Abstract
Nanolipoprotein particles (NLPs) are nanometer-scale discoidal particles that feature a phospholipid bilayer confined within an apolipoprotein “scaffold,” which are useful for solubilizing hydrophobic molecules such as drugs and membrane proteins. NLPs are synthesized either by mixing the purified apolipoprotein with phospholipids and other cofactors or by cell-free protein synthesis followed by self-assembly of the nanoparticles in the reaction mixture. Either method can be problematic regarding the production of homogeneous and monodispersed populations of NLPs, which also currently requires multiple synthesis and purification steps. Telodendrimers (TD) are branched polymers made up of a dendritic oligo-lysine core that is conjugated to linear polyethylene glycol (PEG) on one end, and the lysine “branches” are terminated with cholic acid moieties that enable the formation of nanomicelles in aqueous solution. We report herein that the addition of TD during cell-free synthesis of NLPs produces unique hybrid nanoparticles that have drastically reduced polydispersity as compared to NLPs made in the absence of TD. This finding was supported by dynamic light scattering, fluorescence correlation spectroscopy, and cryo transmission electron microscopy (Cryo-EM). These techniques demonstrate the ability of TDs to modulate both the NLP size (6–30 nm) and polydispersity. The telodendrimer NLPs (TD-NLPs) also showed 80% less aggregation as compared to NLPs alone. Furthermore, the versatility of these novel nanoparticles was shown through direct conjugation of small molecules such as fluorescent dyes directly to the TD as well as the insertion of a functional membrane protein.
Keywords: nanolipoproteins, telodendrimer, cell-free expression, apolipoprotein, nanotechnology, membrane protein, mono-dispersity
Introduction
NLPs are discoidal nanoparticles that form when apolipoproteins and phospholipids (DMPC: 1,2-Dimyristoyl-sn-Glycero-3-Phosphocholine; POPC: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, etc.) self-assemble to form water-soluble particles featuring a phospholipid bilayer that effectively mimics the cell membrane.1–3 NLPs can offer distinct advantages over other membrane model systems in terms of particle size and solubility when compared to liposomes.4 The presence of a circular apolipoprotein scaffolding or “belt”5 constrains the dimensions of the bilayer, helping to maintain particle diameters.6,7 The protein belt may also contribute to improved NLP thermal stability as compared with micelles or liposomes. The size and stability of NLPs has been shown to be useful for a broad range of biotechnology applications,8,9 mainly involving the solubilization of functional membrane proteins.4,10–14 The NLP lipid bilayer provides a hydrophobic patch for the incorporation of membrane proteins,5,12,15 as well as the formulation of hydrophobic drugs.9,16
We recently developed an approach for the synthesis of NLPs via cell-free protein expression in the presence of phospholipids.12 This convenient method results in the spontaneous formation of NLPs as the apolipoprotein is translated in the reaction mixture. The cell-free method is potentially useful for incorporating unnatural amino acids, labels, and tags on NLPs, a strategy not readily available with whole cell systems, and is fast—requiring only a few hours from start to finish.17–20 Cell-free methods have allowed one to go from gene to protein to structure in a single day,21 and accommodates a wide range of additives that augment protein expression, including chaperonins,22 lipids,23 redox factors,20,24 detergents, and protease inhibitors.23,25 Additionally, varying the lipid selection in a cell-free reaction may affect membrane protein function, as has been demonstrated in recent studies with G-protein coupled receptors (GPCR) reconstituted into NLPs using DMPC alone,10 POPC alone,26 or a mixture of POPC/POPG (palmitoyl-oleoyl-phosphoglycerol).11
Micelles self-assembled from amphiphilic PEG-dendritic block copolymers (Telodendrimer, TD) have favorable properties for both cancer imaging and nanoparticle-mediated drug therapy.27,28 The presence of the hydrophilic PEG chain serves to prevent immunogenicity, increase blood circulation time, and minimize biological degradation of nanoparticles in vivo. The presence of cholic acid (CA) moieties imparts amphiphilicity to TDs, enabling them to form nanomicelles upon exposure to the appropriate solvent conditions. In addition, TDs may contain additional functional covalent attachment sites that act as handles for biologically active targeting ligands and cellular uptake molecules. Because TDs are excellent at dissolving hydrophobic compounds for drug formulation applications, they may be amenable to the solubilization of functional membrane proteins. It has been reported that detergent amphiopols could impart solubility to membrane proteins depending on their charge, size, and aqueous solubility.29,30 This effect has never been shown with TD-lipid particles and will be tested. We hypothesize that combining the favorable attributes of TD with NLPs may result in a novel nanoparticle with improved capabilities and broader application potential compared to either nanoparticle, TD, or NLP, composition alone.
In this study, we used cell-free protein expression to generate TD and NLP hybrid nanoparticles. The TD-containing NLPs (TD-NLPs) demonstrated improved nanoparticle homogeneity and the size was tunable by varying the fraction of TD incorporated into the NLPs. We also assessed the versatility of TD-NLPs as a platform technology through the addition of fluorescent molecules and the insertion of a model membrane protein. This assembled nanoparticle represents a novel entity with unique capabilities when compared with either nanomicelles made of TD-lipids or NLP nanocarriers alone.
