Abstract
Enterocolitis caused by Campylobacter jejuni represents an important socioeconomic burden worldwide. The host-specific intestinal microbiota is essential for maintaining colonization resistance (CR) against C. jejuni in conventional mice. Notably, CR is abrogated by shifts of the intestinal microbiota towards overgrowth with commensal E. coli during acute ileitis. Thus, we investigated whether oral transplantation (TX) of ileal microbiota derived from C. jejuni susceptible mice with acute ileitis overcomes CR of healthy conventional animals. Four days following ileitis microbiota TX or ileitis induction and right before C. jejuni infection, mice displayed comparable loads of main intestinal bacterial groups as shown by culture. Eight days following ileitis induction, but not ileal microbiota TX, however, C. jejuni could readily colonize the gastrointestinal tract of conventional mice and also translocate to extra-intestinal tissue sites such as mesenteric lymph nodes, spleen, liver, and blood within 4 days following oral infection. Of note, C. jejuni did not further deteriorate histopathology following ileitis induction. Lack of C. jejuni colonization in TX mice was accompanied by a decrease of commensal E. coli loads in the feces 4 days following C. jejuni infection. In summary, oral ileal microbiota TX from susceptible donors is not sufficient to abrogate murine CR against C. jejuni.
Keywords: Campylobacter jejuni, colonization resistance, fecal transplantation, microbiota, intestinal inflammation, acute ileitis, Toxoplasma gondii, bacterial translocation, susceptibility to infection, E. coli, pathogen–commensal interaction, pathogen–host interaction
Introduction
Campylobacter (C.) jejuni is a very common bacterial pathogen causing gastroenteritis worldwide [1, 2]. The zoonotic pathogen is transmitted from animals to humans via the foodchain by contaminated meat, milk products, or surface water [2, 3]. C. jejuni colonizes the distal parts of the gastrointestinal (GI) tract and subsequently induces an influx of inflammatory cells leading to ulcerations and crypt abscesses [3–6]. Following an incubation period of approximately 2 to 5 days, infected patients suffering from campylobacteriosis present with fever, abdominal cramps, and watery or even bloody diarrhea [5, 6]. Whereas in most cases the disease is self-limited, rarely chronic post-infectious sequelae of campylobacteriosis, such as Guillan–Barré syndrome or reactive polyarthropathy might arise [2, 7].
In vivo studies of molecular mechanisms underlying C. jejuni-induced immunopathology are hampered by the lack of suitable vetebrate models [8]. Conventional laboratory mice, for instance, can, if at all, only sporadically be colonized by the pathogen [1, 2]. Thus, significant immunopathology mimicking human campylobacteriosis cannot be investigated. This is due to the colonization resistance (CR) against C. jejuni exerted by adult mice, a phenomenon caused by the age-specific composition of the conventional intestinal microbiota harbored by mice older than 6 weeks of age, which are commonly used in experimental settings [2]. We have recently shown that this host-specific CR can be overcome by modifying the intestinal microbiota [9]. Conventionally colonized control animals had expelled the pathogen within 48 h following even high dose C. jejuni infection on three consecutive days, whereas the GI tract of gnotobiotic mice lacking any bacteria following antibiotic treatment could be stably colonized by C. jejuni. The pathogen subsequently induced cellular immune and inflammatory responses in the distal intestine [9]. Strikingly, gnotobiotic mice perorally transplanted with complex human, but not conventional murine microbiota could be infected by C. jejuni and displayed immunohistopathological features mimicking human campylobacteriosis [9]. These findings highlight the host specific microbiota composition as pivotal prerequisite in susceptibility to enteropathogens. Furthermore, we clearly showed that murine CR against C. jejuni was abrogated under conditions of acute intestinal inflammation. For instance, mice suffering from acute ileitis or colitis (induced by Toxoplasma (T.) gondii or IL-10 gene deficiency, respectively) could be stably infected with C. jejuni [10–12]. Given that a common feature of acute intestinal inflammation is a tremendous intestinal overgrowth with commensal E. coli [13–16], we artificially elevated the intestinal E. coli load of conventionally colonized mice (lacking intestinal inflammation) by adding live E. coli into the drinking water, and rendered mice susceptible to C. jejuni infection as long as the commensal E. coli burden was kept to supra-physiological levels [10].
In the present study, we investigated whether the CR against C. jejuni displayed by conventionally colonized mice without intestinal inflammation can be overcome by a single peroral fecal transplantation (TX) of the complex microbiota derived from mice suffering from acute ileitis. We further studied the quantitative microbiota and potential histopathological changes in the intestines of TX mice following C. jejuni infection.
Materials and Methods
Ethics statement
All animal experiments were conducted according to the European Guidelines for animal welfare (2010/63/EU) with approval of the commission for animal experiments headed by the “Landesamt für Gesundheit und Soziales” (LaGeSo, Berlin, Germany). Animal welfare was monitored twice daily by assessment of clinical conditions.
Mice
All animals were bred and maintained in the facilities of the “Forschungsinstitut für Experimentelle Medizin” (FEM, Charité-Universitätsmedizin Berlin, Berlin, Germany) under specific pathogen-free (SPF) conditions. Female age matched C57BL/6j wildtype mice between 10 and 12 weeks of age were used.
Acute ileitis induction
For induction of acute ileitis, mice were infected perorally with 100 T. gondii cysts (ME49 strain) from homogenized brains of intraperitoneally infected NMRI mice in a volume of 0.3 mL phosphate-buffered saline (PBS, pH 7.4) by gavage, as described previously [10, 13, 17].
