP/E2 (red line) than at LP (green line), or E2 (blue line) sites (±1.5 kb). EBF differs in having higher coverage at the E2 site than at LP or the LP/E2 site. The lower three panels show higher cell TF coverage for DNA looping factors SMC3, RAD21, and CTCF at LP sites than at E2 or LP/E2 sites.
LP, LP/E2, and E2 sites are associated with RNAPII, P300, and activation-related promoter or enhancer chromatin marks.
Because LP, LP/E2, and E2 sites were highly occupied by cell TFs, the relationship of cell TF occupancies to canonical epigenetic chromatin activation marks at LP, LP/E2, and E2 sites were evaluated using high-quality ENCODE histone mark datasets to impute relative activities (Table 2 and Fig. 3 A and B).
Table 2.
Basal TF cooccupancy and epigenetic marks at LP, LP/E2, or E2 sites
| Histone mark/TF | LP (13,602) | LP/E2 (5,605) | E2 (14,195) | |||
| No. sites | % Total | No. sites | % Total | No. sites | % Total | |
| H3K4me3 | 7,539 | 55 | 3,404 | 61 | 4,176 | 29 |
| H3K27ac | 7,513 | 55 | 4,162 | 74 | 5,687 | 40 |
| H2AZ | 7,412 | 54 | 3,172 | 57 | 4,083 | 29 |
| H3K9ac | 6,905 | 51 | 3,441 | 61 | 4,049 | 29 |
| RNA Pol II | 4,798 | 35 | 2,361 | 42 | 2,148 | 15 |
| H3K4me1 | 2,300 | 17 | 1,960 | 35 | 4,154 | 29 |
| P300 | 1,986 | 15 | 2,233 | 40 | 2,121 | 15 |
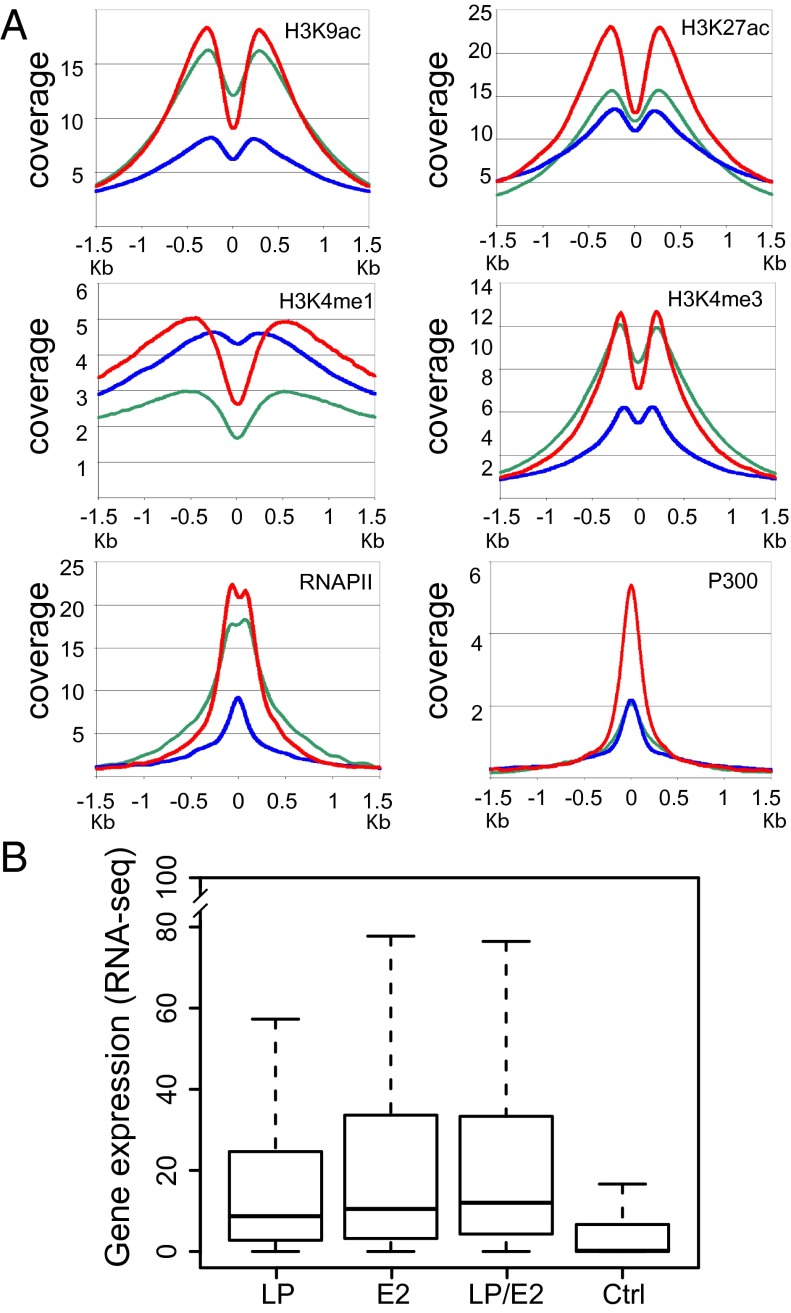
Fig. 3.