Results and Discussion
Cell free production of TD-NLPs
We have used a cell free protein expression method to synthesize TD-NLPs that were different in size and solubility when compared to NLPs or TD-lipid particles. The process to produce NLPs in the presence of additional additives is outlined in Figure 1. The process outlined in Figure 1 represents a novel path for generating NLPs. Typically, the entire process can be completed only in 8 h, compared to potentially taking days to prepare when using traditional methods to form NLPs.3 Several types of TDs were utilized as depicted in Table I and these molecules varied according to the molecular weight of the PEG in daltons and the number of CA per TD monomer unit. This study used the following molecules: PEG2k-CA4, PEG2k-CA8, PEG5k-CA8, and PEG5k-CF4, which contains four cholesterol molecules per TD molecule. Plasmids coding for a His-tagged ApoA1 derivative, Δ49ApoA1, and TD monomers were premixed with DMPC lipids, and incubated with cell free reaction mixtures to produce fully assembled nanoparticles. As controls we mixed TD monomers with DMPC to form TD-lipid particles. NLP and TD-NLP particle formation were first confirmed by native poly acrylamide electrophoresis (PAGE) and then purified by nickel affinity purification, which removed TD-lipid particles. Samples were then analyzed using native gel and size exclusion chromatography (SEC). The 10% PEG5k-CA8 TD-NLP has a size of ∼350 kDa compared to ∼242 kDa of normal NLP (Supporting Information Fig. 1).
Figure 1.
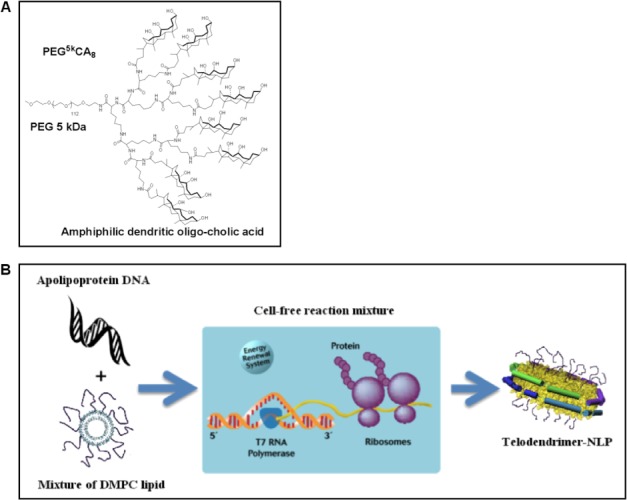
Schematic of TD-NLP assembly process using telodendrimer-based chemistry. Cell-free synthesis of telodendrimer conjugated NLPs. (A) The chemical structure of a telodendrimer comprised of an octamer of cholic acid (CA8) linked to the terminal end of a linear 5 kDa PEG molecule (PEG5K), which is solubilized with DMPC lipid. (B) The TD-lipid mixture is then added to a general cell free reaction, resulting in the self-assembly of TD-NLPs.
Table I.
Telodendrimers Used in Combination with DMPC to Form TD-NLP Complexes
| Telodendrimer molecule | MW (kDa) | Head group | PEG chain (kDa) |
|---|---|---|---|
| PEG2k-CA4 | 4 | 4 × Cholic acid | 2 |
| PEG2k-CA8 | 6 | 8 × Cholic acid | 2 |
| PEG5k-CF4 | 7 | 4 × Cholesterol | 5 |
| PEG5k-CA8 | 9 | 8 × Cholic acid | 5 |
Incorporation of TD enhances soluble NLP yield
We first analyzed the purified yield of the soluble TD-NLP nanoparticles. A total of 1 mL of cell-free reaction mixture was used for particle synthesis with and without TD to compare protein yields and solubility (Fig. 1). A 4–12% SDS–PAGE gel was used to separate the products during and after nickel affinity chromatography (Supporting Information Fig. 2). We noted a 2- to 4-fold increase in production of soluble TD-NLPs produced when compared to NLPs alone. As shown in Supporting Information Figure 2, the PEG5k-CA8 TD reaction yielded ∼750 μg/mL of TD-NLP, as compared to 190 μg/mL of NLP alone, which was a 2.95-fold improvement. The difference in solubility appeared to be a function of TD incorporation. It is fully plausible that the increased yield may have been caused by increased accessibility to the His-tag, which may have been due to changes in the His-tag accessibility that were induced by the TD. This result is particularly interesting because an increase in soluble NLP yield would aid in making cell free methods more cost effective. Moreover, the increase in NLP solubility could potentially help enhance the solubility and yield of a membrane protein that is supported by TD-NLPs by just increasing the efficiency of the purification, which leads to sample enrichment.