Ileitis microbiota transplantation
Fresh samples (free of enteropathogenic bacteria, parasites, and viruses) were collected from the ileal lumen of ten mice suffering from severe T. gondii induced ileitis at day 8 p.i., pooled and dissolved in an equal volume of sterile PBS, and then immediately used for oral re-colonization (transplantation; TX) in a final volume of 0.3 mL by gavage. Right before TX the total as well as the bacterial loads of distinct communities (E. coli, Enterococcus spp., Lactobacillus spp., Bacteroides/Prevotella spp., Clostridium/Eubacterium spp.) were quantified in serious dilutions by cultivation on appropriate solid media as described previously. In addition, colonization efficiency was investigated by cultural analyses of fecal samples at day 4 and day 8 following TX. Between independent experiments, counts of bacterial groups in suspensions used for fecal TX varied less than 0.5 log orders of magnitude.
Campylobacter jejuni infection
Mice were infected with 109 viable colony forming units (CFU) of C. jejuni 43431 ATCC strain by gavage in a total volume of 0.3 mL PBS on three consecutive days starting 4 days following fecal TX or induction of acute ileitis as described earlier in detail [9].
Sampling procedures
Mice were sacrificed by isofluran treatment (Abbott, Germany). Cardiac blood and tissue samples from spleen, liver, mesenteric lymph nodes (MLNs), and luminal contents from the gastrointestinal tract (stomach, duodenum, terminal ileum and colon) were removed under sterile conditions. Ileal samples from each mouse were collected in parallel for microbiological and histopathological analyses. For the latter, ileal samples were immediately fixed in 5% formalin and embedded in paraffin, sections (5 µm) were stained with HE and subjected to a standardized scoring system by two blinded independent investigators (R.P. and M.M.H.) [9, 13, 18].
Quantitative analysis of C. jejuni translocation
Live C. jejuni were detected in MLNs, spleen, liver, and cardiac blood by culture as described earlier [9]. In brief, tissues were homogenized in sterile PBS and analyzed in dilution series on karmali agar (Oxoid, Wesel, Germany) in a microaerophilic atmosphere at 37 °C for at least 48 h. Cardiac blood (0.2 mL) was immediately streaked out on karmali agar plates.
Analysis of the intestinal microbiota
Cultural analyses and biochemical identification of luminal bacterial communities from stomach, duodenum, ileum, and colon were performed as previously described [13–15].
Statistical analysis
Mean values, medians, standard deviations, and levels of significance were determined using appropriate tests as indicated (two-tailed Student’s t-test, Mann–Whitney U test). Two-sided probability (p) values ≤ 0.05 were considered significant. All experiments were repeated at least twice.
Results
Oral transplantation of luminal ileal microbiota derived from mice suffering from acute ileitis into conventional mice
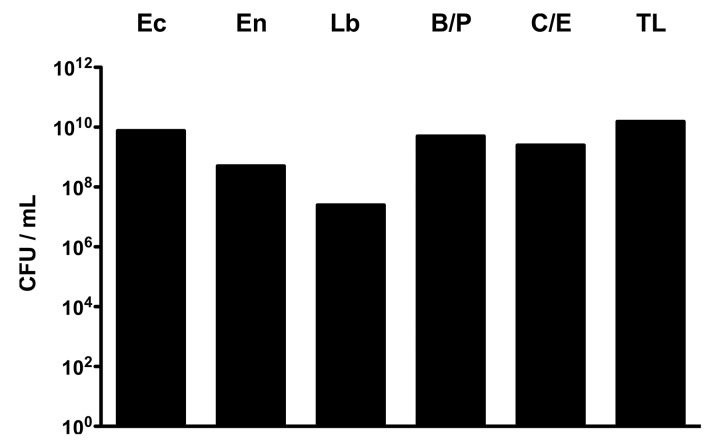
We have recently shown that physiological CR against C. jejuni infection displayed by adult mice harboring a conventional intestinal microbiota (without intestinal inflammation) is abrogated under inflammatory conditions of the intestinal tract such as acute ileitis [10] accompanied by a tremendous overgrowth of the intestinal microbiota with commensal E. coli [13, 14, 18]. We were therefore hypothesizing that oral transplantation of the complex ileal microbiota derived from mice suffering from acute ileitis (i.e. susceptible donor) into conventionally colonized mice (and thus resistant acceptor) might render them susceptible to C. jejuni infection. To address this question, C57BL/6 wildtype mice were perorally infected with 100 cysts of T. gondii ME49 strain in order to induce acute ileitis, sacrificed at day 8 post-infection (p.i.) and fresh luminal ileal content prepared for subsequent oral TX. The quantitative and qualitative bacterial yield of the suspension with “ileitis microbiota” is shown in Fig. 1: Conventional C57BL/6 wildtype mice were challenged perorally with a total load of more than 1010 CFU live bacteria per mL suspension, predominantly consisting of Gram-negative species such as E. coli and obligate anaerobic Bacteroides/Prevotella spp. Gram-positive bacterial groups, such as enterococci, lactobacilli, and anaerobic Clostridium/Eubacterium spp., however, were up to 100 times less abundant.
Fig. 1.
