Introduction
Regulation of gene transcription is a complex process that is critical for the proper development of multi-cellular organisms. In eukaryotic cells, the genome is packaged into chromatin, which consists of DNA and a large number of proteins that often serve regulatory functions. The basic unit of chromatin is the nucleosome, composed of 2 copies each of the core histone H2A, H2B, H3, and H4 surrounded by 147 bases of DNA. Nucleosomes affect gene expression through multiple means that include their positioning and their composition with respect to covalent histone modifications and histone variants. Here, we focus on the role of post-translational modifications (PTMs) of histones, specifically modifications on the amino-terminal tails of H3 and H4, in the regulation of gene expression (Campos and Reinberg 2009).
The addition and removal of histone PTMs is controlled by pairs of enzymes, similar to the addition and removal of phosphates by kinases and phosphatases, respectively. Methyl marks are placed by methyltransferases and removed by demethylases, acetyl marks are placed by histone acetyltransferases and removed by deacetylases, and so on. The occurrence of histone PTMs can thus be highly dynamic, subject to both extra-cellular stimuli such as acetylation of histone H4 lysine 5 (H4K5) and H4K8 after estrogen binding to its receptor, and internal signals that affect the status of chromatin such as phosphorylation of H2AX after DNA damage (Trojer and Reinberg 2006; Kouzarides 2007). In contrast to acetylation and phosphorylation, other histone PTMs such as methylation, are relatively stable and, once established, appear to be actively maintained through DNA replication and cell division. These are the PTMs that are most likely to participate in the epigenetic regulation of gene expression and are the central subject of this review.
In general, histone PTMs affect transcription both directly, through changes in higher-order chromatin structures, and indirectly, by recruitment of downstream effectors. Although the distribution of certain histone PTMs correlates with gene expression levels, the mechanism by which these “marks” affect transcription remains unclear (Barski et al. 2007). Our longstanding goal has been to gain insight into the processes of deposition of certain histone modifications and how they regulate transcription.
In the past, we focused our attention on the basic mechanisms of RNA polymerase II-dependent transcription. Originally, these studies were conducted using conventional biochemical purification and techniques such as chromatographic fractionation and reconstitution experiments, to determine the minimal factors necessary for in vitro transcription on a naked DNA template (Flores et al. 1992). Along with others, we used this approach to identify the general transcription factors and characterize how they regulate RNA polymerase II, and to determine DNA sequence elements required for these processes (Orphanides et al. 1996). The information obtained from these analyses provided a solid foundation to tackle more complex templates consisting of DNA assembled with histones into chromatin, reflective of the in vivo situation. Thus, we set out to study transcription in vitro on a fully reconstituted chromatinized template that necesitates an investigation of the properties of chromatin-modifying enzymes (Pavri et al. 2006).
Chromatin is classified into heterochromatin and euchromatin, based on a combination of both functional and microscopic characteristics (Trojer and Reinberg 2007). Heterochromatin is compact, electron dense (dark under the electron microscope), and transcriptionally repressed. Euchromatin is transcriptionally permissive, less electron dense, and often referred to as being “open” or “loose”, although the exact molecular status corresponding to these terms remains poorly understood. Heterochromatin is functionally categorized as constitutive or facultative, based on whether the repressed state is permanent or conditional, respectively (Margueron et al. 2005). Constitutive heterochromatin silences repetitive elements to prevent genomic instability (Maison and Almouzni 2004), whereas formation of facultative heterochromatin is critical for the transient silencing of tissue-specific genes during differentiation (Trojer and Reinberg 2007). These three forms of chromatin are typically associated with distinct histone modifications. For example, constitutive heterochromatin is enriched for trimethylation of histone H3 lysine 9 (H3K9me3), facultative heterochromatin is enriched for H3K27me3, and euchromatin is punctuated by H3K4me3 and H3K36me3 (Kouzarides 2007).
We originally sought to identify the enzymes that catalyze the deposition of specific histone modifications. Through conventional purification studies, we identified and characterized the Polycomb Repressive Complex 2 (PRC2) and PR-Set7, enzymes responsible for H3K27me3 and H4K20me1, respectively (Kuzmichev et al. 2002; Nishioka et al. 2002). H3K27me3 has a well-established role in facultative heterochromatin. It localizes to developmentally regulated genes, and loss of the enzymatic machinery required for its deposition causes defects in differentiation. In pluripotent embryonic stem cells, H3K27me3 is a component of specialized chromatin regions, known as “bivalent domains”, which mark developmentally regulated genes as being poised for repression or activation in subsequent differentiated lineages (Bernstein et al. 2006a). A universal role for H4K20me1 in transcription has yet to emerge, and we believe that this mark may have different functions according to its genomic localization and timing of deposition. Along these lines, PR-Set7 is essential for development and has also been implicated in processes ranging from X chromosome inactivation to cell cycle regulation (Jorgensen et al. 2007; Tardat et al. 2007; Oda et al. 2009). We extended these studies to MBT-domain containing proteins since one such protein, L3MBTL1, binds H4K20me1, and other members of the family are involved in polycomb mediated repression (Bonasio et al. 2010). We also investigated the Sirtuin family of histone deacetylases, which coordinate multiple silencing pathways during the formation of heterochromatin (Vaquero et al. 2007a).
In the following sections, we present an overview of our findings and current progress in the field of facultative heterochromatin, with respect to PRC2, MBT domain-containing proteins, PR-Set7, and SIRT1. We will then discuss future directions, which focus on understanding how chromatin is regulated spatially within the nucleus.