The size of TD-NLP is controlled by the type of TD used
We next studied how telodendrimers (TD) impacted the size of NLP-based nanoparticles. Dynamic light scattering (DLS) was used to evaluate the size and monodispersity of the TD-lipid particles, NLPs, and TD-NLPs. All three particle types had distinct size differences. The TD-NLP particle sizes were significantly dependent upon the length of the incorporated PEG molecule (Fig. 2). The PEG2K TD-NLPs ranged in mean size from 6 to 13 nm; while the PEG5K TD-NLPs were 23–30 nm in diameter based on DLS traces (Fig. 2, Panel C). The P-value between the mean sizes of PEG2K and the PEG5K TD-NLPs was 0.0001, while it was 0.32 between PEG2K-CA4 and PEG2K-CA8 NLPs, and 0.43 between PEG5K-CA8 and PEG5K-CF4 NLPs. The TD-lipid particles formed larger nanoparticles as compared to those made in the presence of the apolipoproteins (TD-NLPs) and the size of the particles was dependent on the type of telodendrimer used (Supporting Information Fig. 3). In general the more telodendrimer added to the DMPC lipid solution would result in greater TD-lipid particle size increases as well as particle aggregation (data not shown). Previously reported studies have shown that NLPs alone measure ∼8 nm, depending on the apolipoprotein, in solution when properly dispersed through multiple rounds of purification.14 Changing the amount of TD added to the NLP assembly process over a range of 1–10% of total lipid did not significantly alter the overall size of the TD-NLP particles (Fig. 2, Panel A,C). This study demonstrates a new approach to rapidly design and assemble TD-NLPs of desired size to meet specific needs, for example, to potentially support or limit membrane protein complexes of varying sizes that could be carried within the lipid bilayer.
Figure 2.
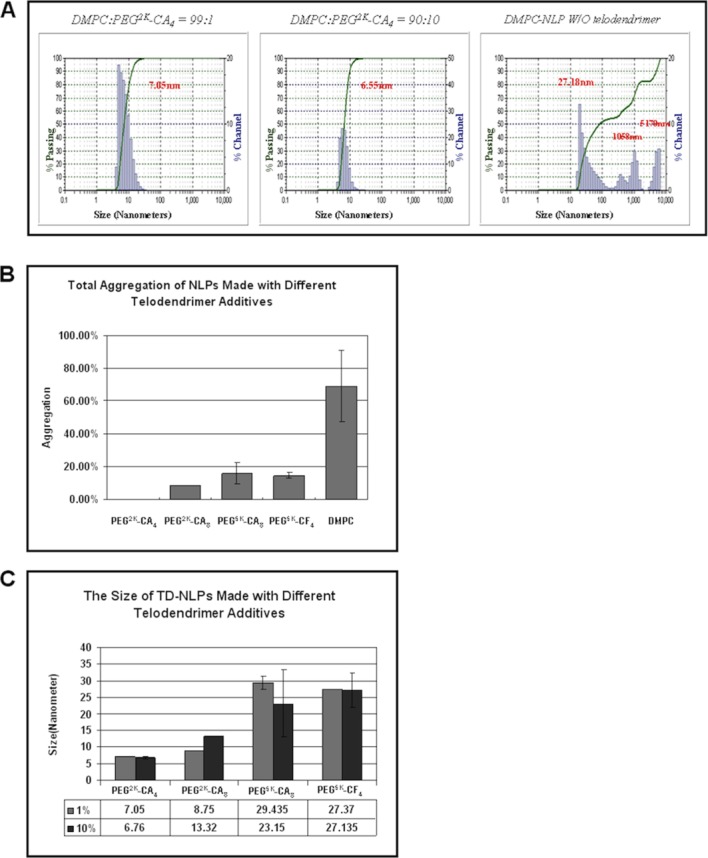
Telodendrimer addition to nanoparticle formation changed aggregation levels and size tenability. Dynamic light scattering (DLS) was used to assess several biophysical properties of the TD-NLP particles. (A) The size distribution of TD-NLPs measured by dynamic light scattering. TD-NLPs demonstrated an improved homogeneity compared to normal DMPC-NLP. (B) Total aggregations of various TD-NLPs are significantly less as compared to normal DMPC-NLP (p < 0.01). (C) PEG tail length and size of the TD-NLPs are significantly correlated (PEG2K vs. PEG5K, p < 0.01). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
TD-NLP shows decreased level of aggregation
Next we assessed the aggregation of TD-NLPs based on DLS measurements. Although not addressed in previous publications, aggregation could complicate experimentation using NLPs. TD-NLP aggregation appears to be size dependent, with larger TD molecules exhibiting higher levels of aggregation. Interestingly, the overall level TD-NLPs aggregation rate was reduced 10–100 times as compared to NLPs assembled only in the presence of DMPC alone (Fig. 2, Panel A,B). Increases in the amount of TD to lipid ratio (>10%) were associated with greater levels of aggregation (Supporting Information Fig. 4). There were no significant changes in NLP size or aggregation when adjusting for TDs containing CA or cholesterol head groups over the 1–10% TD to lipid ratio. Monodispersity of TD-NLPs was also confirmed by SEC (Supporting Information Fig. 5). The much larger TD-lipid particles when isolated were found within the void volume of the SEC fractions (data not shown). It is clear that the addition of CA or cholesterol based TDs to the cell free method can disaggregate NLP-based nanoparticles resulting in a more monodispersed entity.