Bacterial groups within suspension for oral ileitis microbiota transplantation. Luminal ileal contents derived from ten mice with acute T. gondii-induced ileitis were pooled for oral ileal microbiota transplantation as described in Materials and methods. Prior gavage main bacterial groups within the suspension such as E. coli (Ec), Enterococcus spp. (En), Lactobacillus spp. (Lb), Bacteroides/Prevotella spp. (B/P), Clostridium/Eubacterium spp. (C/E), and total bacterial loads (TL) were determined by detection of colony forming units (CFU) per mL on appropriate culture media (see Materials and methods)
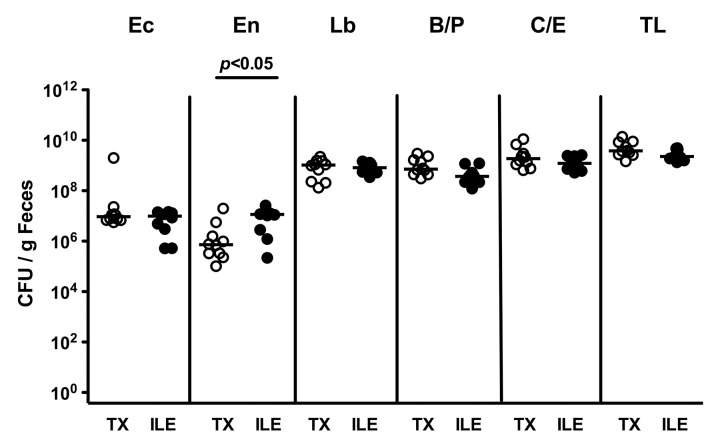
Next, we quantitatively assessed the intestinal colonization efficiency of bacterial species following peroral ileal microbiota TX by culture. Four days following peroral challenge, TX mice harbored approximately 107 CFU E. coli, 106 CFU Enterococcus spp., 109 CFU Lactobacillus spp., Bacteroides/Prevotella spp. and Clostridium/Eubacterium spp. per g feces (Fig. 2; TX: open circles; n = 10). Notably, right before oral C. jejuni infection, the bacterial loads of the respective species in TX mice and in animals at day 4 following acute ileitis induction by T. gondii displaying beginning histopathological changes in their ileal mucosa (Fig. 2; ILE: closed circles; n = 10) were comparable, except for 1.5 orders of magnitude higher intestinal Enterococcus spp. counts in the latter group (p < 0.05). Thus, conventional mice with transplanted ileitis microbiota as well as mice with subtle ileal mucosal histopathology following ileitis induction displayed relatively comparable commensal bacterial species loads in their feces right before C. jejuni infection.
Fig. 2.
Fecal microbiota composition in conventionally colonized mice following oral ileitis microbiota transplantation right before C. jejuni infection. Four days following oral transplantation of ileitis microbiota (TX, open circles; n = 10 mice) into conventional mice and right before oral C. jejuni ATCC 43431 infection, fecal samples were obtained to quantitatively assess E. coli (Ec), Enterococcus spp. (En), Lactobacillus spp. (Lb), Bacteroides/Prevotella spp. (B/P), Clostridium/Eubacterium spp. (C/E), as well as total bacterial loads (TL) by detection of colony forming units (CFU) per mL on appropriate culture media (see Materials and methods) and compared to conventional mice with beginning acute ileitis 4 days following T. gondii infection (ILE, solid circles; n = 10). Medians (black bars) and levels of significance (p-values) as determined by the Mann–Whitney U test are indicated. Data shown are representative for three independent experiments
Transplantation of ileal microbiota derived from mice with acute ileitis does not abrogate CR against C. jejuni
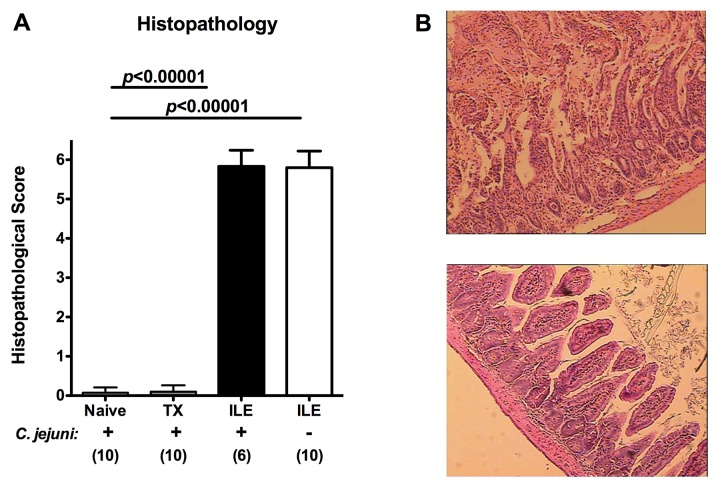
We next infected with ileitis microbiota transplanted mice as well as animals 4 days following ileitis induction perorally with 109 CFU C. jejuni ATCC 43431 strain for three consecutive days, respectively. Whereas 2 days following C. jejuni infection neither TX mice nor naive conventionally colonized control animals displayed histopathological changes within their ileal mucosa (Fig. 3A, B bottom), mice with preceeding ileitis induction 4 days prior C. jejuni infection were severely compromised due to acute transmural small intestinal inflammation as indicated by histopathological scores of 5.7 ± 0.5 (Fig. 3A) and severe ileal necroses (Fig. 3B, top). Of note, C. jejuni infection did not further deteriorate T. gondii-induced acute ileitis as indicated by comparable histopathological scores versus uninfected mice with acute ileitis (Fig. 3A).
Fig. 3.
