Abstract
ASK1, a member of the MAPK Kinase Kinase family of proteins has been shown to play a key role in cancer, neurodegeneration and cardiovascular diseases and is emerging as a possible drug target. Here we describe a ‘replacement-soaking’ method that has enabled the high-throughput X-ray structure determination of ASK1/ligand complexes. Comparison of the X-ray structures of five ASK1/ligand complexes from 3 different chemotypes illustrates that the ASK1 ATP binding site is able to accommodate a range of chemical diversity and different binding modes. The replacement-soaking system is also able to tolerate some protein flexibility. This crystal system provides a robust platform for ASK1/ligand structure determination and future structure based drug design.
Keywords: ASK1, apoptosis signal regulating kinase 1, structure base drug design
Introduction
Apoptosis signal regulating Kinase 1 (ASK1) has been shown to play a key role in cancer, neurodegeneration and cardiovascular diseases.1 ASK1 is a member of the MAPK Kinase Kinase (MKKK) family of kinases, and is activated by stress stimuli including oxidative stress, endoplasmic reticulum stress, and influx of calcium ions2–5 ASK1 activates the JNK and p38 pathways, but not the ERK pathway, and has been shown to directly phosphorylate MKK3, 4, and 6.
ASK1 is a polypeptide chain of ∼1000 residues consisting of a central serine/threonine kinase domain and N- and C-terminal coiled coil domains. ASK1 exists together with other partner proteins as part of a high molecular weight signalosome.6,7 In the signalosome ASK1 is oligomerized through interactions involving the C-terminal and N-terminal coiled coil domain. Upon activation ASK1 is phosphorylated at the key residue Thr845 in the activation loop. Additional ASK1 autophosphorylation sites have also been identified. ASK2 is closely related to ASK1 in sequence identity especially in the kinase domain. ASK1 and ASK2 have a complex relationship with ASK2 associated and stabilised through the C-terminus of ASK1.
The identification of novel ASK1 inhibitors has been reported8–12 and the structure of the kinase domain of human ASK1 has previously been determined, using co-crystallisation techniques, in complex with Staurosporin and more recently with an imidazole (1,2-α)pyridine.12,13 In order to facilitate structure guided approaches robust methods for ligand structure determination would be beneficial. Here we describe a replacement-soaking method for the crystallisation of ASK1 ligand complexes and use the method to characterise inhibitor binding to ASK1 by the determination of 5 different ASK1 liganded structures from 3 different chemotypes. These structures show that the ASK1 ATP binding site is able to accommodate a range of chemical diversity and provide a platform for further structure based drug design.
Results
A replacement soaking method for ASK1 ligand complexes
ASK1 protein for structure determination was expressed and purified as described in the Materials and Methods. Crystallogenesis of ASK1/ligand complexes required considerable optimisation and interplay of additives. Initial crystallisation experiments yielded showers of small crystals under a variety of conditions, principally from a well solution containing 20% peg 3350, 0.2 M sodium acetate (or other salts) and 100 mM BisTrisPropane buffer, pH 6.5. Inclusion of glycerol, which tends to decrease nucleation, did not result in any improvement, however the addition of 0.05% Novexin (NVoy polymer, Expedeon) to the protein and exchange of BisTrisPropane with Bis-Tris buffer in the well solution resulted in a significant reduction in nucleation whilst still allowing crystal growth after seeding. To improve crystal morphology a variety of standard additives, including solvents, were tested and the inclusion of 0.2% isopropyl alcohol appeared to produce reproducible and excellent quality crystals. The final optimised well solution contained 18% Peg3.4K, 0.2 M sodium acetate, 100 mM BisTris, pH 6.5 with 0.2% isopropyl alcohol. Figure 1(a) shows optimal crystal growth, Figure 1(b,c) shows typically nonoptimal growth and over nucleation.
Figure 1.