To further study the shape and aggregation properties of TD-NLP complexes, we performed Cryo-EM experiments. Cryo-EM images illustrated that NLPs are discoidal in shape with height dimensions consistent with previously published images of 10 nm diameter nanodiscs with phospholipid bilayers (Fig. 3). Three assemblies are shown, with DMPC alone (panel A) and with two different types of TDs (Panel B,C). Unlike previous reports, NLPs alone (Panel A) in our study exhibited large clustering rather than stacked particles, or “rouleaux,” which may be attributable to the inclusion of tobacco virus particles in this study. All of the TD-NLPs showed minimum aggregation that is in accordance with the aforementioned DLS results. The disc-like shape of TD-NLPs allows for accessibility from both side of the membrane when supporting membrane protein, which may be an advantage over the traditional liposome and micelle methods.
Figure 3.
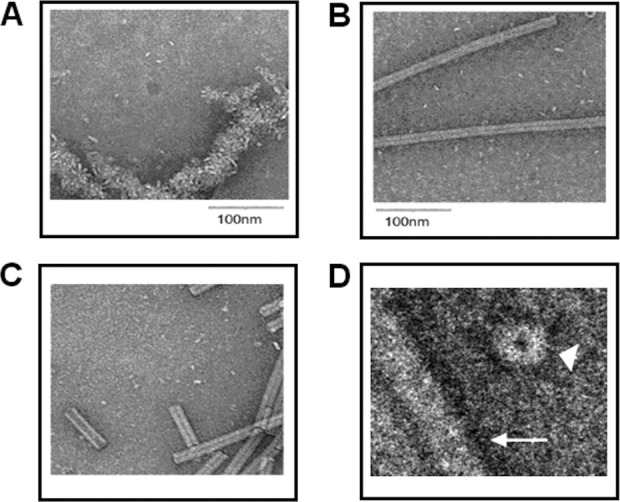
Imaging of TD-NLPs with decreased aggregation compared to NLP. Cryo-EM visualization of NLP preparations with and without telodendrimers is shown to validate reduced aggregation levels of TD-NLPs. (A) NLP assembly using DMPC lipids alone. (B) NLP assembly using DMPC lipids with 10% telodendrimer PEG2K-CA4. (C) NLP assembly using DMPC lipids with 10% telodendrimer PEG2K-CA8. (D) Magnification of NLP with 10% PEG2K-CA8 (arrowhead) from its top view. Tubular structures (white arrow) represent tobacco mosaic viruses for reference.
TD provides a convenient approach for NLP labeling
To investigate the feasibility of using the telodendrimer in NLP labeling as well as measure the solution-based associations between the different molecules, we covalently conjugated FITC fluorophore to the PEG tail of the TD. FITC-TD was then added to the cell-free reactions containing TexasRed-POPC dissolved in DMPC to generate FITC/TexasRed dual labeled TD-NLPs. In comparison, cell-free reactions were used to assemble NLPs tagged with Bodipy®-FL and TexasRed labels. We then performed fluorescent correlation spectroscopy (FCS) analysis14 to topologically confirm TD association and NLP labeling. As seen in Figure 4, both NLP (identified by cross-correlating Bodipy and TexasRed in the complex) and TD-NLP (identified by cross-correlating FITC and Texas Red in the complex) diffused significantly faster than DMPC vesicles alone, but slower than the protein Δ49ApoA1 (apolipoprotein without any DMPC). These results are a clear indication of TD-NLP formation and successful nanoparticle labeling. In theory, our TD-NLP could also allow for the rapid design and production of in vivo targeting, imaging, and therapeutic delivery of biologically active molecules through direct surface conjugation to the TD. Novel labeling and modification strategies as shown by the FITC labeling method used in this study, exhibit the versatility of these vehicles.
Figure 4.
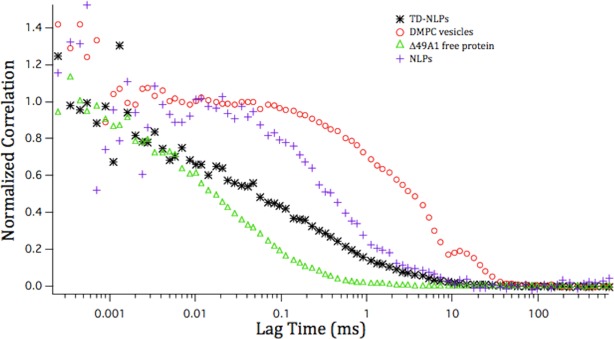
Telodendrimers are associated with nanoparticles as a complex. Diffusion curves of proteins and NLP/TD-NLP complexes as measured by FCS. Green, black, blue, and red curves correspond to Δ49A1, TD-NLP, NLP, and DMPC vesicles in 1× PBS, respectively.
TD-NLP supports functional membrane protein
Self-assembled TD-NLPs will support incorporation of a functional integral membrane protein. It has been previously shown, that bacteriorhodopsin (bR; a seven transmembrane helical protein, from Halobacterium salinarium) can be robustly coexpressed and assembled into NLPs for biophysical characterization.12,14,15 In this study, assembly of the soluble bR-NLP complex was observed within 4 h after addition of plasmids to an E. coli cell-free lysate (Fig. 5, Panel A). Addition of TDs to the cell-free reaction did not affect bR function as indicated by the observable pink coloration of the tubes, which is an indication of proper folding and function.12 Production of similar amounts of total bR protein with and without TDs was also observed (Fig. 5, Panel B). For control, we expressed bR in the presence of TD-lipids without apolipoprotein. This also produced functional bR (Supporting Information Fig. 7, Panel A), but the particles were not soluble when tested for solubility (Supporting Information Fig. 7, Panel B). This solubility difference was distinct, when compared to coexpressed bR TD-NLP. Our data demonstrated that inclusion of the TDs do not inhibit the production of functionally inserted bR protein. This is important because it paves the way for inserting additional receptors for in vivo use. TD-NLPs may be used to deliver nearly any type of membrane protein/receptor or other small molecules. Combined with the labeling technique described above, our soluble TD-NLP complexes could be exploited for multiple applications in cellular targeting, delivery of therapeutics, and membrane protein biochemistry. Moreover, the production of the TD-NLP, which only requires minimum steps to achieve usable material for these biotechnology applications, represents the first unique technique to infer multifunctionality to NLPs.