Facultative heterochromatin
Introduction to PRC2
Polycomb group (PcG) proteins are central players in the formation of facultative heterochromatin. Polycomb genes were originally identified as factors required for the proper maintenance of expression patterns of homeotic genes during development in Drosophila melanogaster (Ringrose and Paro 2004). A major focus of our studies in facultative heterochromatin has been the Polycomb Repressive Complex 2 (PRC2). PRC2 promotes this silencing by methylating H3K27, to form facultative heterochromatin. PRC2 is composed of five core subunits, EED, SUZ12, EZH2, and RBAP46 /48, that are highly conserved. Although the exact contribution of each subunit to the function of the complex has remained elusive, we have made significant progress towards elucidating how PRC2 affects chromatin structure and transcription. We will focus on our recent work concerning the core components of PRC2 including EED and EZH2, as well as the EZH2 homologue EZH1, the role of auxiliary factors such as JARID2 and AEBP2 in regulating the activity and chromatin recruitment of the PRC2 holoenzyme, and the dynamic post-translational regulation of this complex.
PRC2 core components
Together with three other groups (Cao et al. 2002; Czermin et al. 2002; Muller et al. 2002), we discovered that EZH2 catalyzes H3K27me3, but only when part of a complex that we designated PRC2 (Kuzmichev et al. 2002). PRC2 is composed of 5 proteins: EED, SUZ12, EZH2, RbAp46 /48 (Cao et al. 2002; Kuzmichev et al. 2002); (Czermin et al. 2002; Muller et al. 2002)). We also found that PRC2 catalyzes H1bK26me and that its specificity for catalyzing H3K27me2/3 or H1bK26me is dictated by the identity of its EED subunit, which exists in four different isoforms (EED1-4) depending on the translation start site utilized (Kuzmichev et al. 2004; Kuzmichev et al. 2005; Montgomery et al. 2007).
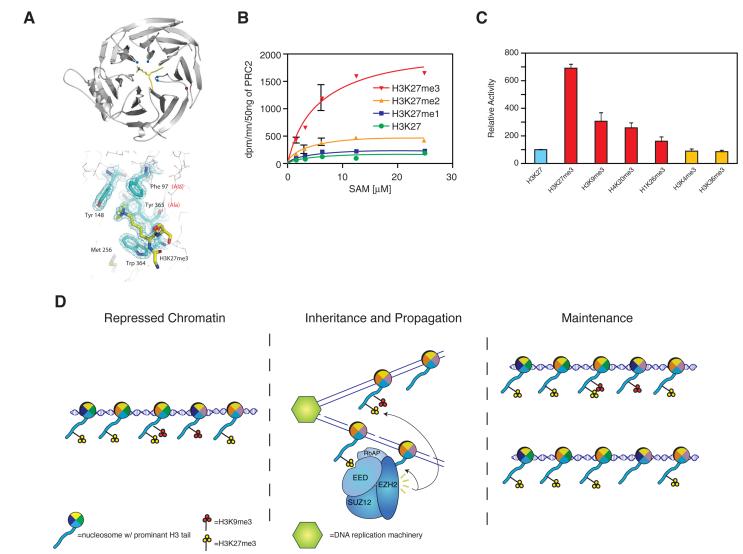
Because of the characteristic distribution of H3K27me3 within silenced developmental genes and the role of the polycomb system in cell identity, it has long been suspected that this histone modification constitutes a bona fide “epigenetic” mark, i.e. it is faithfully transmitted to cell progeny after DNA replication and cell division. However, a molecular mechanism for the transmission of H3K27me3 was lacking. We recently discovered that EED exhibits a novel interaction, the consequences of which may explain how H3K27me3 domains can be epigenetically inherited (Figure 1) (Margueron et al. 2009). EED contains an “aromatic cage” that binds histone H3 peptides trimethylated at lysines 9 and 27 (H3K9me3, H3K27me3), but its binding to H3K27me3 in particular stimulated the enzymatic activity of PRC2 (Figures 1a, b, c). Mutation of the residues in EED responsible for recognition of H3K27me3 did not affect the basal activity of the complex in vitro, yet these mutations did cause a drastic decrease in H3K27me3 levels in vivo, suggesting that EED-mediated PRC2 binding to H3K27me3 regulates the proper recruitment and activity of PRC2, required for maintaining overall H3K27me3 levels (Margueron et al. 2009). Similar feedback loops maintain and propagate other histone modifications. In the case of H3K9me3, the modification is recognized by HP1, which in turn binds to and recruits the histone methyltransferases SUV39H1/2 that catalyze the deposition of H3K9me3 (Stewart et al. 2005). However, the stimulation of EZH2 activity upon binding of its partner, EED, to the product of its activity, H3K27me3, is the first described case of a histone modification allosterically regulating the activity of a histone lysine methyltransferase. This regulatory loop may provide a mechanism for H3K27me3 propagation during DNA replication, in that parental nucleosomes containing H3K27me3 could serve as a template for the establishment of this modification on newly incorporated histone H3 (Figure 1d).
Figure 1.
PRC2 enzymatic activity is regulated by H3K27me3 binding. A) Ribbon representation of the EED–H3K27me3 complex, in which EED is in grey and the histone peptide is in yellow with its methyl-lysine side chain shown in stick representation. Right, depiction of EED aromatic cage residues. B) HKMT assay with PRC2–EZH2, with a titration of the methyl donor (S-adenosyl-methionine) in the presence of H3K27me0/1/2/3 peptides. d.p.m., disintegration per min. C) Relative histone methyltransferase activity in the presence of various peptides as indicated. D) Model for recognition and propagation of H3K27me3 by PRC2 during S phase. Adapted with permission from (Margueron et al. 2009). (Reprinted by permission from Macmillan Publisher’s Ltd: [NATURE] 461(7265): 762-767, 2009).