Anchor plots for cell epigenetic marks and affected gene expression data. (A) Coverage of RNAPII, P300, H3K9ac, H3K27ac, H3K4me1, and H3K4me3 ±1.5 kb of LP (green line), E2 (blue line), and LP/E2 (red line) sites. (B) Box plots of RNA-seq at LP, E2, or LP/E2 annotated peaks for the nearest promoter versus a control (Ctrl) random gene set. LP, E2, and LP/E2 genes were significantly more highly expressed than control genes (P < 2 × 10−16). LP had lower expression than E2 (P < 0.00056) or LP/E2 (P < 2.7 × 10−12), and E2 was lower than LP/E2 (P < 0.05). The box plots indicate the data distribution in percentiles, with the horizontal line being the median. The top of the box represents upper 25% quartile (25% of the data over that value), and the bottom of the box, the lower quartile (25% of the data below that value). The horizontal lines at the ends of the dotted lines are the maximum observed values.
The 13,602 LP-only sites were highly associated with chromatin marks characteristic of promoter-associated, activated transcription effects, including high-level H3K4me3, H3K27ac, H2Az, H3K9ac, as well as RNAPII and P300 signals. The 5,605 LP/E2 sites had even higher promoter-associated H3K4me3, H3K27ac, H2Az, H3K9ac, RNAPII, and P300 coverage, albeit lower H3K4me1, consistent with their promoter localization, whereas the 14,195 E2, mostly enhancer sites, had substantially lower H3K4me3 promoter marks, higher H3K4me1 enhancer activation marks, and lower H3K27ac, H2Az, H3K9ac, RNAPII, and P300 levels. Overall, LP, LP/E2, and E2 sites had active promoter or enhancer epigenetic marks and occupancies by RNAPII and P300 (Table 2), indicative of active transcription.
Consistent with LP’s association with active promoters, LP sites were highly occupied by RNAPII (Fig. 3A, green line) P300, H3K9ac, and H3K27ac, whereas, consistent with E2’s enhancer localization, E2 and LP/E2 sites had higher H3K4me1 levels than LP. LP/E2 sites were overall most coincident with activation-associated histone marks and highest RNAPII and P300 levels (Fig. 3A, red lines). The strong differences in cell TF and epigenetic mark anchor plots at LP, LP/E2, and E2 sites were less evident when LP, E2, and LP/E2 sites in promoter regions were assessed (Fig. S3).
To correlate LP-, E2-, and LP/E2-associated cell TF occupancies and chromatin activation marks with activated transcription, high-quality ENCODE LCL RNA-seq data for genes with LP, LP/E2, or E2 sites (−1/+0.1 kb from a TSS) indicated that LP, LP/E2, and E2 annotated genes are more highly expressed than random genes (P < 1 × 10−16) (Fig. 3B). However, LP-annotated genes were less expressed than E2 (P < 5.6 × 10−4) or LP/E2 (P < 2.7 × 10−12) annotated genes (Fig. 3B), and LP/E2-annotated genes were more highly expressed than E2 annotated genes (P < 0.05), despite substantial differences in cell TF occupancies, as well as RNAPII, P300, and activating histone epigenetic marks (Fig. 3B). These data highlight a complexity of LP and E2 activation of proliferation and survival pathways that remains to be deconstructed using shRNAs for individual LP-, E2-, and LP/E2-associated cell TFs.