Figure 5.
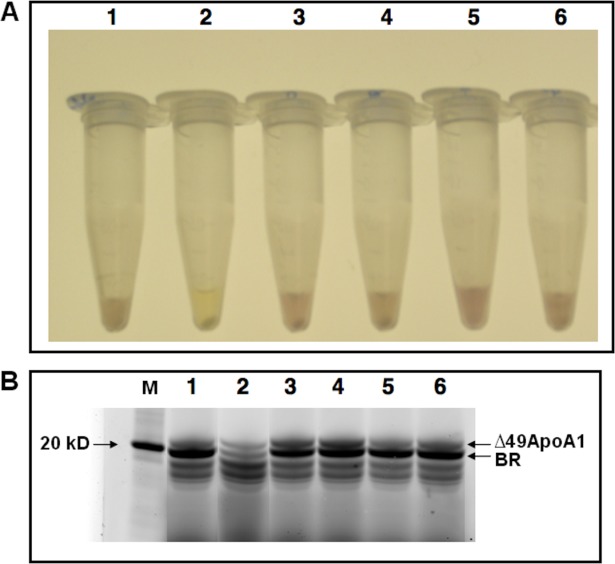
Telodendrimers are compatible with membrane protein production. The cell free reactions are setup with 1 μg/mL pIVEX-2.4d-Δ49ApoA1 and 10 μg/mL pIVEX-2.4d-boP and 2 mg/mL lipid (100% DMPC or 99.5% DMPC and 0.5% TD, molar ratio, etc), BODIPY-FL and 50 μM all trans retinal, 30°C at 990 rpm for 4 h. (A) The pictures of the tubes were taken after the reaction tubes were centrifuged at 14,000 rpm for 10 mins. The purple colored solution indicated soluble production of correctly-folded bR. (B) Denaturing SDS-PAGE gel electrophoresis of cell-free expressed proteins. All samples were loaded along with a molecular weight standard (MW). The pictures were taken with GE-TYPHOON 9410 using laser/filter 488 nm/520 nm. The nonspecific bands below 20 kDa are free BODIPY-FL. The tubes and lanes are as follows: (1) Coexpression of BR and Δ49ApoA1 with DMPC (BR-NLP), (2) Cell free expression of Δ49ApoA1 with DMPC (empty NLP), (3) Coexpression of BR and Δ49ApoA1 with DMPC and 0.5% telodendrimer PEG2k-CA4, (4) Coexpression of BR and ApoA1 with DMPC and 0.5% telodendrimer PEG2k-CA8, (5) Coexpression of BR and Δ49ApoA1 with DMPC and 0.5% telodendrimer PEG5k-CF4, (6) Coexpression of BR and Δ49ApoA1 with DMPC and 0.5% telodendrimer PEG5k-CA8.
Our study is limited by the small number of biophysical and biochemical endpoints used to characterize the TD-NLP molecules. We also limited our experiments to cell-free reactions generated from E. coli lysates. There are other amphiphilic particles (so-called amphiopols) that are well known to form soluble particles and can also act as supports for membrane proteins.31 In fact the use of amphiphiles has been found to be an important approach for determining the structure of some membrane proteins.30 We only used one class of TD in this study, which can form TD-lipid particles and closely resemble other amphiopols. When compared to TD NLPs, the TD-lipid particles had distinct size distributions as well as lacked water solubility when associated with membrane proteins. We focused on demonstrating both the production and potential capabilities of TD combined NLP nanoparticles in what is considered a low cost system for lysate-based expression, which is uniquely different from what has been demonstrated previously with amphiopols. It would be of interest to see how other facile molecules interact or modify apolipoproteins to generate novel nano-scaffolds. This data all indicates that we are obtaining a novel form of nanodisc, which is monodispersed based on the interaction with the TD. Our data also did not identify the molecular mechanism of how TD modify the apolipoprotein to lipid interaction. It is well known that varying the protein to lipid ratio can directly affect NLP diameter.7 It is possible that the branching of TDs presents a steric structure, which could limit the apolipoprotein to lipid interaction. However, it is clear that the TDs have a favorable impact on the NLP aggregation levels. Also, the TDs clearly facilitate unique labeling strategies for direct application to the NLP as demonstrated by the fluorescent labeling. Future studies will be required to evaluate the specific molecular mechanisms by which TDs offer size tunability.