In mammals, there are two homologs of the D. melanogaster E(z) gene, EZH1 and EZH2. EZH2 was identified as a component of PRC2 by conventional biochemical purifications, but the function of EZH1 in mammals had not been characterized. EZH1 is ubiquitously expressed, whereas EZH2 expression is restricted to highly proliferating cells (Margueron et al. 2008). EZH1 forms a complex with the same components as the canonical PRC2, clearly establishing EZH1 as an alternate core component of PRC2 (Shen et al. 2008). Despite the fact that EZH1 and EZH2 contain largely homologous SET domains (responsible for their methyltransferase activity), PRC2-EZH2 and PRC2-EZH1 exhibit a major difference is their level of methyltransferase activity, with PRC2-EZH2 being considerably more active towards H3K27 than PRC2-EZH1, both in vivo and in vitro (Margueron et al. 2008). We have also shown by genome-wide analysis that EZH1 and EZH2 share a common set of promoter binding sites. Yet, independent of its SET domain, EZH1 acts uniquely to repress transcription by directly compacting chromatin, as assessed by electron microscopy (Margueron et al. 2008). PRC2-EZH1 may also indirectly regulate PRC2-EZH2 activity and H3K27me3 levels by decreasing available pools of SUZ12, EED, and RBAP46/48. We envision these two PRC2-mediated silencing pathways, via EZH1 or EZH2, as having unique roles in quiescent versus proliferating cell lineages, respectively.
Ancillary PRC2 subunits
PRC2 core components interact with several proteins that regulate its activity and, possibly, its recruitment to chromatin (Cao et al. 2008; Sarma et al. 2008; Li et al.). Although other interacting proteins have been identified, we will focus our discussion on three of these factors: PHF1, JARID2, and AEBP2. In the absence of PHF1, PRC2 mostly catalyzes deposition of H3K27me2 in vitro, but when PHF1 is added, PRC2 is able to catalyze deposition of H3K27me3 (Sarma et al. 2008). Similar observations were made with the D. melanogaster homologue of PHF1, polycomb-like (Nekrasov et al. 2007). PHF1 contains two PHD fingers and a tudor domain, two motifs that often function as recognition modules for methyl-lysine and methyl-arginine, respectively. The presence of these domains suggests that PHF1 may function as a chromatin adaptor in vivo, linking different methylation states with a functional outcome by modulating PRC2 activity.
As with PHF1, w, e recently found that JARID2 stimulates PRC2 enzymatic activity (Li et al. 2010), although two other reports claimed that JARID2 inhibits PRC2 (Peng et al. 2009; Shen et al. 2009). This discrepancy may be explained by differences in the assay conditions. Our studies showed that JARID2 stimulates PRC2 when added at equimolar concentrations, conditions that more accurately reflect the stoichiometry of the complex in vivo. We also found that the stimulatory effect of JARID2 on PRC2 is allosteric and nucleosome-specific (J.S., R.M., and D.R., unpublished results). Despite this controversy regarding the effect of JARID2 on the enzymatic activity of PRC2 (Peng et al. 2009; Shen et al. 2009), all studies agree that JARID2 is important for recruiting PRC2 to its target genes, and this may be mediated through the DNA binding domain of JARID2 (Li et al. 2010). Indeed JARID2 was found in isolation to bind to DNA with preference to G:C rich sequences ((Peng et al. 2009) and see below). Interestingly, JARID2 is a member of a family of histone demethylases, yet it lacks any demethylase activity itself and may function by modulating the activity of other enzymes in vivo. JARID2 is essential for proper embryogenesis, highlighting its importance in development (Takeuchi et al. 1995).
Another protein that interacts with PRC2 and stimulates its enzymatic activity is AEBP2, which also contains a DNA binding motif (Kim et al. 2009). We found that AEBP2 regulates the Km for the co-substrate S-adenosyl methionine (SAM) of PRC2. We also characterized how JARID2 and AEBP2 cooperate in regulating PRC2 activity in vitro. Both factors stimulate the enzymatic activity of PRC2 and act synergistically (J.S., R.M., and D.R., unpublished results). Furthermore, this joint stimulation is enhanced further by the presence of an H3K27me3 peptide that is recognized by the EED component of PRC2 resulting in increased PRC2 activity (see above). Thus JARID2, AEBP2, and the H3K27me3 modification itself appear to modulate the PRC2 complex through distinct mechanisms (J.S., R.M., and D.R., unpublished results). This interplay suggests a dynamic regulation of PRC2 activity in vivo.
Phosphorylation of PRC2
Core components of PRC2 are regulated by post-translational modifications (Cha et al. 2005). Using mass spectrometry analyses, we found that EZH2 is phosphorylated in vivo at two novel sites by CDK1. Neither of these two sites, T345 and T487, is within a characterized domain of EZH2. Using phospho-specific antibodies directed against these two residues, we demonstrated that this phosphorylation is cell-cycle regulated, with phosphorylation at T345 occurring during mitosis and phosphorylation at T487 decreasing during S-phase. To determine the functional importance of these phosphorylations, we generated independent mutants of each site by substitution with either alanine (loss of phosphorylation) or aspartic acid (phosphorylation mimic). We reconstituted mutant PRC2 complexes containing recombinant proteins expressed in insect cells, but we did not observe any major effects on enzymatic activity. However, phosphorylation of EZH2 may regulate binding of accessory factors, chromatin targeting, or other unknown aspects of PRC2 biology, and that it varies during the cell cycle suggests that further investigation is warranted. Recently, we have demonstrated that phosphorylation effects ncRNA binding suggesting an important role in recruitment (S.K. and D.R., unpublished results).