LP Sites Clusters Differed in Cell TF Composition and Transcription Effects.
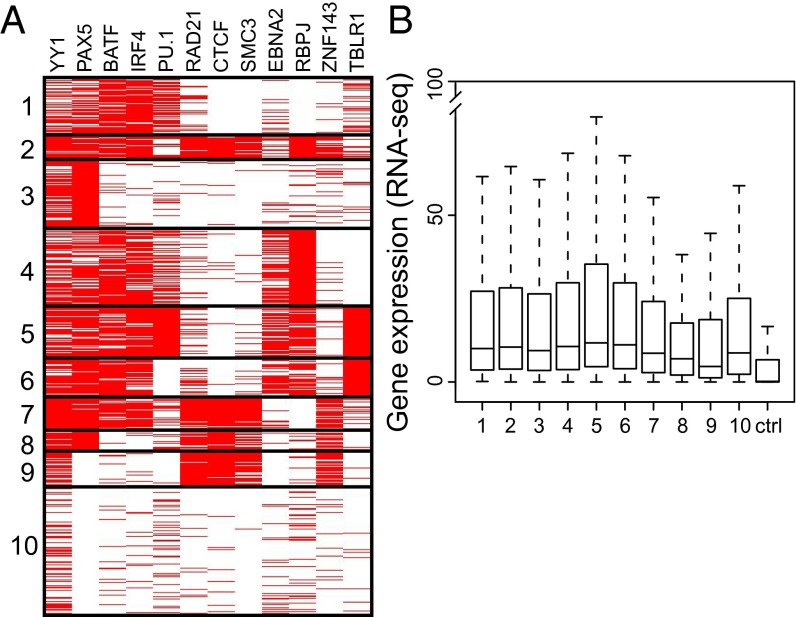
To better understand the range of LP site cell TF occupancies, a K-means clustering segregation analysis of LP sites was undertaken (Fig. 4). Clusters 1–7 were highly occupied by B-cell developmental TFs, YY1, PAX5, BATF, IRF4, and PU.1. Cluster 2 had more YY1, PAX5, BATF, and IRF4, less PU.1, and high RAD21, CTCF, SMC3, E2, RBPJ, and ZNF143 levels, TFs characteristic of B-cell enhancers and promoter looping factors. Cluster 3 was uniformly PAX5, an EBF-induced early B-cell transcription activator, and ∼50% YY1. Cluster 4 was uniformly RBPJ and almost 50% E2. Cluster 5 was solidly PU.1, an activating B-cell developmental TF, and TBLR1, an NCOR/SMRT repressive complex component that may be an LP target. Cluster 5 was enriched for E2 and RBPJ, which enhance MYC and MYC-driven cell survival gene expression. Cluster 6 was uniformly TBLR1, which associates with NCOR, YY1, PAX5, BATF, IRF4, RBPJ, and E2 (17). Cluster 7 was uniformly YY1, PAX5, BATF, and IRF4 occupied, consistent with B-cell developmental transcription activation. Cluster 7 was also high in RAD21, CTCF, SMC3, and ZNF143; ZNF143 is a potential alternative mediators of E2 or Notch interaction with DNA. Cluster 8 was uniformly PAX5, an activating B-cell TF, and RAD21, CTCF, and SMC3, looping factors that mediate enhancer interactions with promoters. Cluster 9 was YY1, CTCF, RAD21, SMC3, and ZNF143 occupied, whereas cluster 10 included ∼25% of LP sites that had substantially less prevalent cell TF occupancies.
Fig. 4.
LP site-associated cell TFs separated into 10 K-means clusters and gene expression. (A) Cell TFs YY1, PAX5, BATF, IRF4, and PU.1 were prominent components of most clusters, consistent with their 30–59% LP site association. Notably, E2, RBPJ, and TBLR1 clustered separately from looping factors CTCF, SMC3, and RAD21. (B) Box plots of RNA-seq gene expression from LP-affected promoters. All LP-affected genes were more highly expressed than random control genes (P < 2 × 10−16). The box plots indicate the data distribution in percentiles, with the horizontal line being the median. The top of the box represents upper 25% quartile (25% of the data over that value), and the bottom of the box, the lower quartile (25% of the data below that value). The horizontal lines at the ends of the dotted lines are the maximum observed values, excluding the outliers.