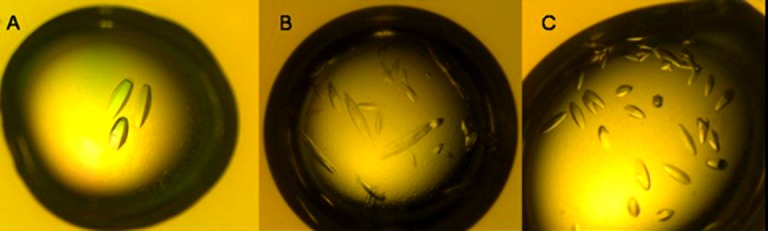
Crystals of ASK1 complexed with Compound 1. All crystallisation experiments were carried out in LINBRO trays by the hanging drop vapour diffusion method with 1 mL of well solution, drops were 1 μL of well solution and 1 μL protein solution. A: Ordered crystals (300–400 μM x 120 μM) grown from a well solution of 18% PEG 3.4K (w/v), 0.2M sodium acetate, 0.1M Bis-Tris buffer pH 6.5 and 0.2% isopropanol (v/v). B: Defective growth with thin elongated edge crystals grown from the well solution containing 3% glycerol instead of isopropanol. C: Over-nucleated drop with crystals grown from the well solution containing 4% peg400 as additive instead of isopropanol.
Replacement-soaking ASK1 crystals with compounds to replace staurosporin proved problematic due to irreproducible exchange and crystal instability, so instead cocrystals were obtained with a lower affinity and more soluble indole-carboxamide (Compound 1, Table II). A replacement-soaking approach using ASK1/Compound 1 crystals was then adopted with compounds of interest and has allowed the rapid determination of more than 50 structures covering a range of ligand chemotypes. Figure 2 shows 2Fo-Fc electron density maps for the four replacement-soaked compounds described in this paper and illustrates that complete replacement of compound 1 (see Supporting Information Fig. S1) could be achieved using this method.
Table II.
Binding Data for Compounds to Ask1 Crystallographic and Full Length Constructs
| pIC50 | ||||
|---|---|---|---|---|
| Chemical structure | ASK1 660–978 T838E N-terminal Flag tag | ASk1 Full length N-terminal GST tag | ASk1 Full length N-terminal 6HisAvi tag | |
| Compound 1 | 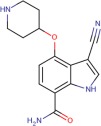 |
6.2 ± 0.5 | 6.6 ± 0.4 | 6.2 ± 0.1 |
| Compound 2 | 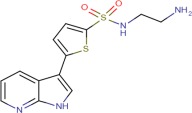 |
6.6 ± 0.1 | 7.3 ± 0.4 | 7.1 ± 0.03 |
| Compound 3 | 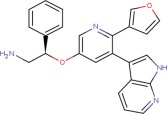 |
7.9 ± 0.1 | 8.1 ± 0.1 | 8.2 ± 0.1 |
| Compound 4 | 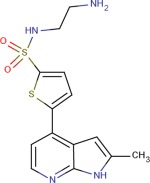 |
6.1 ± 0.1 | 6.6 ± 1.1 | 6.8 ± 0.4 |
| Compound 5 | 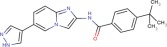 |
7.9 ± 0.1 | 8.2 ± 0.1 | 8.3 ± 0.1 |
Figure 2.
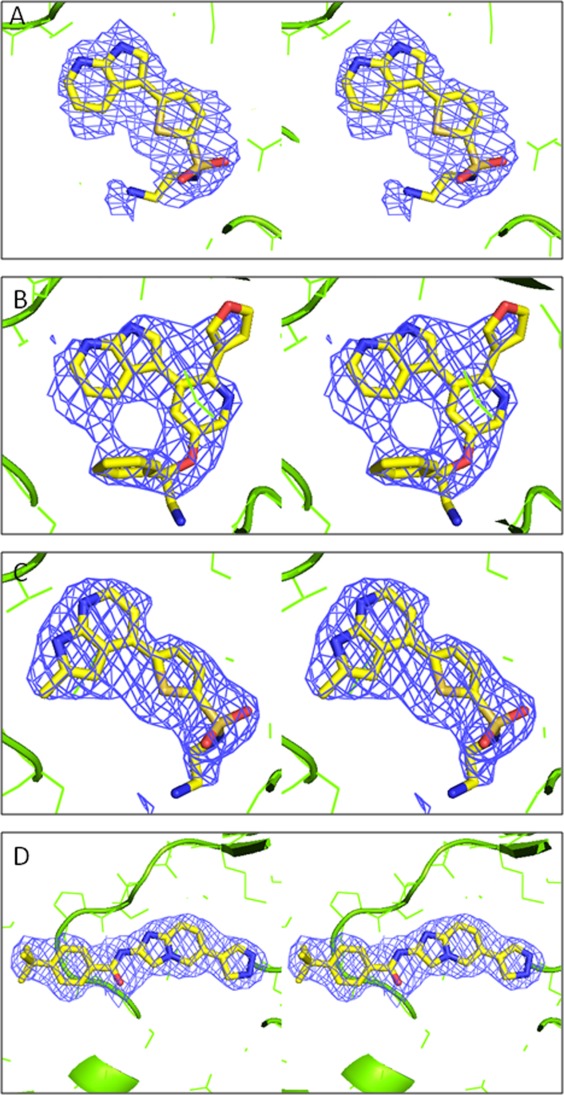
Stereo diagrams showing 2Fo-Fc electron density contoured at 1σ for the four compounds soaked into Ask1/compound 1 co-crystals. A: compound 2, B: compound 3, C: compound 4, D: compound 5. To calculate the electron density maps the structure was refined using the programme Buster21 in the absence of the ligand. Fc and phases were calculated for the structure with the ligand not included. The figure was generated with PyMol (Delano Scientific). For compound 3 the electron density (2b) allows the azaindole group to be placed unambiguously.