Recruitment of PRC2
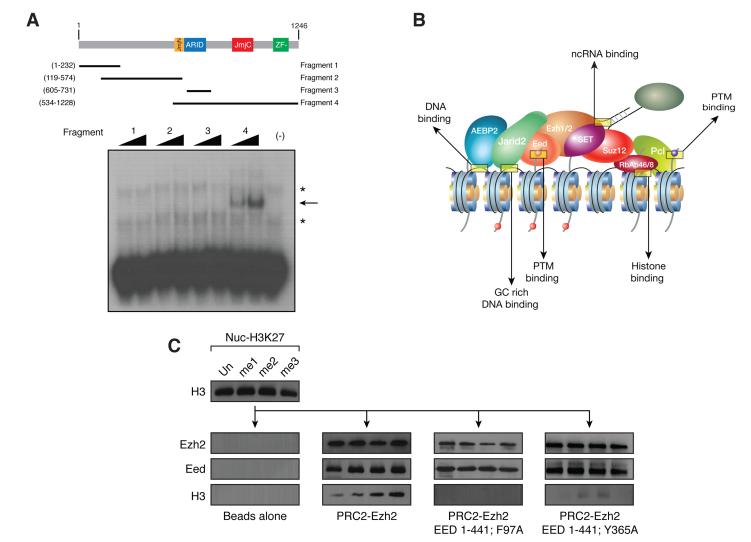
Recruitment of PRC2 to target genes and regulation of its enzymatic function are two critical steps in polycomb silencing, yet little is known regarding how PRC2 is recruited to its genomic targets in vertebrates. In D. melanogaster, specific DNA sequences called polycomb response elements (PREs) are sufficient to induce polycomb-mediated repression, but the existence of a similar DNA sequence-based mechanism in other organisms remains controversial (Ringrose and Paro 2004). One recent report found genomic elements involved in recruiting the PRC1 complex (Woo et al.), but it remains unclear whether they also recruit PRC2. As of yet, there has not been compelling evidence for a single definitive recruitment mechanism for PRC2 in mammals. We believe that PRC2 recruitment may depend on multiple agents, each contributing a low binding affinity, but together synergizing to attain stable binding (Figure 2b). These factors would include JARID2 and AEBP2 through their DNA binding domains, RBAP46/48 that binds H4 tails, and EED that binds to H3K27me3 (Figure 2c) (Margueron et al. 2009). In support of this model, JARID2 targets are enriched for a GC-rich motif (Peng et al. 2009), which is consistent with a computationally predicted, putative PRC2 recognition element (Figure 2a) (Ku et al. 2008). We speculate that these factors may function together as a PRC2 targeting mechanism.
Figure 2.
PRC2 is recruited to chromatin through multiple binding events. A) Schematic representing the known JARID2 domains. Gel shift assay in the presence of increasing amounts of each of the JARID2 fragments using radiolabeled DNA probes selected by SELEX. The asterisks indicate nonspecific signals present in the free probes. The arrow indicates the specific band shift. B) Schematic depicting the diversity of binding events targeting PRC2 to chromatin. C) Pull down experiments to analyze the interaction between EED, PRC2-EZH2 or reconstituted EEDTyr365Ala, lacking H3K27me3 binding, and H3K27 modified chromatin. Adapted with permission from (Margueron et al. 2009; Li et al. 2010). (Reprinted by permission from Macmillan Publisher’s Ltd: [NATURE] 461(7265): 762-767, 2009 and forthcoming Nature review).
Although we have uncovered some aspects of PRC2 regulation, many issues remain unresolved. These include how depletion of each subunit of the PRC2 complex affects its genome-wide distribution and activity. Furthermore, post-translational modifications, such as phosphorylation, of the core components may affect chromatin binding or ancillary subunit composition. Also, non-coding RNAs (ncRNAs) have emerged as potential elements for PRC2 recruitment, although their precise role remains unclear.
Introduction to MBT domains
Histone modification-binding proteins, or “readers,” are downstream effectors of histone post-translational modifications. These proteins share common structural motifs that recognize certain modification types in a context dependent manner, often with a remarkable degree of accuracy. Central among these motifs are the chromo-, bromo-, and tudor domains along with PHD fingers. A less well studied, but not less important, histone modification binding module is the Malignant Brain Tumor (MBT) domain, which predominantly binds mono- and di-methylated lysines by means resembling those of chromodomains and WD40 domains, although the overall folds are unrelated. MBT domains are found in polycomb proteins such as SCM and SFMBT, suggesting a role in the formation of facultative heterochromatin. Furthermore, the Drosophila MBT domain-containing protein, L(3)MBT, is a tumor suppressor that functions as a transcriptional corepressor (Bonasio et al. 2010).
The human MBT protein family comprises nine members (Bonasio et al. 2010). These proteins contain either two, three, or four MBT domains arranged in tandem and organized in an interlocked superstructure (Sathyamurthy et al. 2003; Wang et al. 2003; Eryilmaz et al. 2009; Guo et al. 2009). Yet, MBT proteins appear to bind a histone methyl-lysine residue in only one of their MBT domains (Trojer et al. 2007) (Li et al. 2007; Taverna et al. 2007) (Figure 3b). When in the binding pocket, the methylated lysine is caged by aromatic residues, with a limited number of interactions involving histone residues flanking the methylated lysine. MBT domains exhibit a preference in binding for mono- and di-methyl modifications (Bonasio et al. 2010). Unlike other histone-methyl binding modules, such as chromodomains and PHD fingers, target site selection appears promiscuous for MBT domains in vitro (Taverna et al. 2007). Importantly, the MBT proteins studied to date have all been implicated in transcriptional repression. Owing to genetic studies in D. melanogaster and biochemical studies in mammalian models, we know that repression mediated by MBT-domain proteins intersects with that of PcG proteins.
Figure 3.