Not surprisingly, the genome-wide distribution of most LP clusters was similar to LP overall (Fig. S4). However, cluster 3 (PAX5) was >57% promoter associated, substantially higher than LP. At the other extreme, cluster 9 was only 16% promoter associated, had higher intron and intergenic localization, and was highly occupied with YY1, CTCF, RAD21, SMC3, and ZNF143.
YY1, PAX5, and BATF/ETS/IRF4 were abundant components of most LP clusters and are essential B-cell developmental TFs that affect cell growth and gene expression (Table 1 and Fig. 4A) (43, 45). DNA looping factors CTCF, RAD21, and SMC3 were characteristic of LP clusters 2, 7, 8, and 9, which were also rich in ZNF143, E2, and RBPJ. Clusters 2, 4, 5, and 6 were rich in TBLR1, a ubiquitin ligase that is likely activated by LP-mediated NCOR removal leading to transcription derepression (46). LP cluster 5 includes the hes1 locus, which is less NCOR occupied and derepressed when LP is expressed in BJAB B-lymphoma cells (9) (Fig. S5A). Interestingly, PKCδ (prckd), the protein kinase that activates TBLR1 to degrade NCOR is up-regulated 1.9-fold in LCLs [P < 0.05 (47)]. Furthermore, LP/E2 occupied three sites in the prckd locus (Fig. S5B), indicative of a role for LP/E2 in regulating prckd expression.
To investigate the relationship between gene transcription and LP site clusters, LCL RNA-seq data annotated to LP, LP/E2, or E2 promoter sites were used. All LP-affected clusters were significantly more highly expressed than random control genes (P < 2 × 10−16) (Fig. 4B). Cluster 8, which included YY1, PAX5, CTCF, RAD21, SMC3, and ZNF143 and cluster 9, which included YY1, CTCF, RAD21, SMC3, and ZNF143, had relatively lower expression levels, compared with other clusters (Fig. 4B).
Overall, LP positively affected genes with CTCF sites (Fig. S6), likely by removing repressors from these sites (Fig. S6). Comparison of RNA-seq expression data from genes having a promoter-associated CTCF sites without LP with those having a promoter-associated CTCF sites with LP, revealed genes with overlapping CTCF/LP sites to be significantly more highly expressed than genes having CTCF sites without LP (P < 2 × 10−16). These data indicate that LP localized with CTCF at promoter sites increases transcription. LP-associated transcription increases are most likely mediated by LP dismissal of CTCF-, SMC3-, or RAD21-associated NCOR or HDACs and may also be affected by long-distance enhancer interaction with CTCF, RAD21, and SMC3 at CTCF/LP sites. Overall, LP, localized with CTCF at or near promoters had derepressive effects comparable to E2’s activating effects (9, 10, 24, 48, 49).
To illustrate LP and associated TFs roles in up-regulating biologically important genes for LCL proliferation and survival, relevant ChIP-seq tracks are presented for the MYC-regulated cell cycle entry gene ccnd2, the MYC proliferation-associated cell survival genes, bcl2 and igf2r, the MYC-induced cell senescence genes, cdkn2a and cdkn2b, and 1 of the 23 LP affected mediator components, med26 (Fig. S7 A–E) (50–58).
Thus, the data presented here position LP as a key component of EBV’s control of the cell transcription, proliferation, and survival-related gene transcription (Fig. S7 A–E). The genome-wide approaches used to generate these data enabled the discovery of unique aspects of LP roles, including functional associations with B-cell TFs to affect key pathways in B-cell growth, survival, and gene expression.
Fig. S7A shows the bcl2, promoter and enhancer, which has LP at the promoter and at two distal enhancers, with E2, RBPJ, TBLR1, ZNF143, CTCF, YY1, IRF4, BATF, ETS1, PU.1, EBF, PAX5, SP1, NFκB, H3K27ac, RNAPII, H3K4me1, H3K9ac, and H3k4me3.
Fig. S7B shows the igf2r locus, with strong LP signals at the promoter along with YY1, ETS1, PAX5, SP1, H3K27ac, RNAPII, H3K4me1, H3K9ac, and H3K4me3.
Fig S7C shows the ccnd2 promoter with strong LP signals, weak E2, ZNF143, CTCF, RAD21, SMC3, YY1, BATF, ETS1, SP1, NFκB, H3K27ac, RNAPII, H3K9ac, and H3K4me3.