Here we focus on the binding of three chemotypes, the indole-carboxamides (Compound 1), the 7-azaindoles (Compound 2, Compound 3 and Compound 4), and the imidazopyridines (Compound 5). Four of the compounds were identified through GlaxoSmithKline's kinase system-based research approach.14 Compound 2 and Compound 4 were identified through screening of a kinase focussed set, whereas Compound 1 and Compound 3 were found by cross-screening compounds made for other kinase programs. We identified compound 5 as a close analogue of inhibitors published in reference 15 and synthesised it as a tool compound for crystallography. Tables I and II show the crystallographic statistics and pIC50 values for the compounds studied. There is good agreement between the binding affinities of the compounds in this study for full length ASK1 protein and the kinase domain construct used for crystallography.
Table I.
Data Collection and Refinement Statistics
| Inhibitor | Staurosporin | Compound 1 | Compound 2 | Compound 3 | Compound 4 | Compound 5 |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P6522 | P6522 | P6522 | P6522 | P6522 | P6522 |
| Cell dimensions | ||||||
| a, b, c (Å) | 78.2, 78.2, 421.3 | 77.9, 77.9, 418.4 | 78.6, 78.6, 420.0 | 77.9, 77.9, 420.1 | 78.4, 78.4, 419.90 | 77.5, 77.5, 423.8 |
| Resolution (Å) | 2.11 (2.11–2.15) | 2.43 (2.43–2.56) | 2.62 (2.62–2.67) | 2.80 (2.80–2.85) | 2.36 (2.36–2.49) | 2.30 (2.30–2.34) |
| aRmerge | 0.082 (0.508) | 0.087 (0.437) | 0.064 (0.390) | 0.127 (0.413) | 0.068 (0.452) | 0.104 (0.410) |
| I/σI | 18.3 (2.2) | 10.2 (2.9) | 32.0 (6.8)b | 8.0 (3.1) | 11.6 (2.7) | 15.3 (2.4) |
| Completeness (%) | 92.7 (97.8) | 99.9 (100.0) | 99.1 (100.0) | 93.6 (94.5) | 94.7 (94.6) | 94.7 (96.5) |
| Redundancy | 4.0 (4.3) | 4.5 (4.7) | 9.1 (10.3) | 4.4 (4.6) | 4.7 (4.2) | 2.8 (2.9) |
| Refinement | ||||||
| No. reflections | 42333 | 29686 | 24197 | 18622 | 30722 | 33093 |
| Rwork/Rfree | 0.206/0.246 | 0.200/0.239 | 0.207/0.265 | 0.194/0.259 | 0.210/0.252 | 0.195/0.235 |
| Overall B-factors (Å2) | 53.1 | 52.4 | 62.9 | 75.9 | 50.6 | 49.9 |
| r.m.s. deviations | ||||||
| Bond length (Å) | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 |
| Bond angles (°) | 1.06 | 1.16 | 1.18 | 1.20 | 1.23 | 1.19 |
Values in parentheses are for the highest resolution shell.