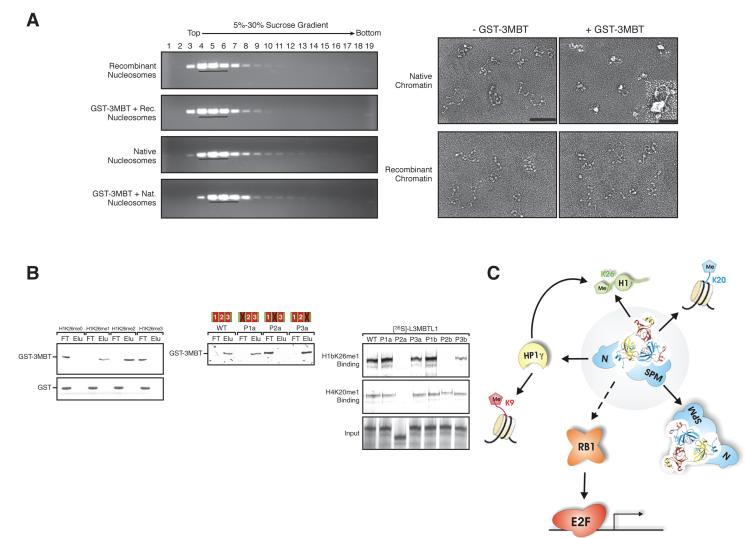
L3MBTL1 represses transcription through multiple means including chromatin compaction. A) Left, sucrose gradient fractions, containing either native or recombinant chromatin, and GST 3MBT, analyzed by agarose gel electrophoresis and ethidium bromide staining. Right, sucrose gradient peak fractions were subjected to electron microscopy (EM) analysis. Scale bar is 100 nm, and bar in the insert is 25 nm. B) Left, GST or GST-3MBT binding to peptides comprising histone H1b-K26me0/1/2/3 were analyzed by silver stain. Center, silver stain of interactions between immobilized H1b-K26me2 peptides and recombinant GST-3MBT either wild-type or with point mutations P1a, P2a, and P3a in the first, second, and third MBT domain, respectively. Right, autoradiography of interactions between immobilized H1b-K26me1 or H4-K20me1 peptides and full-length in-vitro-translated [35S]-labeled L3MBTL1, either wild-type or with indicated mutants. C) The malignant brain tumor (MBT) domain containing protein L3MBTL1 (center, light grey background) heterodimerizes (via its Sexcomb-Polyhomeotic-MBT (SPM) domain) and binds directly to chromatin (mono- and di methylated histone H4 lysine 20 and linker histone H1 isotype b lysine 26 (H4K20me1/2; H1bK26me1/2)) with its second MBT-domain. Moreover, L3MBTL1 associates with a number of proteins including Heterochromatin Protein 1 isoform gamma (HP1γ; via its N-terminus), Retinoblastoma (RB1) and the ETS family transcription factor TEL. Adapted with permission from (Trojer and Reinberg 2008).
Chromatin readers, including MBT proteins, are common components of the polycomb group (PcG) complexes and contribute to their function (Taverna et al. 2007). For instance, the founding member of the group is Polycomb (Pc) a PRC1 subunit that recognizes H3K27me3 via its chromodomain (Cao et al. 2002; Bernstein et al. 2006b) (Kuzmichev et al. 2002). MBT proteins are also physically associated with PcG complexes and potentially serve a similar role. For instance, SCM, a protein with two MBT domains, co-purified from D. melanogaster and human cells as a substoichiometric subunit of PRC1 (Levine et al. 2002) (Saurin et al. 2001) (Shao et al. 1999). Moreover, SCM and the PcG protein Polyhomeotic interact via their sterile alpha motif (SAM) domains and might form copolymers along chromatin, thus promoting the spreading of PcG proteins (Kim et al. 2005). In addition, Drosophila SFMBT was purified as a component of the PhoRC PcG complex (Klymenko et al. 2006). MBT domain-containing proteins seem to be important in integrating existing histone modifications and histone-modifying enzymes.
L3MBTL1 complex
Recently, we set out to determine the molecular mechanism of L3MBTL1-mediated transcriptional repression. Toward this end, we isolated L3MBTL1 associated polypeptides. L3MBTL1 interacts with Rb, HP1γ, H1bK26me2, and H4K20me1 (Figure 3c). We demonstrated that the three MBT domains are required for L3MBTL1-mediated repression of transcription, and that the second MBT domain interacts with H4K20me1 and H1K26me2. Most importantly, we demonstrated that L3MBTL1 compacts chromatin in an H4K20me1-dependent manner (Figure 3b) (Trojer et al. 2007). In vivo, L3MBTL1 represses transcription through a targeting mechanism involving Rb. Repression is likely mediated through local compaction that precludes binding of regulatory factors (Trojer and Reinberg 2008) (Figure 3a).
We also determined that L3MBTL1 specifically interacts with PR-Set7, the enzyme that catalyzes H4K20me1, which is only present during mitosis, early G1, and late G2 (Oda et al. 2009). Two domains in L3MBTL1 independently interact with PRSet7: the N terminus and the MBT domains. Rice and co-workers also observed a similar interaction between L3MBTL1 and PR-Set7, however they indicated that L3MBTL1 also binds to H3K9me1 (Kalakonda et al. 2008). Additionally, studies by Allis, Patel and colleagues (Li et al. 2007) indicated that L3MBTL1 binds to mono- and di-methylated lysine residues, but with little specificity. However, in our experiments the interaction between PR-Set7 and L3MBTL1-MBT domains is competed by peptides carrying H4K20me1 or H1K26me2, but not by peptides carrying H3K9me1, K3K4me1, or H4K20me3. We hypothesize that L3MBTL1 binding to H4K20me1 helps recruit PRSet7 to its own mark (Trojer and Reinberg 2006). If validated, this scenario would be similar to the feedback loops discussed above for PRC2 and SUV39H1/2.