Fig. S7D shows the cdkn2a and 2b promoters with strong LP, ZNF143, CTCF, RAD21, SMC3, YY1, SP1, signals, ZNF143, weak and strong YY1, SP1, NFκB, H3K27ac, RNAPll, H3K9ac, and H3K4me3.
Fig. S7E shows the med26 locus with promoter-associated LP, ZNF143, RAD21, SMC3, YY1, ETS1, strong PAX5, SP1, H3K27ac, RNAPll, H3K4me1, and H3K4me3 signals at the promoter. Notably, stronger signals are also apparent at the med26 enhancer for LP, E2, RBPJ, TBLR1, RAD21, SMC3, YY1, IRF4, BATF, ETS1, PU.1, EBF, PAX5, SP1, H3K27ac, RNAPII, H3K4me1, H3K9ac, and H3K4me3.
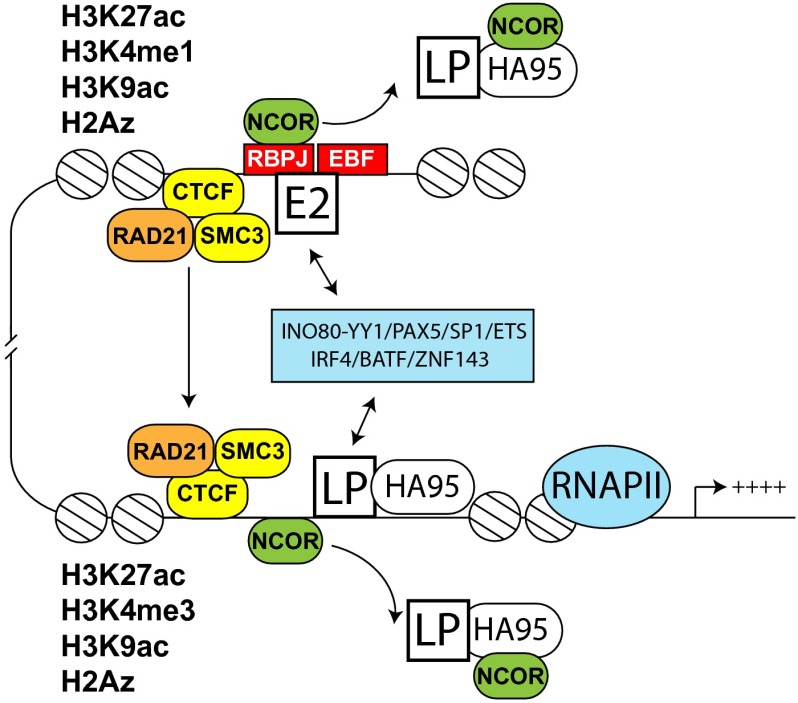
These results support the model shown in Fig. 5, that LP, predominantly at or near promoters, and E2 at enhancer sites, cooperatively affect LCL gene expression. LP’s presence at DNA sites dismisses repressive NCOR complexes. Affected genes are up-regulated by LP-mediated derepression and long-distance DNA interactions through CTCF, RAD21, and SMC3.
Fig. 5.
Model. LP occupies mostly promoters, whereas E2 is found at enhancers (17). Enhancers regulate transcription via long-distance DNA interactions mediated by CTCF, RAD21, and SMC3. Several transcription factors are associated with LP sites and E2 sites and likely help mediate the interactions between LP and E2 (blue box). The genes associated with LP sites are highly expressed and rich in activation histone marks (Fig. 3). LP and HA95 regulate the dismissal of the NCOR repressive complex (9, 59).
Materials and Methods
ChIP-seq was performed as described (17). IB4 cells were grown in RPMI medium, 10% (vol/vol) FBS. Detailed methods for analysis of ChIP-seq data, dataset access, and ChIP-seq protocol are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
This research was supported by Grants R01CA131354, R01CA170023, and R01CA047006 from the National Cancer Institute of the National Institutes of Health of the US Public Health Service.
Footnotes
The authors declare no conflict of interest.
Data deposition: All information regarding access to data is included in SI Materials and Methods.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317608110/-/DCSupplemental.