In summary, we present a novel nanoparticle using apolipoproteins and lipids combined with TD. The end product is rapidly produced and results in homogeneous NLPs that are monodispersed. In addition, varying TD can be selected to tune disc size between 6 and 30 nm, thus obviating the need for optimizing the lipid to protein ratio and conducting multiple rounds of SEC. Lastly, TD add to the versatility of the NLP as they can serve as chemical handles for direct surface conjugation of multiple biologically active molecules. Future studies will be performed to evaluate the utility of this vehicle for the delivery of imaging, targeting, or therapeutic agents.
Materials and Methods
Plasmids
The truncated form of Apo A1 (Δ1-49) or Δ49ApoA1 was cloned into pIVEX2.4d using NdeI and SmaI restriction sites. This vector contained a His-tag used for nickel affinity purification as previously described.12
Preparation of the telodendrimers
We have previously published on the use of TD for as a nanodelivery tool.27 PEG5k-CA8 and related cholesterol or CA based amphiphilic polymers, were prepared according to the published methods. For the TD-lipid mixtures a total of 25 mg/mL DMPC and each individual polymer were mixed at different molar ratios at an ∼0.5–10%. The mixtures of DMPC/polymer were then sonicated for 15 min, or until optical clarity was achieved. Samples were kept on ice during the entire sonication process. After the sonication, samples were centrifuged at 13,000 RCF for 1–2 min to remove any metal contamination from the probe tip.
DMPC/polymer preparation
Small unilamellar vesicles of DMPC (Avanti Polar Lipids, Alabaster, AL) were prepared by probe sonication of a 25 mg/mL aqueous solution of DMPC until optical clarity was achieved; typically 15 min was sufficient. Samples were kept on ice during the sonication process. After the sonication, the samples were centrifuged at 13,000 RCF for 2 min to remove metal contamination from the probe tip. TDs lipid mixtures (TD-lipid) were created with the above method with a total of 25 mg/mL DMPC and each individual polymer mixed at varying molar ratios between ∼0.5–10%. If a fluorescent label on the NLP complex was desired, a 0.1% Texas Red-POPC (Life Technologies Corporation, Carlsbad, CA) was added to the DMPC solution before sonication. TD-lipid mixtures were purified by spin column size fractionation and SEC fractionation as described below.
Cell-free Reaction
Small-scale reactions (100 μL) or large scale (1 mL) were carried out using Expressway Maxi kit (Life Technologies, Carlsbad, CA). Reaction components (lysate, reaction buffer, T7 polymerase, amino acid mix, and methionine) were combined as specified by the manufacturer. Ten microgram of Δ49ApoA1 DNA was added to each 1 mL reaction. A total of 2 mg DMPC/polymer mixture was then added. The reactions were incubated at 30°C, with shaking at 990 rpm for 2–8 h or over night in a thermomixer.
Affinity purification of NLP complexes
Immobilized metal affinity chromatography was used to isolate the protein of interest (Δ49ApoA1) from the cell-free reaction mixture. Two milliliter of 50% slurry nickel-nitrilotriacetic acid Superflow resin (Qiagen, Hilden, Germany) was equilibrated with PBS (50 mM Na2HPO4, 300 mM NaCl, pH 8.0) under native conditions in a 10 mL capped column. The total cell free reaction (1 mL) was mixed with the equilibrated resin, and was incubated/nutated on ice for 2 h. One milliliter of slurry and 5 mL capped column were used for the purification from small-scale reactions. The column was then washed with increasing concentrations of imidazole (10, 20, and 50 mM) in the mentioned PBS buffer. Two column volumes (CV) of each wash buffer were used for a total of 6 CVs of washing. The His-tagged proteins of interest were eluted in six 1 mL fractions of 400 mM imidazole, PBS buffer. All elution were combined, dialyzed against PBS for 18 h at 4°C with stirring. After that, the combined elution was concentrated using a Vivaspin 6 100K MWCO molecular weight sieve filters (GE Healthcare, Waukesha, WI) to a volume of ∼200 μL.
SDS PAGE
Ten microliter aliquots of the purified NLPs or lipid micelles were mixed with 10 µL 2× LDS sample buffer with reducing agents (Life Technologies, Carlsbad, CA), heat denatured and loaded on to a 4–12% gradient premade bis-Tris gel (Life Technologies, Carlsbad, CA) along with the molecular weight standard NovexSharp (Life Technologies, Carlsbad, CA). The running buffer was 1× MES-SDS (Life Technologies, Carlsbad, CA). Samples were electrophoresed for 38 min at 200 V. Gels were stained with Coomassie Brilliant Blue.
Native PAGE
Ten microliter aliquots of the purified NLPs or lipid micelles were mixed with 2× native gel sample buffer (Life Technologies, Carlsbad, CA) and loaded onto 4–12% gradient premade Tris-glycine gels (Life Technologies, Carlsbad, CA). Samples were electrophoresed for 2 h at 125 V. After electrophoresis, gels were imaged using the laser (488 nm) of a Typhoon 9410 (GE Healthcare, Waukesha, WI) with a 520 nm bandpass 30 filter for the detection of the produced NLPs with incorporated FITC labeled polymer. For detection of the produced NLPs with incorporated TexasRed-POPC, the laser (532 nm) with a 610 nm bandpass 30 filter is used. Molecular weights were determined by comparing migration versus log molecular weight of standard proteins found in the NativeMark standard (Life Technologies, Carlsbad, CA).