Ileal histopathology in C. jejuni infected mice following oral ileitis microbiota transplantation or induction of acute ileitis. (A) Luminal ileal contents derived from ten mice with acute T. gondii-induced ileitis were pooled for oral ileitis microbiota transplantation into mice harbouring a conventional microbiota by gavage as described in Materials and methods (TX). Four days following transplantation, TX mice were perorally infected with C. jejuni strain ATCC 43431 (as indicated on x-axis: + C. jejuni) and compared to conventional mice without TX (Naive). In addition, conventionally colonized animals were orally infected with T. gondii in order to induce acute ileitis. At day 4 following ileitis induction (the time point when beginning histopathological changes of the ileal mucosa can be detected; ILE) mice were orally infected with C. jejuni strain ATCC 43431 (as indicated on x-axis: + C. jejuni; black bar) and compared to mice without C. jejuni infection (as indicated on x-axis: – C. jejuni; white bar). Histopathological changes of the ileal mucosa were assessed in ileum paraffin sections at day 8 following ileitis induction applying a standardized histopathological score. Numbers of analyzed animals are given in parentheses. Means, standard deviations and levels of significance (p-values) determined by Student’s t-test are indicated. Data shown are representative for three independent experiments. (B) Representative photomicrographs of HE-stained ileal paraffin sections (×200 magnification) of mice following acute T. gondii-induced ileitis irrespective of C. jejuni infection (top). Micrograph shows severe necrosis affecting all layers and entirely disturbed villus structures. Conventional mice with transplanted ileitis microbiota (TX) displayed an intact ileal mucosa 4 days following C. jejuni comparable to non-transplanted conventional (Naive) mice (bottom)
Lack of C. jejuni colonization and translocation in conventionally colonized mice following ileitis microbiota transplantation
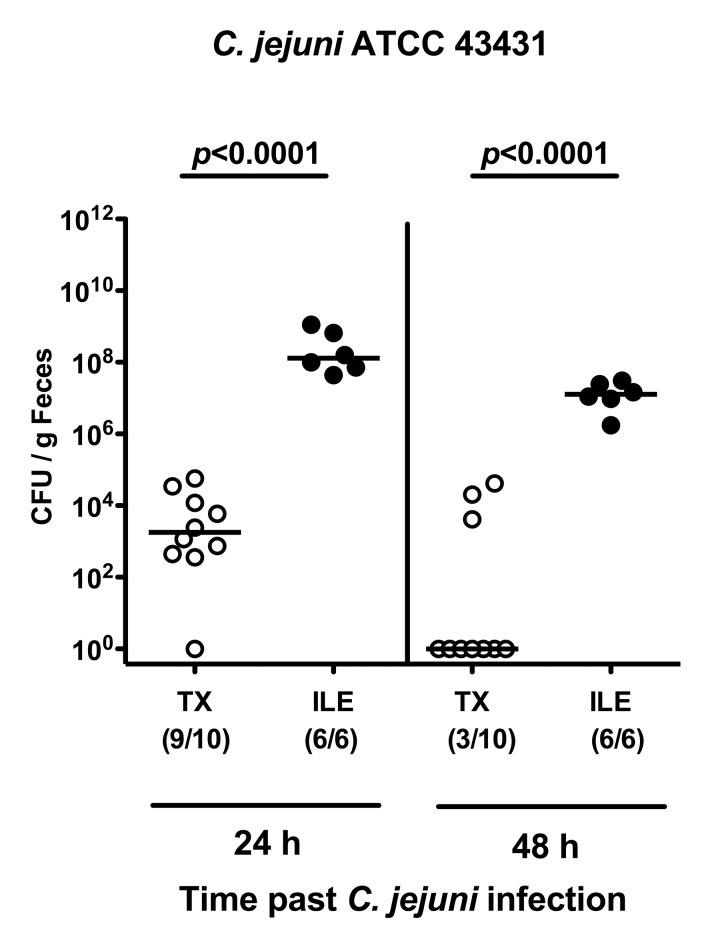
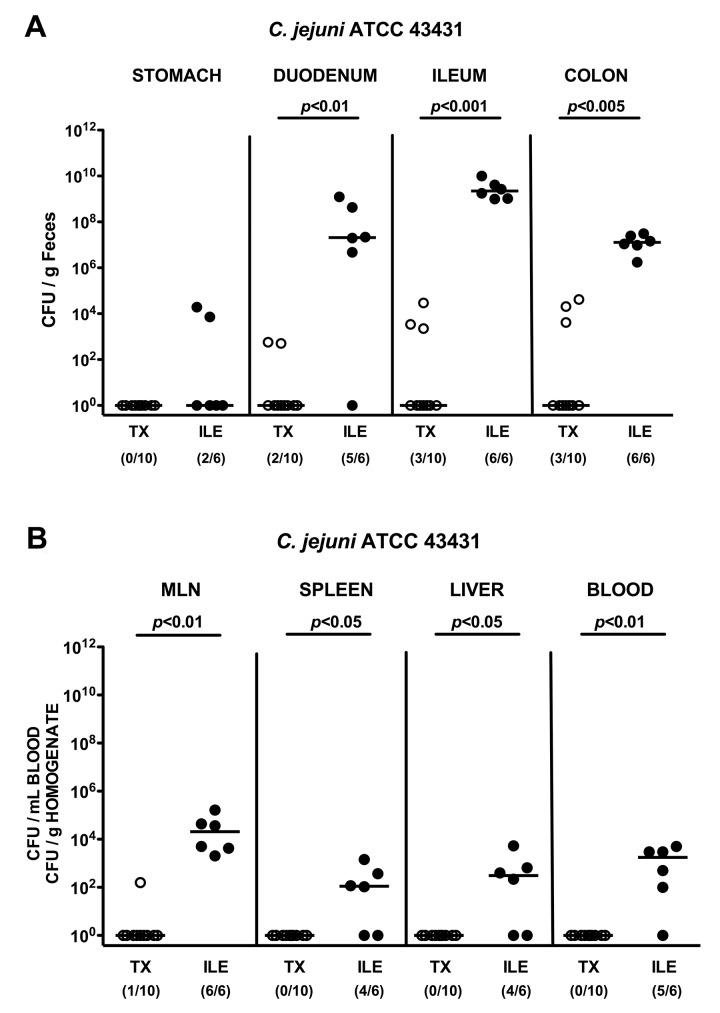
We next assessed the pathogen loads in the GI tract following C. jejuni infection. Whereas TX mice had expelled the pathogen within the first 2 days following the latest infection, animals with T. gondii-induced ileitis were stably infected by C. jejuni (Fig. 4; p < 0.0001). Furthermore, 2 days following infection, live C. jejuni could be cultured at high levels from proximal and distal parts of the intestinal tract in virtually all mice suffering from acute ileitis with maximum loads in the inflamed ileum of up to 1010 CFU/g luminal content (Fig. 5A), whereas in only 20–30% of ileal microbiota transplanted animals C. jejuni could be detected at low numbers in the duodenal, ileal, and colonic lumen (Fig. 5A). Moreover, C. jejuni did not translocate to extra-intestinal tissue sites in transplanted animals, but could be cultured from mesenteric lymph nodes (MLNs; 100%), spleens (66.7%), livers (66.7%), and even cardiac blood (83.3%) of mice with acute ileitis (Fig. 5B).