References
- 1.Cohen JI, Kieff E. An Epstein-Barr virus nuclear protein 2 domain essential for transformation is a direct transcriptional activator. J Virol. 1991;65(11):5880–5885. doi: 10.1128/jvi.65.11.5880-5885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen JI, Wang F, Kieff E. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J Virol. 1991;65(5):2545–2554. doi: 10.1128/jvi.65.5.2545-2554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen JI, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86(23):9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mannick JB, Cohen JI, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65(12):6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dambaugh T, Hennessy K, Chamnankit L, Kieff E. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc Natl Acad Sci USA. 1984;81(23):7632–7636. doi: 10.1073/pnas.81.23.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dambaugh T, et al. Expression of the Epstein-Barr virus nuclear protein 2 in rodent cells. J Virol. 1986;59(2):453–462. doi: 10.1128/jvi.59.2.453-462.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F, Petti L, Braun D, Seung S, Kieff E. A bicistronic Epstein-Barr virus mRNA encodes two nuclear proteins in latently infected, growth-transformed lymphocytes. J Virol. 1987;61(4):945–954. doi: 10.1128/jvi.61.4.945-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling PD, et al. Mediation of Epstein-Barr virus EBNA-LP transcriptional coactivation by Sp100. EMBO J. 2005;24(20):3565–3575. doi: 10.1038/sj.emboj.7600820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Portal D, et al. EBV nuclear antigen EBNALP dismisses transcription repressors NCoR and RBPJ from enhancers and EBNA2 increases NCoR-deficient RBPJ DNA binding. Proc Natl Acad Sci USA. 2011;108(19):7808–7813. doi: 10.1073/pnas.1104991108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portal D, Rosendorff A, Kieff E. Epstein-Barr nuclear antigen leader protein coactivates transcription through interaction with histone deacetylase 4. Proc Natl Acad Sci USA. 2006;103(51):19278–19283. doi: 10.1073/pnas.0609320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han I, et al. Protein kinase A associates with HA95 and affects transcriptional coactivation by Epstein-Barr virus nuclear proteins. Mol Cell Biol. 2002;22(7):2136–2146. doi: 10.1128/MCB.22.7.2136-2146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han I, et al. EBNA-LP associates with cellular proteins including DNA-PK and HA95. J Virol. 2001;75(5):2475–2481. doi: 10.1128/JVI.75.5.2475-2481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harada S, Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol. 1997;71(9):6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181(2):595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 15.Mannick JB, Tong X, Hemnes A, Kieff E. The Epstein-Barr virus nuclear antigen leader protein associates with hsp72/hsc73. J Virol. 1995;69(12):8169–8172. doi: 10.1128/jvi.69.12.8169-8172.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng CW, et al. Hsp72 up-regulates Epstein-Barr virus EBNALP coactivation with EBNA2. Blood. 2007;109(12):5447–5454. doi: 10.1182/blood-2006-08-040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B, et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci USA. 2011;108(36):14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91(16):7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henkel T, Ling PD, Hayward SD, Peterson MG. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265(5168):92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 20.Tong X, Wang F, Thut CJ, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69(1):585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol. 1995;15(9):4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikitin PA, et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe. 2010;8(6):510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng R, Tan J, Ling PD. Conserved regions in the Epstein-Barr virus leader protein define distinct domains required for nuclear localization and transcriptional cooperation with EBNA2. J Virol. 2000;74(21):9953–9963. doi: 10.1128/jvi.74.21.9953-9963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng R, Moses SC, Tan J, Kremmer E, Ling PD. The Epstein-Barr virus EBNA-LP protein preferentially coactivates EBNA2-mediated stimulation of latent membrane proteins expressed from the viral divergent promoter. J Virol. 2005;79(7):4492–4505. doi: 10.1128/JVI.79.7.4492-4505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137(7):1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkenschlager M, Odom DT. CTCF and cohesin: Linking gene regulatory elements with their targets. Cell. 2013;152(6):1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 27.Remeseiro S, Cuadrado A, Gómez-López G, Pisano DG, Losada A. A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 2012;31(9):2090–2102. doi: 10.1038/emboj.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuadrado A, Remeseiro S, Gómez-López G, Pisano DG, Losada A. The specific contributions of cohesin-SA1 to cohesion and gene expression: Implications for cancer and development. Cell Cycle. 2012;11(12):2233–2238. doi: 10.4161/cc.20318. [DOI] [PubMed] [Google Scholar]