Dynamic light scattering (DLS)
The measurements were performed on a Nanotrac Particle Size Analyzer (Microtrac, Montgomeryville, PA) following the manufacturer's instruction menu.
Cryo transmission electron microscopy (Cryo-EM)
All the samples are preserved as frozen hydrated specimen in the presence of saturated ammonium molybdata and examined with a JEOL JEM-2100F transmission electron microscope (JOEL USA, Peabody, MA) at magnification of 80,000× under liquid nitrogen temperature. Clusters of NLPs were found with plain NLP sample, while only a few clusters of NLPs were found with TD-NLPs. The majority of the NLPs are shown inside views. A few of them appeared in their top view. Tobacco mosaic virus (TMV) was added as reference to indicate the quality of Cryo-EM preparation, as well as the internal calibration of microscope magnification.
Solution phase characterization using fluorescent correlation spectroscopy (FCS)
The methods in this study have been described in detail previously.14 Characterization of nanoparticles and their dynamic shape and association in solution remains a challenge, which we have addressed using FCS performed on a MicroTime 200 single molecule fluorescence lifetime measurement system (PicoQuant GmbH, Berlin, Germany). FCS is capable of measuring molecular diffusion statistics in solution with sensitivity for single molecule fluorescence. This allows us to rapidly and accurately determine the hydrodynamic radii of the newly formed nanocomplexes in an aqueous environment. Complimentary techniques were performed, such as SEC using a Superdex 200 HR 10/300 column (GE Healthcare, Waukesha, WI) that was expected to efficiently separate different sized NLP constructs.
Acknowledgments
The authors thank Dr. Tiffany Scharadin for critical reading of the manuscript.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Jonas A. Reconstitution of high-density lipoproteins. Methods Enzymol. 1986;128:553–582. doi: 10.1016/0076-6879(86)28092-1. [DOI] [PubMed] [Google Scholar]
- 2.Bayburt TH, Grinkova YV, Sligar SG. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 3.Chromy BA, Arroyo E, Blanchette CD, Bench G, Benner H, Cappuccio JA, Coleman MA, Henderson PT, Hinz AK, Kuhn EA, Pesavento JB, Segelke BW, Sulchek TA, Tarasow T, Walsworth VL, Hoeprich PD. Different apolipoproteins impact nanolipoprotein particle formation. J Am Chem Soc. 2007;129:14348–14354. doi: 10.1021/ja074753y. [DOI] [PubMed] [Google Scholar]
- 4.Sunahara H, Urano Y, Kojima H, Nagano T. Design and synthesis of a library of BODIPY-based environmental polarity sensors utilizing photoinduced electron-transfer-controlled fluorescence ON/OFF switching. J Am Chem Soc. 2007;129:5597–5604. doi: 10.1021/ja068551y. [DOI] [PubMed] [Google Scholar]
- 5.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 6.Blanchette CD, Law RW, Benner H, Pesavento B, Cappuccio JA, Walsworth V, Kuhn EA, Corzett M, Chromy BA, Segelke BW, Coleman MA, Bench G, Hoeprich PD, Sulchek TA. Quantifying size distributions of nanolipoprotein particles with single-particle analysis and molecular dynamic simulations. J Lipid Res. 2009;49:1420. doi: 10.1194/jlr.M700586-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Blanchette CD, Fischer NO, Corzett M, Bench G, Hoeprich PD. Kinetic analysis of his-tagged protein binding to nickel-chelating nanolipoprotein particles. Bioconjug Chem. 2010;21:1321–1330. doi: 10.1021/bc100129s. [DOI] [PubMed] [Google Scholar]
- 8.Ryan RO. Nanodisks: hydrophobic drug delivery vehicles. Expert Opin Drug Deliv. 2008;5:343–351. doi: 10.1517/17425247.5.3.343. [DOI] [PubMed] [Google Scholar]
- 9.Ryan RO. Nanobiotechnology applications of reconstituted high density lipoprotein. J Nanobiotech. 2010:8–28. doi: 10.1186/1477-3155-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Whorton MR, Bokoch MP, Rasmussen SGF, Huang B, Zare RN, Kobilka B, Sunahara RK. Monomeric G protein-coupled receptor isolated in a high-density lipo-protein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cappuccio JA, Blanchette CD, Sulchek TA, Arroyo ES, Kralj JM, Hinz AK, Kuhn EA, Chromy BA, Segelke BW, Rothschild KJ, Fletcher JE, Katzen F, Peterson TC, Kudlicki WA, Bench G, Hoeprich PD, Coleman MA. Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Mol Cell Proteomics. 2008;7:2246–2253. doi: 10.1074/mcp.M800191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker SE, Hopkins RC, Blanchette CD, Walsworth VL, Sumbad R, Fischer NO, Kuhn EA, Coleman M, Chromy BA, Létant SE, Hoeprich PD, Adams MW, Henderson PT. Hydrogen production by a hyperthermophilic membrane-bound hydrogenase in water-soluble nanolipoprotein particles. J Am Chem Soc. 2009;131:7508–7509. doi: 10.1021/ja809251f. [DOI] [PubMed] [Google Scholar]
