Abstract
Chronic myeloid leukemia (CML) and some acute lymphoblastic leukemias are characterized by the t(9;22) that encodes the BCR/ABL oncogene. Multiple mouse models of CML express BCR/ABL at high levels from non-Bcr promoters, resulting in the development of leukemias. In contrast, a significant fraction of healthy humans have been found to have BCR/ABL positive hematopoietic cells. To bridge the gap between the information derived from current mouse models and the non-leukemic humans with the BCR/ABL oncogene, we generated a knockin model with BCR/ABL p210 expressed from the Bcr locus. Unlike previous models, expression of BCR/ABL from the knockin allele did not induce leukemia. BCR/ABL mutant cells did exhibit favorable bone marrow engraftment compared to wild-type cells. These data suggest BCR/ABL expression alone is insufficient to induce disease. This new model allows for inducible spatial and temporal control of BCR/ABL expression for analysis of early steps in the pathogenesis of BCR/ABL-expressing leukemias.
Introduction
The molecular hallmark of chronic myeloid leukemia (CML) is the BCR/ABL-expressing t(9;22) Philadelphia chromosome (Groffen et al., 1984; Rowley, 1973; Rudkin et al., 1964). The discovery of this mutation led to the development of tyrosine kinase inhibitors (Druker et al., 2001a; Druker et al., 2001b; Kantarjian et al., 2006; Kantarjian et al., 2002; Kantarjian et al., 2010; Talpaz et al., 2006) that are now standard of care for patients with CML (Ernst and Hochhaus, 2012; Zhang and Rowley, 2011). BCR/ABL is believed to be sufficient for disease development, as expression of BCR/ABL in mouse models leads to rapid development of hematopoietic neoplasias (Daley et al., 1990; Elefanty et al., 1990; Honda and Hirai, 2001; Honda et al., 1998; Huettner et al., 2003; Jaiswal et al., 2003; Kelliher et al., 1990; Koschmieder et al., 2005; Voncken et al., 1995; Wong and Witte, 2001). The current understanding of BCR/ABL-induced disease stems from several experimental settings, including the injection of cell lines or primary cells from CML patients into immune-deficient mice, transduction of bone marrow-derived cells with retroviruses that express BCR/ABL (Daley et al., 1990; Elefanty et al., 1990; Kelliher et al., 1990; Wong and Witte, 2001), and expression of BCR/ABL in transgenic mice (Honda and Hirai, 2001; Honda et al., 1998; Huettner et al., 2003; Jaiswal et al., 2003; Koschmieder et al., 2005; Voncken et al., 1995). All of these mice express BCR/ABL from active promoters and multiple copies of the oncogene.
Although data from these mouse models suggest that BCR/ABL is sufficient to induce leukemia, some reports have indicated that only high levels of BCR/ABL induce disease in humans (Barnes et al., 2005; Gaiger et al., 1995). On the other hand, low levels of BCR/ABL expression have been linked to persistence of CML in the presence of the tyrosine kinase inhibitors, such as imatinib, due to a lack of oncogene dependency (Kumari et al., 2012). Thus extremes of oncogene expression may each lead to our inability to eradicate CML. To address whether a single copy of the BCR/ABL oncogene when expressed from the endogenous locus leads to disease development and persistence, we have constructed a knockin model with expression of human BCR/ABL p210 from the mouse Bcr locus. This model has allowed us to determine if expression of BCR/ABL as a single copy allele is sufficient to induce neoplasia.