Rmerge = Σ|Ij - <Ij>|/Σ<Ij>.
I/σI is high for this dataset because the resolution was limited by the distance of the detector.
Indole-carboxamide binding
The indole-carboxamide Compound 1 is bound in the ATP binding site through hydrogen bonding to the hinge, specifically between the carboxamide moiety and Val757 main chain nitrogen and carbonyl oxygen atoms [Fig. 3(b)]. A salt bridge is formed between the piperadine of the ligand and Asp822, although a high energy axial connectivity to the piperadine is adopted to facilitate this interaction. The nitrile group occupies space close to the gatekeeper residue, Met754.
Figure 3.
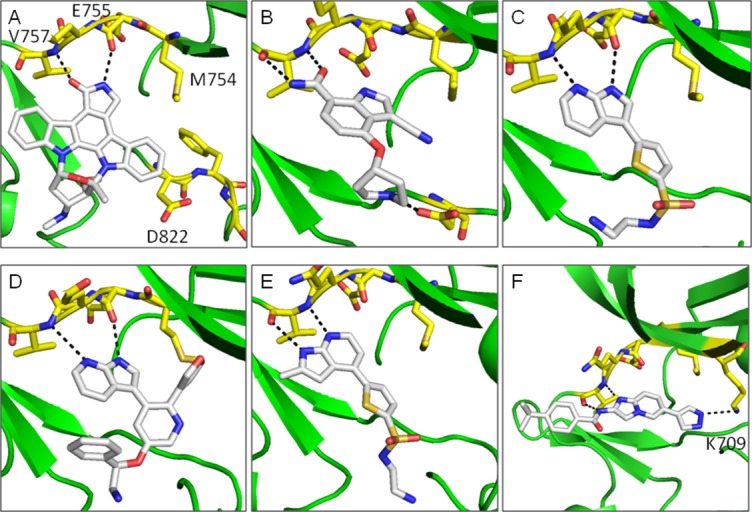
Compound binding to ASK1. A: Staurosporin, B: Compound 1, C: Compound 2, D: Compound 3, E: Compound 4, F: Compound 5. Compounds are shown in white stick representation and key side chain residues are shown as yellow sticks. Hydrogen bonds are shown as black dashed lines. The figure was generated with PyMol (Delano Scientific).
7-Azaindole binding
The crystal structures of ASK1 complexed with Compound 2 and Compound 3 share a common hydrogen bonding pattern to the hinge [Fig. 3(c,d)]. In both cases the pyridine nitrogen hydrogen bonds to the backbone NH of Val757 and the pyrrole NH hydrogen-bonds to the backbone carbonyl of Glu755. In contrast, the structure of ASK1 complexed with Compound 4 shows that the aza-indole binding mode has “flipped” [Fig. 3(e)]. In this case the pyridine nitrogen maintains its hydrogen bond to the backbone NH of Val757, but the pyrrole NH now interacts with the carbonyl of Val757. The reason for this “flipping” seems to be that there is insufficient space available next to the gatekeeper residue (Met754) to accommodate the methyl substituent of the azaindole in the orientation observed for the other ligands. A similar binding mode to Compound 4 is observed in the X-ray structure of a closely related compound complexed with JNK1 (Fig. 1. in Ref. 16). The pIC50 for the azaindoles binding to full length ASK1 vary from pIC50 = 6.6 for Compound 4 to pIC50 = 8.1 for Compound 3. This provides a number of options for exploring chemistry using this chemical template.
Imidazopyridine binding
To investigate the binding of a different chemical template, the structure of ASK1 complexed with Compound 5 was determined. As shown in Figure 3f, Compound 5 was found to bind along the hinge, perpendicular to the binding orientation observed for the aza-indole compounds and in agreement with the previously reported structure.12 Similarly to Compound 4 and Compound 1, Compound 5 hydrogen bonds to Val757 mainchain nitrogen and carbonyl oxygen atoms. The backbone NH interacts with the imidazopyridine of the ligand whilst the backbone carbonyl forms a hydrogen bond with the amide nitrogen. The pyrazole group of Compound 5 forms a hydrogen bond with the catalytic lysine 709. The potency of Compound 5 for full length ASK1 (pIC50 = 8.2) is similar to the azaindole Compound 3 (pIC50 = 8.1) indicating that similar potency can be achieved by compounds with two distinct binding modes (Table II).