Fig. 4.
Kinetic analysis of C. jejuni colonization in conventional mice following oral ileitis microbiota transplantation. Mice harboring a conventional intestinal microbiota were orally infected with T. gondii in order to induce ileitis (ILE; filled circles). At day 4 following ileitis induction (the time point when beginning histopathological changes of the ileal mucosa can be detected) mice were orally infected with C. jejuni strain ATCC 43431 as described in Materials and methods. In addition, conventionally colonized mice were subjected to oral transplantation of luminal ileum flora derived from mice with acute ileitis (TX; open symbols) and after 4 days orally infected with C. jejuni strain ATCC 43431. The pathogen densities in the feces were quantified 24 and 48 h following infection by cultural analysis of live C. jejuni (CFU, colony forming units) per g feces. Numbers of animals harboring C. jejuni out of the total number of analyzed animals are given in parentheses. Medians (black bars) and levels of significance (p-values) determined by Mann–Whitney U test are indicated. Data shown are representative for three independent experiments
Fig. 5.
C. jejuni colonization along the gastrointestinal tract and translocation in mice following oral ileitis microbiota transplantation. Conventional mice were orally infected with T. gondii in order to induce ileitis (ILE; filled circles). At day 4 following ileitis induction (the time point when beginning histopathological changes of the ileal mucosa become overt) mice were orally infected with C. jejuni strain ATCC 43431 as described in Materials and methods. In addition, conventional mice were subjected to oral transplantation of luminal ileum flora derived from mice with acute ileitis (TX; open symbols) and also orally infected with C. jejuni strain ATCC 43431 after 4 days of TX. The pathogen densities along the gastrointestinal tract (stomach, duodenum, ileum, and colon; A) as well as in homogenates of mesenteric lymph nodes (MLN), spleen, liver and cardiac blood (B) were determined 48 h following infection by cultural analysis of live C. jejuni (CFU, colony forming units). Numbers of animals harboring C. jejuni out of the total number of analyzed animals are given in parentheses. Medians (black bars) and levels of significance (p-values) as determined by Mann–Whitney U test are indicated. Data shown are representative for three independent experiments
Microbiota changes following ileal microbiota transplantation, induction of acute ileitis and C. jejuni infection
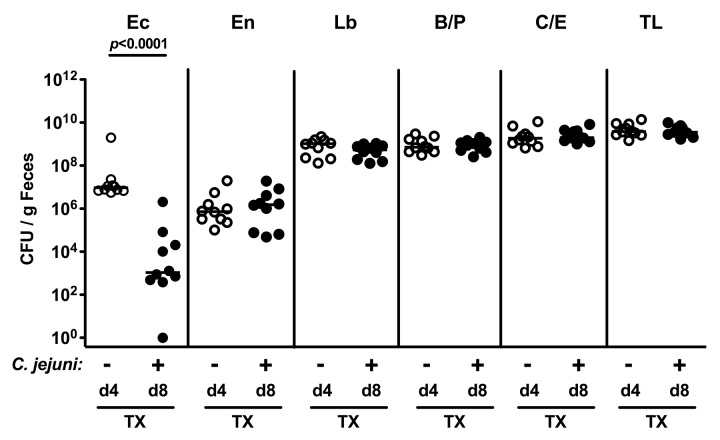
Given that following C. jejuni infection, the pathogen was expelled in with ileitis microbiota-transplanted animals, but not in conventional mice with induced acute ileitis, we were wondering whether this might be due to changes in the fecal microbiota composition in the course of the C. jejuni and/or T. gondii infection. To address this question, we performed quantitative cultural analyses of fecal samples derived from mice 4 days after TX and another 4 days following C. jejuni infection (Fig. 6). Whereas total bacterial loads as well as counts of Enterococcus spp., Lactobacillus spp., Bacteroides/Prevotella spp., and Clostridium/Eubacterium spp. remained virtually unchanged, intestinal E. coli loads dropped by four orders of magnitude within 4 days upon C. jejuni infection (Fig. 6).
Fig. 6.
Fecal microbiota composition of conventional mice following oral ileitis microbiota transplantation and C. jejuni infection. Four days (d4, open circles; n = 10 mice) following oral ileitis microbiota transplantation of conventional mice (TX) and right before oral C. jejuni ATCC 43431 infection (– C. jejuni) as well as 4 days following C. jejuni ATCC 43431 infection and thus 8 days after TX (+ C. jejuni, d8, closed circles; n = 10 mice), fecal samples were obtained to quantitatively assess E. coli (Ec), Enterococcus spp. (En), Lactobacillus spp. (Lb), Bacteroides/Prevotella spp. (B/P), Clostridium/Eubacterium spp. (C/E), as well as total bacterial loads (TL) by detection of colony forming units (CFU) per mL on appropriate culture media (see Materials and methods). Medians (black bars) and levels of significance (p-values) as determined by the Mann–Whitney U test are indicated. Data shown are representative for three independent experiments
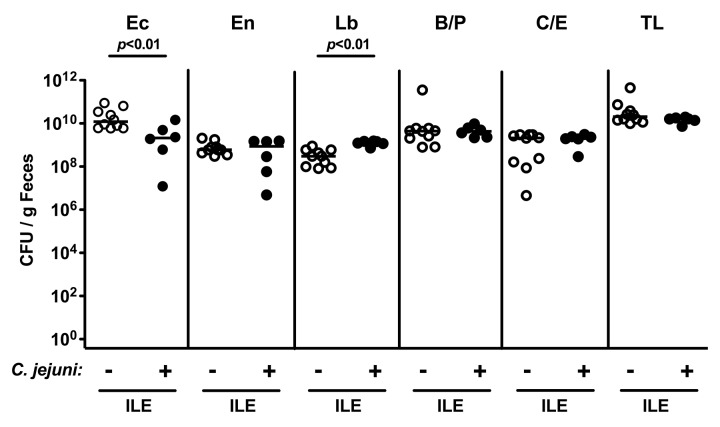
Interestingly, C. jejuni infected mice with acute ileitis displayed slightly lower fecal E. coli and subtle higher Lactobacillus spp. loads of less than one order of magnitude as compared to uninfected control mice 8 days following acute ileitis induction (Fig. 7; p < 0.01). Numbers of the remaining bacterial groups as well as the total bacterial load were comparable (Fig. 7).