L3MBTL2 complex
Similar to the case of L3MBTL1, we identified those polypeptides associated with L3MBTL2, including HP1γ, E2F6, CBX3, and PcG proteins RING1A, RING1B, and PCGF6. Based on the presence of the last three proteins, we termed this complex Polycomb Repressive Complex 1-like 3 (PRC1L3). We demonstrated that PRC1L3 catalyzes the deposition of monoubiquitin onto H2AK119ub1. L3MBTL2 binds chromatin independent of histone modifications and is required for the repressive function of the PRC1L3 complex. Genome-wide chromatin immunoprecipitation identified several hundred genes simultaneously bound by L3MBTL2 and E2F6, preferentially around transcriptional start sites. Importantly, these genes are distinct from those targeted by other E2Fs or the L3MBTL1 complex (P.T. and D.R., unpublished results).
Introduction to PR-Set7
Facultative heterochromatin is also marked in part by H4K20me1. We isolated and identified PR-Set7 as the sole enzyme responsible for catalyzing H4K20me1 (Nishioka et al. 2002). The role of H4K20me1 in transcription remains unclear, although it is enriched on the inactive X chromosome, a classic example of facultative heterochromatin. PR-Set7 has been shown to repress transcription, potentially through its role in synthesizing the necessary precursor for H4K20me3 (Karachentsev et al. 2005) (Oda et al. 2009). In turn, H4K20me3 is highly correlated with gene silencing and is found at regions of constitutive heterochromatin (Schotta et al. 2004).
PR-Set7 is required for proper cell cycle progression and its deletion in mice is embryonic lethal at the 4-cell stage(Oda et al. 2009). Both PR-Set7 expression and H4K20me1 are cell-cycle regulated. PR-Set7 is absent during S phase, and reaches its peak during mitosis when it binds to chromosomes (Rice et al. 2002) and interacts with L3MBTL1 (Kalakonda et al. 2008) and PCNA (Oda et al. 2010) an important regulator of DNA replication and the DNA damage response (Jorgensen et al. 2007; Tardat et al. 2007; Huen et al. 2008) (Figure 4a). Many unresolved questions remain surrounding the function of PR-Set7 and H4K20me1. In the next section, we review our latest results on the role of PR-Set7 during DNA damage, and its regulation by degradation during DNA damage and the cell cycle.
Figure 4.
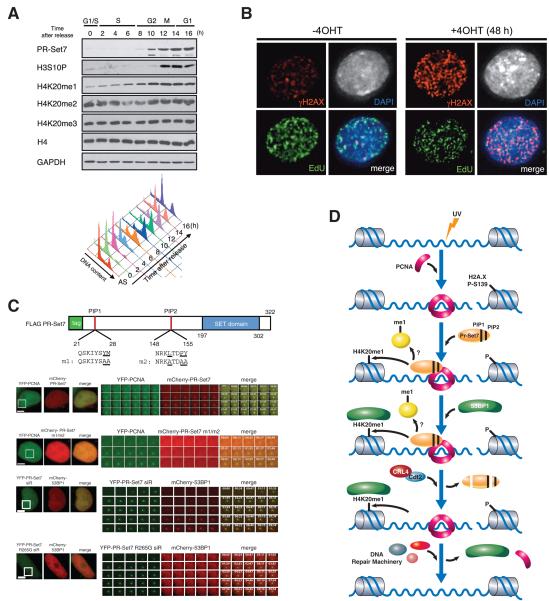
PR-Set7 is an H4K20 mono-methyltransferase that regulates genomic integrity. A) Western analysis of histone H3 and H4 modifications and PR-Set7 protein in synchronized HeLa cells. Cells released from G1 arrest after a double-thymidine block were analyzed every 2 h. Cell cycle phases were confirmed by FACS analyses. B) Deletion of PR-Set7 in PR-Set7 flox/-; CreERT, by addition of 4OHT for 48hr cells causes increased γ-H2AX foci. C) PR-Set7 domain structure depicting the two PIP domains. PR-Set7 is recruited to areas of laser induced DNA damage (white box) and this recruitment is dependent on the PIP domains (PR-Set7 m1/m2). 53BP1 is subsequently recruited to chromatin and this recruitment is dependent upon PR-Set7 catalytic activity (PR-Set7 R265G). D) Model depicting the recruitment and function of PR-Set7 after DNA damage. Adapted with permission from (Oda et al. 2009; Oda et al. 2010).
PR-Set7 and PCNA during DNA damage
Because of the interaction between PR-Set7 and PCNA, a role for PR-Set7 during DNA replication had been proposed (Jorgensen et al. 2007; Tardat et al. 2007), challenging our initial observation that PR-Set7 expression is mitosis-specific. To clarify this issue, we used live cell imaging and verified that PR-Set7 was indeed absent during S phase. We also observed the appearance of spontaneous foci containing PR-Set7 and PCNA within the nucleus of cells at early G1 and late G2 and these foci corresponded to DNA damage sites (Oda et al. 2010). These findings together with our previous observation that ES cells lacking PR-Set7 displayed increased DNA damage, support a role for PR-Set7 in the DNA damage response (Oda et al. 2009) (Figure 4b).
We next examined whether the interaction between PCNA and PR-Set7 is functionally relevant in the context of DNA damage Using time-lapse microscopy, we found that after laser irradiation PR-Set7 and PCNA co-localized at the irradiated foci. This co-localization required an intact PCNA Interaction Protein (PIP) domain within PR-Set7 (Figure 4c, d). The kinetics of recruitment for PR-Set7 and PCNA were similar, but PCNA remained associated with the DNA damage sites for a longer time. 53BP1, an important DNA damage checkpoint protein, was also recruited to these foci, but with much slower kinetics than that of PR-Set7, PCNA, or γH2AX. 53BP1 was reported to bind to the product of PR-Set7 activity, H4K20me1 and strikingly, PR-Set7 enzymatic activity is required for 53BP1 recruitment to sites of DNA damage (Figure 4c, d) (Oda et al. 2010). These results potentially explain the increased DNA damage detected in PRSet7−/− ES cells.