Overall structure of the kinase domain
For the ASK1/ligand structures described in this article the overall structure of the kinase domain closely resembles the previously reported structures of ASK1, 2CLQ, and 3VW6 (for Ask1/Staurosporin complex the RMSd = 0.39 Å2 on all Cα atoms from Chain A), with the Cα-helix in a catalytically non-productive conformation.12, 13 In agreement with structure 3VW6, the DFG loop adopts a “DFG-in” conformation for all ligand complexes (Fig. 3a).
Superposition of all structures described in this paper show changes in secondary structure of the ASK1 nucleotide binding loop, a feature known to be common for protein kinases (Fig. 4). The largest difference is observed between ASK1/staurosporin and ASK1/Compound 5 structures where a difference of greater than 1 Å is observed in the Cα positions of residues Gly689 to Tyr691. This comparison shows that the crystal form and replacement-soaking methodology described in this paper is able to accommodate small changes in nucleotide binding loop conformation which could be important for facilitating a diversity of inhibitor binding.
Figure 4.
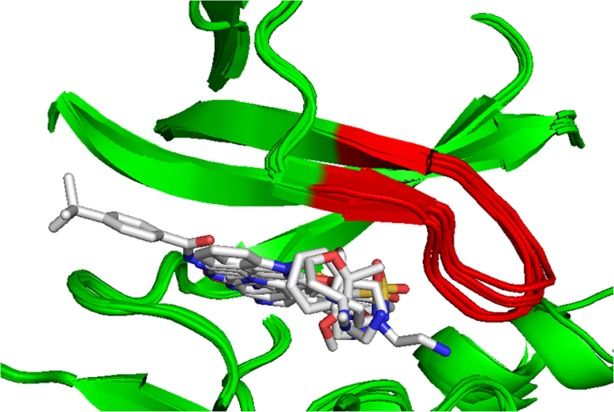
Superposition of all six structures described in this article. Superposition was carried out on all Cα atoms from molecule A in the asymmetric unit. The nucleotide binding loop, residues Gly687 to Val694, is shown in red. The figure was generated with PyMol (Delano Scientific).
Discussion
We have developed a reproducible crystallisation system where the crystals routinely diffract to better than 2.5 Å resolution. The ASK-1 crystals are grown with an uncommon additive, novexin with a trace amount of isopropanol. Both of these affect protein solubility, reduce aggregation and control crystal growth. The additives also appeared to increase the crystal size and uniformity. The crystals were initially optimised as staurosporin complexes, however we found this inhibitor difficult to replace with low molecular weight ligands. A simpler replacement-soaking method using co-crystals of ASK1 with the indole-carboxamide Compound 1 was established for the determination of ASK1/inhibitor complexes. This compound was selected on the basis that it was reasonably potent for its size, soluble and could be co-crystallised with ASK1.
Five crystal structures from three different chemical templates, the indole-carboxamides (Compound 1), the 7-azaindoles (Compound 2, Compound 3, and Compound 4) and the imidazopyridines (Compound 5) have been determined. All the chemotypes make well-defined interactions with the kinase hinge, however the binding geometries, the nature of the interaction with the hinge and the areas of the active site accessed differ substantially between chemotypes. Comparison of the binding modes of the 7-azaindoles with the imidazopyridine clearly show how distinct compound binding modes can be accommodated by ASK1, while the structures of Compound 3 and Compound 4 illustrate that, even within the azaindole class, modest changes to the ligand structure are seen to affect ligand binding geometry. Taken together these structures demonstrate that we have developed a robust crystallography system for ASK1 that can accommodate different chemotypes and changes in protein conformation. This crystal system can be used to drive future structure-based design efforts on ASK1.