Fig. 7.
Fecal microbiota composition of conventional mice following acute ileitis induction and C. jejuni infection. Four days following acute ileitis induction by T. gondii infection (ILE), conventional mice were orally infected with C. jejuni ATCC 43431 (+ C. jejuni; closed circles, n = 6) and fecal samples obtained 4 days thereafter (8 days following ileitis induction) to quantitatively assess E. coli (Ec), Enterococcus spp. (En), Lactobacillus spp. (Lb), Bacteroides/Prevotella spp. (B/P), Clostridium/Eubacterium spp. (C/E), as well as total bacterial loads (TL) by detection of colony forming units (CFU) per mL on appropriate culture media (see Materials and methods) and compared to mice at day 8 following ileitis induction without C. jejuni infection (– C. jejuni; open circles, n = 10). Medians (black bars) and levels of significance (p-values) as determined by the Mann–Whitney U test are indicated. Data shown are representative for three independent experiments
Taken together, murine CR against C. jejuni is abrogated by ileitis induction, but not by ileal microbiota transplantation. In mice suffering from acute ileitis, C. jejuni readily colonized the GI tract and translocated to extra-intestinal tissue sites due to barrier defects set by the preceeding T. gondii infection. The lack of C. jejuni colonization in TX mice was accompanied by a marked decrease of commensal E. coli loads in the feces 4 days following C. jejuni infection.
Discussion
Reliable experimental data regarding intestinal colonization capacities of C. jejuni, mechanisms of pathogen–host interactions, and immunopathological responses in the course of C. jejuni infections in vertebrates are scarce. Adult mice without intestinal inflammation harboring a conventional intestinal microbiota display CR against C. jejuni infection and are therefore not suitable as valid vertebrate model for the study of C. jejuni infection in vivo [9]. In the past, we have successfully overcome these limitations by modifying the commensal intestinal microbiota [9, 10, 19]. Given that mice suffering from acute ileitis following oral T. gondii infection display a tremendous overgrowth of their intestinal microbiota with commensal E. coli [13, 14, 18], we were wondering whether oral transplantation of the complex ileal microbiota derived from mice with acute ileitis (i.e. susceptible donor) into conventionally colonized animals (i.e. resistant acceptor) might result in abrogation of CR against C. jejuni. Interestingly, TX mice expelled the pathogen from their GI tract within 2 days upon infection as seen in conventionally colonized adult mice from previous studies [9, 10, 12, 20], whereas conventional mice suffering from acute ileitis harbored C. jejuni in their small and large intestines at high levels. Of note, live translocated C. jejuni could be cultured from extra-intestinal locations such as MLNs, spleen, liver, and even blood pointing toward bacteremia and sepsis. Interestingly, C. jejuni infection did not further deteriorate ileal histopathology following acute ileitis induction. Furthermore, C. jejuni-infected mice with acute ileitis harbored slightly lower E. coli and higher Lactobacillus spp. loads (one order of magnitude difference; p < 0.05). Whether this might be attributed to direct pathogen–commensal interaction or considered an epiphenomenon without biological impact needs to be further unraveled.
When quantitatively comparing main intestinal bacterial goups harbored by mice right before C. jejuni infection, TX mice displayed slightly lower Enterococcus spp. numbers (approximately 1.5 log) as compared to animals with induced ileitis following T. gondii infection, whereas E. coli numbers were comparable at approximately 107 CFU per g feces. The total bacterial loads as well as the burden of Lactobacillus spp. and obligate anaerobic species did not differ between ileitis microbiota transplanted and ileitis-induced mice upon C. jejuni infection. From previous experiments, where we artificially raised the intestinal E. coli load by adding the commensal into the drinking water, we know, however, that stable C. jejuni colonization in the GI tract of conventional mice is facilitated when the intestinal commensal E. coli loads are kept above a certain threshold of at least 108 CFU per g feces [10]. As soon as the E. coli load goes below this threshold, however, e.g. after withdrawal of the E. coli suspension, then C. jejuni becomes expelled from the GI tract within a few days [10]. This might have been the case in the present study given that at the time point of C. jejuni infection, the intestinal E. coli burden was slightly (one order of magnitude) below this threshold. In addition, 4 days following C. jejuni infection, E. coli loads in TX mice had further dropped by approximately four orders of magnitude to 103 CFU per g feces whereas the loads of the other main bacterial groups had remained stable. This raises the question whether the microbiota transplantation procedure in our study (e.g. with just one single oral gavage of luminal ileal content derived from mice with acute ileitis) was sufficient to guarantee a stable and long-term quantitative and qualitative establishment of the transplanted species within the acceptor mice where the conventional microbiota competes for nutrients and niches.
As shown in our recolonization experiments of gnotobiotic mice generated by antibiotic treatment, we need to take into consideration that CR against C. jejuni is caused by a multi-facetted interplay of the complex microbiota composition, physiological intraluminal milieu of the intestine or so far unknown factors. CR can thus not simply be attributed to absolute loads or relative distribution of single bacterial species [9, 19].