PR-Set7 degradation during DNA damage and the cell cycle
The interaction of PR-Set7 with PCNA is not only required for PR-Set7 recruitment to sites of DNA damage, but also promotes its degradation, both during DNA damage and specific stages of the cell cycle. In the case of DNA damage, our data support a model in which PR-Set7 is recruited to DNA damage sites through its interaction with PCNA, and the generation of H4K20me1 by PR-Set7 subsequently recruits 53BP1 (Figure 4d). PR-Set7 is then degraded via a PCNA dependent process involving the CRL4cdt2 ubiquitylation complex.
Regulation of PR-Set7 during the cell cycle, in particular its rapid disappearance in S phase, also occurs post-translationally (Nishioka et al. 2002). We found that PR-Set7 is polyubiquitinylated and that proteasome inhibitors impair its degradation. To determine the enzymatic machinery required for its degradation, we analyzed a series of E3 ligases and found that only two interact with PR-Set7, APCCdc20 (a member of the complex) and CRL4cdt2 (H.O. and D.R., unpublished results) (Oda et al. 2010). This result suggests that levels of PR-Set7 protein are controlled by at least two independent degradation pathways, one operating early in G1 through APCCdc20 and the other mediated by CRL4cdt2 via a PCNA-dependent mechanism. There are likely multiple pathways regulating PR-Set7 expression levels, given that previous studies have also implicated SCFSkp2 in the degradation of PR-Set7 (Yin et al. 2008). We propose that such tight regulation of PR-Set7 levels throughout the cell cycle, controlled in part by its targeted degradation, is critical to preserve genome integrity.
SIRT1
Histone deacetylases (HDACs) constitute a class of histone modifying enzymes that contribute to transcriptional silencing through deacetylation of histone lysine residues (Kouzarides 2007). Sirtuins are class III HDACs that use NAD+ as a co-factor and have an evolutionarily conserved role in transcriptional repression. Early evidence from the budding yeast S. cerevisiae demonstrated that the founding member of the Sirtuin family, Sir2p, deacetylates histone H4 at lysine 16, a critical step in the formation of facultative heterochromatin (Vaquero et al. 2007b). We have shown that the closest mammalian homologue, SIRT1, not only functions in transcriptional repression through histone deacetylation, but also through its targeting of SUV39H1, the histone methyltransferase required for deposition of H3K9me3. SirT1 binds, recruits, and activates SUV39H1 through its deacetylation of a lysine residue within the SET domain of SUV39H1. Loss of SirT1 causes misregulation of H3K9me3, which in turn causes loss of HP1α binding (Vaquero et al. 2007a). Thus, through these overlapping silencing mechanisms, SirT1 helps ensure that target genes are silenced.
Future directions
Although we have dedicated most of our recent efforts towards understanding the biochemical specificity and regulation of histone methyltransferases, we believe that spatial organization of the nucleus is an important, yet unresolved issue in chromatin biology. Pioneering studies have already shown that chromosomes occupy predetermined positions within the nucleus, with gene-rich chromosomes being localized preferentially toward the nuclear interior (Cremer et al. 2006) (Chubb et al. 2002). Repositioning of genes, either through looping or long-range movements, has been demonstrated in multiple systems to correlate with changes in gene expression and chromatin (Brown et al. 1997; Osborne et al. 2004). Moreover, the nucleus contains nuclear bodies composed of protein and nucleic acid that exemplify compartmentalization of function (Spector 2003). Despite the wealth of evidence for a functional role of nuclear organization, the mechanisms for spatial coordination of the genome and formation of nuclear bodies have remained elusive. As a first step towards understanding its contribution to gene regulation, one of our future goals is to identify how nuclear spatial organization is established.
Novel methodologies, such as DAM-ID and 3C-6C, have proven to be powerful tools to investigate nuclear organization, yet they can only probe protein-DNA and DNADNA interactions, respectively (van Steensel et al. 2001; Dekker 2006). The complex network of protein-protein interactions that take place in biochemically inaccessible compartments of the nucleus remain for the most part unexplored due to limitations in conventional biochemical techniques.
Conventional biochemical approaches have been invaluable in determining the network of physical interactions that directs and regulates gene expression in eukaryotic cells (Orphanides et al. 1996). Even in the absence of enzymatic assays, nuclear extraction followed by affinity purification and mass spectrometry can identify stable interactions between proteins, instructing the design of functional in vivo studies (Li et al.). However, a large number of nuclear proteins resist such extraction under the mild conditions required for preserving protein function. Up to 10% of total nuclear protein remain insoluble even after extraction with 2 M salt, 1% Triton X-100, and nucleases and, in fact, these are the proteins that constitute the elusive and controversial “nuclear matrix” (Albrethsen et al. 2008). Protein-protein interactions among these un-extractable proteins, as well as low affinity interactions, cannot be revealed with existing methodologies, yet they may be of fundamental importance in gene regulation, as suggested by the many studies that link nuclear organization with gene expression, development, and cellular memory. We are attempting to break this barrier using a novel chemical biology strategy as well as a genetic screen.
Our novel chemical biology approach is based on recruiting a photoactivatable probe to a protein of interest (bait) via high affinity recognition of the probe by a binding domain fused to the bait. The probe is then activated by UV light, creating a reactive species that forms a covalent bond with any protein in the vicinity of, and thus likely to interact with, the bait. After formation of the covalent adducts, labeled proteins are recovered under highly denaturing conditions that leave no insoluble structures left behind, but do not disrupt the covalent adducts.