Methods
Cloning, expression, and purification
Preparation of FLAG-TEV-ASK1 660-978 T838E
The amino acids 660-978 of human ASK1 were PCR amplified from codon-optimized ASK1 (Genscript) using two oligonucleotides (5′-CGC GGA TCC ATG GAT TAC AAG GAT GAC GAC GAT AAG GAA AAC CTG TAT TTT CAG GGC TCC ACC GAG GAG GGT GAC TGC GAG TCC-3′ and 5′-GCG TCT AGA TCA GGA GGT GTC CTC CAC CAG CAC GGG CAC G-3′), adding an N-terminal FLAG tag and TEV protease cleavage site. This was inserted into pFASTBac1 (Invitrogen, San Diego, CA) using BamHI and XbaI restriction endonuclease sites. The T838E mutation was introduced by site directed mutagenesis (using the oligonucleotides 5′-CCCTTGCACCGAGGAGTTCACCGGCACCC-3′ and 3′-GGGTGCCGGTGAACTCCTCGGTGCAAGGG-5′). Expression of FLAG TEV ASK1 660-978 T838E was performed using SF9 insect cells. Using Excell 420 media (SAFC Biosciences), sufficient baculovirus stock was amplified and subsequently used to express ASK1 by synchronous baculoviral infection in either a bioreactor (Applikon) or a wave culture system (Wave Biotech). The infected cells were harvested using a continuous centrifuge.
For purification, two cell pellets 129g (Run A) and 119g (Run B) were individually processed in the same manner by mixing with 650 mL of 10 mM Hepes, 500 mM NaCl, 1 mM EDTA, 0.2 mM PMSF, 1 mM benzamidine, 1 μg/mL BLAP (a protease inhibitor cocktail consisting of 1 mg/mL each of bestatin, leupeptin, aprotinin and 2 mg/mL pepstatin), 0.1% Triton X-100, pH 7.4 and left to stir for 30 min at 4°C. The resulting suspension was lysed by dounce homogenisation and then centrifuged for 60 min at 100,000g. The lysate supernatant was harvested and then contacted with 100 mL Anti-FLAG M2 resin (Sigma) which was washed backed to baseline with buffer A. (Buffer A : 10 mM Hepes, 500 mM NaCl, 1 mM EDTA, 0.2 mM PMSF, 1 mM benzamidine, 1 μg/mL BLAP, pH 7.4). Bound ASK1 protein was eluted with 4 column volumes of buffer B (Buffer B: 10 mM Hepes, 150 mM NaCl, 1 mM EDTA, 0.2 mM PMSF, 1 mM benzamidine, 1 μg/mL BLAP, pH 7.4) containing 100 μg/mL Flag peptide (Sigma F3290). This yielded 82.4 mg (Run A) and 120 mg (Run B) of total protein. Each pool was diluted down to 5m/Scm using 20 mM Tris, 1 mM EDTA, 0.2 mM PMSF, 1 mM benzamidine, pH 8.5 (Buffer A) and loaded directly onto and further purified by a 20 mL Source15Q anion exchange. Elution of the ASK1 was carried using a NaCl linear gradient (0-100% over 8Cvs) using 20 mM Tris, 1M NaCl, 1 mM EDTA, 0.2 mM PMSF, 1 mM bBenzamidine, pH 8.5 (Buffer B). Post 15Q fractions were pooled yielding 23.7 mg (Run A) and 14.8 mg (Run B). Post 15Q pools run A and run B were combined together and further purified with 100 mL Anti-FLAG M2 resin (Sigma), using buffers already detailed. Post Flag fractions were pooled yielding 19.3 mg of total protein which was polished by gel filtration (Superdex 75, 120 ml Cv) giving a final yield of 10.5 mg in 20 mM Tris, 150 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 5% Glycerol, pH 8.0. The molecular weight of the protein was determined by LC-MS and indicated a mass difference consistent with a protein modification of one single acetylation.