In a few studies, TX of fecal microbiota has been successfully performed in animals already populated with intestinal microbiota without compromising CR by antibiotic treatment before [21]. These studies had made use of both gavage and cohousing, given that rodents are coprophagous and thus continuously reinoculate themselves with microbiota by eating feces from their littermates or themselves [22–27]. In humans, fecal microbiota transplantation has been taken into consideration as a potential treatment option in severe cases of antibiotic-associated diarrhea, i.e. pseudomembranous colitis caused by Clostridium difficile toxin, inflammatory bowel diseases, such as ulcerative colitis, irritable bowel syndrome and constipation, or diabetes and obesity applying fecal samples from healthy donors via rectal enemas or nastrogastric tubes [28–35].
Taken together, in the presented study, a single oral transplantation of microbiota derived from susceptible mice with acute ileitis into conventional resistant mice is not sufficient to abrogate the physiological CR against C. jejuni. Mice harboring ileitis microbiota plus displaying beginning histopathologic changes of the ileum mucosa following T. gondii infection, however, were highly susceptible to C. jejuni infection.
In conclusion, future studies need to be undertaken to further unravel the interplay between intestinal pathogens, intestinal microbiota and intraluminal mileu, as well as the innate and adaptive immune system.
Acknowledgements
This work was supported by grants from the German Research Foundation (DFG) to UBG (GO363/12-1, CampyGerm; SFB633, TP A7), SB and AF (SFB633, TP A7), MMH (SFB633, TP B6), and from the German Federal Ministery of Education and Research (BMBF) to SB (“Lab in a hanky” projects TP 1.1 and TP 8.2). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Michaela Wattrodt and the staff of the animal research facility for excellent technical assistance and breeding of mice, respectively.
Contributor Information
M. M. Heimesaat, Charité-University Medicine Berlin, CC5, Department of Microbiology and Hygiene, Campus Benjamin Franklin, Hindenburgdamm 27, D-12203 Berlin, Phone: +49-30-8445-2194, FAX: +49-30-450-524-902, Email: markus.heimesaat@charite.de, Germany.
R. Plickert, Department of Microbiology and Hygiene, Charité, University Medicine Berlin, Berlin, Germany
A. Fischer, Department of Microbiology and Hygiene, Charité, University Medicine Berlin, Berlin, Germany
U. B. Göbel, Department of Microbiology and Hygiene, Charité, University Medicine Berlin, Berlin, Germany
S. Bereswill, Department of Microbiology and Hygiene, Charité, University Medicine Berlin, Berlin, Germany
References
- 1.Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. Experimental Campylobacter jejuni infection in humans. The Journal of Infectious Diseases. 1988 Mar 1;157(3) doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- 2.Kist M, Bereswill S. Campylobacter jejuni. Contributions to Microbiology. 2001;8:150-65. doi: 10.1159/000060405. [DOI] [PubMed] [Google Scholar]
- 3.Bereswill S, Kist M. Recent developments in Campylobacter pathogenesis. Current Opinion in Infectious Diseases. 2003 Oct 1;16(5) doi: 10.1097/01.qco.0000092833.20498.c5. [DOI] [PubMed] [Google Scholar]
- 4.van Spreeuwel JP, Duursma GC, Meijer CJ, Bax R, Rosekrans PC, Lindeman J. Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut. 1985 Sep 1;26(9) doi: 10.1136/gut.26.9.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker RI, Caldwell MB, Lee EC, Guerry P, Trust TJ, Ruiz-Palacios GM. Pathophysiology of Campylobacter enteritis. Microbiological Reviews. 1986 Mar 1;50(1) doi: 10.1128/mr.50.1.81-94.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ketley JM. Pathogenesis of enteric infection by Campylobacter. Microbiology (Reading, England) 1997 Jan 1;143 (Pt 1) doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- 7.Tsang RS. The relationship of Campylobacter jejuni infection and the development of Guillain-Barr syndrome. Current Opinion in Infectious Diseases. 2002 Jun 1;15(3) doi: 10.1097/00001432-200206000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Dorrell N, Wren BW. The second century of Campylobacter research: recent advances, new opportunities and old problems. Current Opinion in Infectious Diseases. 2007 Oct 1;20(5) doi: 10.1097/QCO.0b013e3282a56b15. [DOI] [PubMed] [Google Scholar]
- 9.Bereswill S, Fischer A, Plickert R, Haag LM, Otto B, Kühl AA, Dasti JI, Zautner AE, Muñoz M, Loddenkemper C, Gross U, Göbel UB, Heimesaat MM. Novel murine infection models provide deep insights into the "ménage à trois" of Campylobacter jejuni, microbiota and host innate immunity. PloS One. 2011 Jun 15;6(6): doi: 10.1371/journal.pone.0020953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haag LM, Fischer A, Otto B, Plickert R, Kühl AA, Göbel UB, Bereswill S, Heimesaat MM. Intestinal microbiota shifts towards elevated commensal Escherichia coli loads abrogate colonization resistance against Campylobacter jejuni in mice. PloS One. 2012 May 1;7(5): doi: 10.1371/journal.pone.0035988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haag LM, Fischer A, Otto B, Plickert R, Kühl AA, Göbel UB, Bereswill S, Heimesaat MM. Campylobacter jejuni induces acute enterocolitis in gnotobiotic IL-10-/- mice via Toll-like-receptor-2 and -4 signaling. PloS One. 2012 Jul 10;7(7): doi: 10.1371/journal.pone.0040761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto B, Haag LM, Fischer A, Plickert R, Kühl AA, et al. Campylobacter jejuni induces extra-intestinal immune responses via Toll-like-receptor-4 signaling in conventional IL-10 deficient mice with chronic colitis. Eur J Microbiol Immunol. 2012;2:210–219. doi: 10.1556/EuJMI.2.2012.3.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, Niebergall J, Jahn HK, Dunay IR, Moter A, Gescher DM, Schumann RR, Göbel UB, Liesenfeld O. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. Journal of Immunology (Baltimore, Md. : 1950) 2006 Dec 15;177(12) doi: 10.4049/jimmunol.177.12.8785. [DOI] [PubMed] [Google Scholar]