We are also using a microscopy-based assay to determine factors required for nuclear spatial organization. We are performing a high-content genome-wide RNAi screen for master regulators of nuclear architecture and for factors influencing the formation of specific nuclear bodies. This type of high content screening has been successfully employed to determine regulators of such complex processes as DNA double-strand break repair, mitosis, and viral mediated infection (Doil et al. 2009; Stewart et al. 2009) (Neumann et al. 2010) (Karlas et al. 2010). The system allows for unbiased investigation of changes in the spatial distribution of proteins in an automated manner (Roukos et al.). This screen has the potential to identify architectural and regulatory factors that will expand our knowledge of the determinants of the highly compartmentalized nucleus (Dechat et al. 2008). After identifying these factors, we will be better able to determine how nuclear positioning affects transcription and chromatin dynamics.
Concluding Remarks
Although we have made great strides toward a more comprehensive mechanistic understanding of transcription and transcriptional regulation, many aspects still remain unclear. Recent work has demonstrated that chromatin is controlled through a complex interplay of enzymatic machinery that is dynamically regulated in response to many signals including differentiation, growth factors, and stress. Despite this wealth of experimental observations, how this molecular interplay is coordinated and integrated within the chromatin environment and the underlying nuclear landscape is still an open question. We are currently attempting to combine our original in vitro studies with in vivo models to better correlate transcriptional regulation with physiological functions and phenotype.
Acknowledgements
We are grateful to Stephanie Kim, Deborah Hernandez, and Kettly Cabane for technical assistance and the members of the Reinberg labfor helpful discussions. We thank Lynne Vales and Philipp Voigt for critical reading of the manuscript. We would like to thank our collaborators Drs. Michael Huebner, Maria-Elena Torres-Padilla, Edith Heard, Steve Gamblin, and David Spector. This work was supported by grants from NIH (GM64844 to D.R.) and the HHMI.
References
- Albrethsen J, Knol J, Jimenez C. Unravelling the nuclear matrix proteome. Journal of Proteomics. 2008:11. doi: 10.1016/j.jprot.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006a;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006b;26(7):2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Lecona E, Reinberg D. MBT domain proteins in development and disease. Semin Cell Dev Biol. 2010;21(2):221–230. doi: 10.1016/j.semcdb.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91(6):845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol. 2008;28(5):1862–1872. doi: 10.1128/MCB.01589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310(5746):306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12(6):439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories – a functional nuclear landscape. Curr Opin Cell Biol. 2006;18(3):307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker D, Solimando L, Goldman R. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes & Development. 2008;22(7):832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Meth. 2006;3(1):17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 Binds and Amplifies Ubiquitin Conjugates on Damaged Chromosomes to Allow Accumulation of Repair Proteins. Cell. 2009;136(3):435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Eryilmaz J, Pan P, Amaya MF, Allali-Hassani A, Dong A, Adams-Cioaba MA, Mackenzie F, Vedadi M, Min J. Structural studies of a four-MBT repeat protein MBTD1. PLoS One. 2009;4(10):e7274. doi: 10.1371/journal.pone.0007274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O, Lu H, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Identification and characterization of factor IIH. J Biol Chem. 1992;267(4):2786–2793. [PubMed] [Google Scholar]
- Guo Y, Nady N, Qi C, Allali-Hassani A, Zhu H, Pan P, Adams-Cioaba MA, Amaya MF, Dong A, Vedadi M, et al. Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2. Nucleic Acids Res. 2009;37(7):2204–2210. doi: 10.1093/nar/gkp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283(17):11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179(7):1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27(31):4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19(4):431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463(7282):818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- Kim CA, Sawaya MR, Cascio D, Kim W, Bowie JU. Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer. J Biol Chem. 2005;280(30):27769–27775. doi: 10.1074/jbc.M503055200. [DOI] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009;37(9):2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20(9):1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14(2):183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102(6):1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22(17):6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24(4):368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell. 2007;28(4):677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5(4):296–305. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(7265):762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32(4):503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15(2):163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Montgomery SA, Magnuson T. Molecular and functional mapping of EED motifs required for PRC2-dependent histone methylation. J Mol Biol. 2007;374(5):1145–1157. doi: 10.1016/j.jmb.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Muller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26(18):4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464(7289):721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9(6):1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the Histone H4 Monomethylase PR-Set7 by CRL4(Cdt2)-Mediated PCNA-Dependent Degradation during DNA Damage. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29(8):2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10(21):2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Osborne C, Chakalova L, Brown K, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell J, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36(10):1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125(4):703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16(17):2225–2230. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Roukos V, Misteli T, Schmidt CK. Descriptive no more: the dawn of high-throughput microscopy. Trends Cell Biol. doi: 10.1016/j.tcb.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28(8):2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyamurthy A, Allen MD, Murzin AG, Bycroft M. Crystal structure of the malignant brain tumor (MBT) repeats in Sex Comb on Midleg-like 2 (SCML2) J Biol Chem. 2003;278(47):46968–46973. doi: 10.1074/jbc.M306469200. [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412(6847):655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18(11):1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98(1):37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. The Dynamics of Chromosome Organization and Gene Regulation. Annu Rev Biochem. 2003;72(1):573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136(3):420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25(7):2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9(10):1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol. 2007;179(7):1413–1426. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. L3MBTL1, a histone methylation-dependent chromatin lock. Cell. 2007;129(5):915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125(2):213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- - Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28(1):1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- - Beyond histone methyl-lysine binding: how malignant brain tumor (MBT) protein L3MBTL1 impacts chromatin structure. Cell Cycle. 2008;7(5):578–585. doi: 10.4161/cc.7.5.5544. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27(3):304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007a;450(7168):440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007b;26(37):5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- Wang WK, Tereshko V, Boccuni P, MacGrogan D, Nimer SD, Patel DJ. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure. 2003;11(7):775–789. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 140(1):99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Yu VC, Zhu G, Chang DC. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell Cycle. 2008;7(10):1423–1432. doi: 10.4161/cc.7.10.5867. [DOI] [PubMed] [Google Scholar]