Enzyme assay
The ASK1 FP assay was configured in an assay buffer containing 50 mM Hepes, pH7.5, 150 mM NaCl, and 1 mM CHAPS and carried out in low volume, black 384-well microtitre plates (Greiner BioOne, 784076) in a final reaction volume of 10 μL. This consisted of a 5 μL ligand addition containing 2.5 nM of an ATP competitive fluoroprobe (FP ligand), which is FAM, fluorophore labelled, to assay wells containing 100 nL test compound, dissolved in 100% DMSO, and 5 μL ASK1. The concentrations of ASK1 used were dependent on the construct and the Kd for the FP ligand, as all assays were configured at 2Kd. The concentration of ASK1 660-978 T838E used was 50 nM (Kd = 25 nM), full length GST-ASK1 and His-Avi ASK1 both used 10 nM (Kd = 25 nM). After addition of reagents to the microtitre plates the assays were incubated for 60 min before reading on a Perkin Elmer Envision. Fluorescence polarisation (FP) was measured using a PerkinElmer Envision plate reader (PerkinElmer, Waltham, MA). Briefly, the FP ligand was excited at 485 nm and the resultant emission measured at 535 nm using both S (parallel) and P (perpendicular) filters. The following equation was used to convert parallel and perpendicular intensities into polarization (mP) data:
Polarization (mP) = 1000 × (S − P)/(S + P)
For IC50 determination, test compounds were serially diluted in DMSO (1% final assay concentration) and subsequently IC50 values were calculated using a 4 parameter logistic equation.
Crystallization
ASK1 protein [8–10 mg/mL, in 20 mM Tris, 150 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 5% Glycerol, pH 8.0 and stored at −80°C] was incubated with 1 mM Compound 1 and 0.05% Novexin (Nvoy, Expedeon Protein Solutions) and then 1–2 μL of the solution was mixed with equal volume of optimised crystallisation solution [18% PEG 3.4K (w/v), 0.2M sodium acetate, 0.1M Bis-Tris buffer pH 6.5 and 0.2% isopropanol (v/v)]. The crystallisation drops were streak seeded, this produced crystals which grew over 3–4 weeks to sufficient size. Replacement soaking involved incubating Ask1/compound1 cocrystals with new ligands at 10 mM (max. 5% DMSO final concentration) in crystallization solution for 2–3 days. Finally, the crystals were immersed into the cryo-cooling solution (crystallisation solution containing 10 mM compound and with 20% glycerol) before fast freezing with liquid nitrogen.
Structure determination
X-ray data for Ask1 complexed with staurosporin, Compound 1, Compound 2, and Compound 4 was collected at the ESRF on beamline ID23-1, data for ASK1/Compound 3 at the ESRF beamline ID14-4 and data for ASK1/Compound 5 at Diamond beamline i02. For structures liganded with staurosporin, Compound 2, Compound 3 and Compound 5 data were processed and scaled with the programmes DENZO and SCALEPACK,17 for structures complexed with Compound 1 and Compound 4 data were processed and scaled with mosflm and scala.18 All structures were solved by Molecular Replacement with the programme PHASER19 using structure 2CLQ as a search model with the staurosporin and water molecules removed. For all structures initial rounds of building and refinement were carried out with COOT20 and REFMAC.18 Final rounds of refinement were carried out with BUSTER.21 In the final structures the following residues are disordered in the electron density: residues 660–670 at the N-terminus, residues 940 at the C-terminus, residues 830–837 from the activation loop and residues 714–720. Structures have been deposited into the PDB with the following codes; 4bf2 (Ask1/Staurosporin), 4bhn (Ask1/Compound5), 4bib (Ask1/Compound1), 4bic (Ask1/Compound2), 4bid (Ask1/compound3), 4bie (Ask1/Compound4).