- 14.Heimesaat MM, Fischer A, Jahn HK, Niebergall J, Freudenberg M, Blaut M, Liesenfeld O, Schumann RR, Göbel UB, Bereswill S. Exacerbation of murine ileitis by Toll-like receptor 4 mediated sensing of lipopolysaccharide from commensal Escherichia coli. Gut. 2007 Jul 1;56(7) doi: 10.1136/gut.2006.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heimesaat MM, Fischer A, Siegmund B, Kupz A, Niebergall J, Fuchs D, Jahn HK, Freudenberg M, Loddenkemper C, Batra A, Lehr HA, Liesenfeld O, Blaut M, Göbel UB, Schumann RR, Bereswill S. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PloS One. 2007 Jul 25;2(7): doi: 10.1371/journal.pone.0000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erridge C, Duncan SH, Bereswill S, Heimesaat MM. The induction of colitis and ileitis in mice is associated with marked increases in intestinal concentrations of stimulants of TLRs 2, 4, and 5. PloS one. 2010 Feb 9;5(2): doi: 10.1371/journal.pone.0009125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bereswill S, Muñoz M, Fischer A, Plickert R, Haag LM, Otto B, Kühl AA, Loddenkemper C, Göbel UB, Heimesaat MM. Anti-inflammatory effects of resveratrol, curcumin and simvastatin in acute small intestinal inflammation. PloS one. 2010 Dec 3;5(12): doi: 10.1371/journal.pone.0015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muñoz M, Heimesaat MM, Danker K, Struck D, Lohmann U, Plickert R, Bereswill S, Fischer A, Dunay IR, Wolk K, Loddenkemper C, Krell HW, Libert C, Lund LR, Frey O, Hölscher C, Iwakura Y, Ghilardi N, Ouyang W, Kamradt T, Sabat R, Liesenfeld O. Interleukin (IL)-23 mediates Toxoplasma gondii-induced immunopathology in the gut via matrixmetalloproteinase-2 and IL-22 but independent of IL-17. The Journal of experimental medicine. 2009 Dec 21;206(13) doi: 10.1084/jem.20090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bereswill S, Plickert R, Fischer A, Kühl AA, Loddenkemper C, et al. What you eat is what you get: Novel Campylobacter models in the quadrangle relationship between nutrition, obesity, microbiota and susceptibility to infection. J Microbiol Immunol. 2011;1:237–248. doi: 10.1556/EuJMI.1.2011.3.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haag LM, Fischer A, Otto B, Grundmann U, Kühl AA, et al. Campylobacter jejuni infection of infant mice: acute enterocolitis is followed by asymptomatic intestinal and extra-intestinal immune response. Eur J Microbiol Immunol. 2012;2:2–11. doi: 10.1556/EuJMI.2.2012.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damman CJ, Miller SI, Surawicz CM, Zisman TL. The microbiome and inflammatory bowel disease: is there a therapeutic role for fecal microbiota transplantation? The American journal of gastroenterology. 2012 Oct 1;107(10) doi: 10.1038/ajg.2012.93. [DOI] [PubMed] [Google Scholar]
- 22.Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, Guigo R, Knight R, Guarner F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome research. 2010 Oct 1;20(10) doi: 10.1101/gr.107987.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007 Oct 5;131(1) doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell host & microbe. 2010 Sep 16;8(3) doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011 May 27;145(5) doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell host & microbe. 2008 Oct 16;4(4) doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009 Oct 30;139(3) doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eiseman B, Silen W, Bascom GS, Kauvar AJ. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery. 1958 Nov 1;44(5) [PubMed] [Google Scholar]
- 29.Bowden TA, Jr, Mansberger AR, Jr, Lykins LE. Pseudomembraneous enterocolitis: mechanism for restoring floral homeostasis. The American Surgeon. 1981 Apr 1;47(4) [PubMed] [Google Scholar]
- 30.Schwan A, S Sjölin, Trottestam U, Aronsson B. Relapsing clostridium difficile enterocolitis cured by rectal infusion of homologous faeces. Lancet. 1983 Oct 8;2(8354) doi: 10.1016/s0140-6736(83)90753-5. [DOI] [PubMed] [Google Scholar]
- 31.Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nature reviews. Gastroenterology & hepatology. 2011 Dec 20;9(2) doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- 32.Borody TJ, Cole P, Noonan S, Morgan A, Lenne J, Hyland L, Brandl S, Borody EG, George LL. Recurrence of duodenal ulcer and Campylobacter pylori infection after eradication. The Medical journal of Australia. 1989 Oct 16;151(8) doi: 10.5694/j.1326-5377.1989.tb101251.x. [DOI] [PubMed] [Google Scholar]
- 33.Gustafsson A, Berstad A, Lund-Tønnesen S, Midtvedt T, Norin E. The effect of faecal enema on five microflora-associated characteristics in patients with antibiotic-associated diarrhoea. Scandinavian journal of gastroenterology. 1999 Jun 1;34(6) doi: 10.1080/003655299750026038. [DOI] [PubMed] [Google Scholar]
- 34.van Nood E, Speelman P, Kuijper EJ, Keller JJ. Struggling with recurrent Clostridium difficile infections: is donor faeces the solution? Euro surveillance : bulletin europeen sur les maladies transmissibles = European communicable disease bulletin. 2009 Aug 27;14(34): doi: 10.2807/ese.14.34.19316-en. [DOI] [PubMed] [Google Scholar]
- 35.Vrieze A, Holleman F, Zoetendal EG, de Vos WM, Hoekstra JB, Nieuwdorp M. The environment within: how gut microbiota may influence metabolism and body composition. Diabetologia. 2010 Apr 1;53(4) doi: 10.1007/s00125-010-1662-7. [DOI] [PMC free article] [PubMed] [Google Scholar]