- 14.Gao T, Blanchette CD, He W, Bourguet F, Ly S, Katzen F, Kudlicki WA, Henderson PT, Laurence TA, Huser T, Coleman MA. Characterizing diffusion dynamics of a membrane protein associated with nanolipoproteins using fluorescence correlation spectroscopy. Prot Sci. 2011;20:437–447. doi: 10.1002/pro.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katzen F, Fletcher JE, Yang J, Kang D, Peterson TC, Cappuccio JA, Blanchette CD, Sulchek T, Chromy BA, Hoeprich PD, Coleman MA, Kudlicki W. Insertion of membrane proteins into discoidal membranes using a cell-free protein expression approach. J Proteome Res. 2008;7:3535–3542. doi: 10.1021/pr800265f. [DOI] [PubMed] [Google Scholar]
- 16.Rensen PC, Schiffelers RM, Versluis AJ, Bijsterbosch MK, Van Kuijk-Meuwissen ME, Van Berkel TJ. Human recombinant apolipoprotein E-enriched liposomes can mimic low-density lipoproteins as carriers for the site-specific delivery of antitumor agents. Mol Pharmacol. 1997;52:445–455. doi: 10.1124/mol.52.3.445. [DOI] [PubMed] [Google Scholar]
- 17.Coleman MA, Lao VH, Segelke BW, Beernink PT. High-throughput, fluorescence-based screening for soluble protein expression. J Proteome Res. 2004;3:1024–1032. doi: 10.1021/pr049912g. [DOI] [PubMed] [Google Scholar]
- 18.Kigawa T, Yabuki T, Yoshida Y, Tsutsui M, Ito Y, Shibata T, Yokoyama S. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 1999;442:15–19. doi: 10.1016/s0014-5793(98)01620-2. [DOI] [PubMed] [Google Scholar]
- 19.Doi N, Takashima H, Kinjo M, Sakata K, Kawahashi Y, Oishi Y, Oyama R, Miyamoto-Sato E, Sawasaki T, Endo Y, Yanagawa H. Novel fluorescence labeling and high-throughput assay technologies for in vitro analysis of protein interactions. Genome Res. 2002;12:487–492. doi: 10.1101/gr.218802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang JP, Cirico T, Katzen F, Peterson TC, Kudlicki W. Cell-free synthesis of a functional G protein-coupled receptor complexed with nanometer scale bilayer discs. BMC Biotechnol. 2011;11:57. doi: 10.1186/1472-6750-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keppetipola S, Kudlicki W, Nguyen BD, Meng X, Donovan KJ, Shaka AJ. From gene to HSQC in under five hours: high-throughput NMR proteomics. J Am Chem Soc. 2006;128:4508–4509. doi: 10.1021/ja0580791. [DOI] [PubMed] [Google Scholar]
- 22.Frydman J, Hartl FU. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science. 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- 23.Klammt C, Schwarz D, Fendler K, Haase W, Dötsch V, Bernhard F. Evaluation of detergents for the soluble expression of alpha-helical and beta-barrel-type integral membrane proteins by a preparative scale individual cell-free expression system. FEBS J. 2005;272:6024–6038. doi: 10.1111/j.1742-4658.2005.05002.x. [DOI] [PubMed] [Google Scholar]
- 24.Jewett MC, Swartz JR. Rapid expression and purification of 100 nmol quantities of active protein using cell-free protein synthesis. Biotechnol Prog. 2004;20:102–109. doi: 10.1021/bp0341693. [DOI] [PubMed] [Google Scholar]
- 25.Mori M, Miura S, Tatibana M, Cohen PP. Cell-free synthesis and processing of a putative precursor for mitochondrial carbamyl phosphate synthetase I of rat liver. Proc Natl Acad Sci U S A. 1979;76:5071–5075. doi: 10.1073/pnas.76.10.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 27.Luo J, Xiao K, Li Y, Lee JS, Shi L, Tan YH, Xing L, Holland Cheng R, Liu GY, Lam KS. Well-defined, size-tunable, multifunctional micelles for efficient paclitaxel delivery for cancer treatment. Bioconjug Chem. 2010;21:1216–1212. doi: 10.1021/bc1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao K, Luo J, Fowler WL, Li Y, Lee JS, Xing L, Cheng RH, Wang L, Lam KS. A self-assembling nanoparticle for paclitaxel delivery in ovarian cancer. Biomaterials. 2009;30:6006–6016. doi: 10.1016/j.biomaterials.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenow MA, Magee CL, Williams JC, Allen JP. The influence of detergents on the solubility of membrane proteins. Acta Cryst. 2002;D58:2076–2081. doi: 10.1107/s0907444902016736. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Ma X, Ward A, Hong WX, Jaakola VP, Stevens RC, Finn MG, Chang G. Designing facial amphiphiles for the stabilization of integral membrane proteins. Angew Chem Int Ed Engl. 2007;46:7023–7025. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- 31.Charvolin D, Perez JB, Rouvière F, Giusti F, Bazzacco P, Abdine A, Rappaport F, Martinez KL, Popot JL. The use of amphipols as universal molecular adapters to immobilize membrane proteins onto solid supports. Proc Natl Acad Sci. 2009;106:405–410. doi: 10.1073/pnas.0807132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