- 29.Degner SC, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci USA. 2011;108(23):9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: Developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182(1):44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herold M, Bartkuhn M, Renkawitz R. CTCF: Insights into insulator function during development. Development. 2012;139(6):1045–1057. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- 32.Chaumeil J, Skok JA. The role of CTCF in regulating V(D)J recombination. Curr Opin Immunol. 2012;24(2):153–159. doi: 10.1016/j.coi.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillen AE, Harris A. The role of CTCF in coordinating the expression of single gene loci. Biochem Cell Biol. 2011;89(5):489–494. doi: 10.1139/o11-040. [DOI] [PubMed] [Google Scholar]
- 34.Rhodes JM, et al. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol. 2010;344(2):637–649. doi: 10.1016/j.ydbio.2010.05.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorsett D, Merkenschlager M. Cohesin at active genes: A unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol. 2013;25(3):327–333. doi: 10.1016/j.ceb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorsett D. Cohesin: Genomic insights into controlling gene transcription and development. Curr Opin Genet Dev. 2011;21(2):199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci USA. 2008;105(51):20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20(17):2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai Y, et al. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol. 2007;14(9):872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- 40.Lin YC, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13(12):1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2011;108(36):14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halbig KM, Lekven AC, Kunkel GR. The transcriptional activator ZNF143 is essential for normal development in zebrafish. BMC Mol Biol. 2012;13:3. doi: 10.1186/1471-2199-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li P, et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490(7421):543–546. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amouyal M. Gene insulation. Part II: Natural strategies in vertebrates. Biochem Cell Biol. 2010;88(6):885–898. doi: 10.1139/O10-111. [DOI] [PubMed] [Google Scholar]
- 45.Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460(7251):128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perissi V, et al. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol Cell. 2008;29(6):755–766. doi: 10.1016/j.molcel.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price AM, et al. Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-κB activation. J Virol. 2012;86(20):11096–11106. doi: 10.1128/JVI.01069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng CW, Zhao B, Kieff E. Four EBNA2 domains are important for EBNALP coactivation. J Virol. 2004;78(20):11439–11442. doi: 10.1128/JVI.78.20.11439-11442.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng CW, et al. Direct interactions between Epstein-Barr virus leader protein LP and the EBNA2 acidic domain underlie coordinate transcriptional regulation. Proc Natl Acad Sci USA. 2004;101(4):1033–1038. doi: 10.1073/pnas.0307808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mushinski JF, et al. (1999) Myc-induced cyclin D2 genomic instability in murine B cell neoplasms. Curr Top Microbiol Immunol 246:183–189; discussion 190–192. [DOI] [PubMed]
- 51.Bouchard C, et al. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001;15(16):2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinclair AJ, Palmero I, Holder A, Peters G, Farrell PJ. Expression of cyclin D2 in Epstein-Barr virus-positive Burkitt’s lymphoma cell lines is related to methylation status of the gene. J Virol. 1995;69(2):1292–1295. doi: 10.1128/jvi.69.2.1292-1295.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zinkel S, Gross A, Yang E. BCL2 family in DNA damage and cell cycle control. Cell Death Differ. 2006;13(8):1351–1359. doi: 10.1038/sj.cdd.4401987. [DOI] [PubMed] [Google Scholar]
- 54.Harris LK, Westwood M. Biology and significance of signalling pathways activated by IGF-II. Growth Factors. 2012;30(1):1–12. doi: 10.3109/08977194.2011.640325. [DOI] [PubMed] [Google Scholar]
- 55.Lewis BA, Reinberg D. The mediator coactivator complex: Functional and physical roles in transcriptional regulation. J Cell Sci. 2003;116(Pt 18):3667–3675. doi: 10.1242/jcs.00734. [DOI] [PubMed] [Google Scholar]
- 56.Maruo S, et al. Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc Natl Acad Sci USA. 2011;108(5):1919–1924. doi: 10.1073/pnas.1019599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130(8):1715–1725. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sinclair AJ, Palmero I, Peters G, Farrell PJ. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 1994;13(14):3321–3328. doi: 10.1002/j.1460-2075.1994.tb06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, et al. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006;20(18):2566–2579. doi: 10.1101/gad.1455006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.