Acknowledgments
The authors thank placement students Mark White, Jing Yang Liu as well as Dr. Alex Wohlkonig for help with crystallization, John Darker for chemistry support and the staff at Diamond and the ESRF for help with data collection.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Hattori K, Naguro I, Runchel C, Ichijo H. The roles of ASK family proteins in stress responses and diseases. Cell Commun Signal. 2009;7:9. doi: 10.1186/1478-811X-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ichijo H, Nishida E, Irie K, TenDijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 3.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, Hori S, Kakizuka A, Ichijo H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeda K, Matsuzawa A, Nishitoh H, Tobiume K, Kishida S, Ninomiya-Tsuji J, Matsumoto K, Ichijo H. Involvement of ASK1 in Ca2+-induced p38 MAP kinase activation. EMBO Rep. 2004;5:161–166. doi: 10.1038/sj.embor.7400072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tobiume K, Saitoh M, Ichijo H. Activation of apoptosis signal-regulating kinase 1 by the stress-induced activating phosphorylation of pre-formed oligomer. J. Cell Physiol. 2002;191:95–104. doi: 10.1002/jcp.10080. [DOI] [PubMed] [Google Scholar]
- 7.Noguchi T, Takeda K, Matsuzawa A, Saegusa K, Nakano H, Gohda J, Inoue J, Ichijo H. Recruitment of tumor necrosis factor receptor-associated factor family proteins to apoptosis signal-regulating kinase 1 signalosome is essential for oxidative stress-induced cell death. J Biol Chem. 2005;280:37033–37040. doi: 10.1074/jbc.M506771200. [DOI] [PubMed] [Google Scholar]
- 8.Okamoto M, Saito N, Kojima H, Okabe T, Takeda K, Ichijo H, Furuya T, Nagano T. Identification of novel ASK1 inhibitors using virtual screening. Bioorg Med Chem. 2011;19:486–489. doi: 10.1016/j.bmc.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Volynets G, Bdzhola V, Kukharenko O, Iarmoliuk S. Identification of low-molecular inhibitors of proteinase ASK1. Ukr Biokhim Zh. 2010;82:41–50. [PubMed] [Google Scholar]
- 10.Volynets G, Chekanov MO, Synyugin AR, Golub AG, Kukharenko OP, Bdzhola VG, Yarmoluk S. Identification of 3H-naphtho[1,2,3-de]quinoline-2,7-diones as inhibitors of apoptosis signal-regulating kinase 1 (ASK1) J Med Chem. 2011;54:2680–2686. doi: 10.1021/jm200117h. [DOI] [PubMed] [Google Scholar]
- 11.Volynets G, Bdzhola V, Golub A, Synyugin A, Chekanov M, Kukharenko O, Yarmoluk S. Rational design of apoptosis signal-regulating kinase 1 inhibitors: discovery novel structural scaffold. Eur J Med Chem. 2012;S0223-5234:00561–00562. doi: 10.1016/j.ejmech.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 12.Terao Y, Suzuki H, Yoshikawa M, Yashiro H, Takekawa S, Frujitani Y, Okada K, Inoue Y, Yamamoto Y, Nakagawa H, Yao S, Kawamoto, Uchikawa O. Design and biological evaluation of imidazo[1,2-a]pyridines as novel and potent ASK1 inhibitors. Bioorg Med Chem Lett. 2012;22:7326–7329. doi: 10.1016/j.bmcl.2012.10.084. [DOI] [PubMed] [Google Scholar]
- 13.Bunkoczi G, Salah E, Filippakopoulos P, Fedorov O, Muller S, Sobott F, Parker S, Zhang H, Min W, Turk B, Knapp S. Structural and functional characterization of the human protein kinase ASK1. Structure. 2007;15:1215–1226. doi: 10.1016/j.str.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bamborough P, Brown M, Christopher J, Chung C, Mellor J. Selectivity of kinase inhibitor fragments. J Med Chem. 2011;54:5131–5143. doi: 10.1021/jm200349b. [DOI] [PubMed] [Google Scholar]
- 15. Takeda. W020080161131(A1)
- 16.Liddle J, Bamborough P, Barker MD, Campos S, Chung CW, Cousins RP, Faulder P, Heathcote ML, Hobbs H, Holmes DS, Ioannou C, Ramirez-Molina C, Morse MA, Osborn R, Payne JJ, Pritchard JM, Rumsey WL, Tape DT, Vicentini G, Whitworth C, Williamson RA. 4-Phenyl-7-azaindoles as potent, selective and bioavailable IKK2 inhibitors demonstrating good in-vivo efficacy. Bioorg Med Chem Lett. 2012;22:5222–5226. doi: 10.1016/j.bmcl.2012.06.065. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z. Data collection and processing. In: Sawyer L, editor. Warrington, UK: SERC Daresbury Laboratory; 1993. pp. 56–62. Proceedings of CCP4 Study Weekend. [Google Scholar]
- 18.The CCP4 Suite. Programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 19.McCoy AJ, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.Bricogne G. Direct phase determination by entropy maximization and likelihood ranking: status report and perspectives. Acta Cryst. 1993;D49:37–60. doi: 10.1107/S0907444992010400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