Methods
Generation of Conditional BCR/ABL Knockin Mice
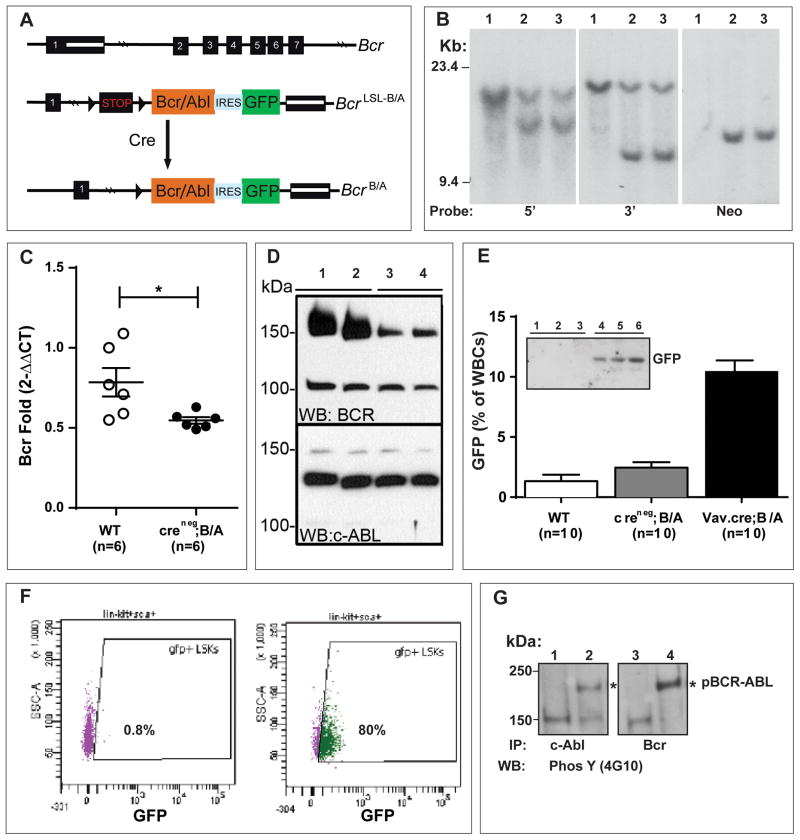
The targeting vector was constructed to generate a knockin mouse model that conditionally expresses BCR/ABL and eGFP (Figure 1A, Extended Experimental Procedures and Figure S1).
Figure 1. Conditional Knockin of BCR/ABL into the Murine Bcr Locus.
A. Schematic of the first 7 of 23 exons of the murine Bcr genomic locus (Bcr+), the targeted BcrLSL-BCR/ABL knockin allele generated by homologous recombination, and the allele with the deleted stop cassette (BcrBCR/ABL) following Cre-mediated recombination. The following features are indicated: stop cassette bracketed by loxP recombination sequences (LSL), human BCR/ABL-ires-GFP cDNA, and genomic hybridization probes. See Figure S1 for details of vector construction.
B. Southern blot analysis using 5′, 3′, and neo probes of SpeI-digested DNA from ES cell clones showing the wild-type (WT) Bcr and mutant knockin alleles. Lane 1, wild type Bcr and lanes 2 and 3, Bcr+/LSL-BCR/ABL clones that were used to generate two different targeted lines of mice.
C. Bcr message levels were significantly diminished in the spleens of Cre-negative heterozygous Bcr+/LSL-BCR/ABL adult mice (denoted Creneg;B/A). *p<0.05. Error bars denote SEM.
D. Anti-Bcr and anti-ABL polyclonal antibodies were used to detect the endogenous mouse proteins in brain extracts from Cre-negative Bcr+/LSL-BCR/ABL and wild-type mice as indicated. Lanes 1 and 2 are from wild-type mice, and lanes 3 and 4 are from knockin mice.
E. Vav-Cre expression in the hematopoietic system leads to recombination of the BcrLSL-BCR/ABL allele and GFP expression in peripheral blood WBCs. WT, Creneg;B/A, and Vav-Cre;B/A mice were bled at 6 weeks of age, and WBCs were subjected to flow analysis for GFP fluorescence. The expression of GFP was significantly higher in the Vav-Cre;B/A mice (p<0.0001). GFP was quantified as the percentage of WBCs that exhibit a lateral shift in fluorescent signal intensity relative to a wild-type baseline (denoted GFP(% of WBCs)). Inset: Mouse spleen extracts were blotted with an anti-GFP monoclonal antibody (Sigma). Lanes 1–3 are from three Creneg;B/A mice and lanes 4–6 are from three Vav-Cre;B/A mice.
F. Representative FACS plot of GFP expression in bone marrow stem and progenitor LSK (Lin-Sca+c-Kit+) populations from control (left) or Vav-Cre;B/A (right) mice. The majority (80%) of LSK cells from Vav-Cre;B/A mice were GFP positive (green) compared none (0.8%) in the wild type mice. The wild type LSK cells were almost all GFP negative (purple).
G. Immunoprecipitation and western blot analysis of spleen extracts from Vav-Cre;B/A mice for expression and constitutive tyrosine phosphorylation of BCR/ABL (4G10). Lane 1 and 3 are extracts from wild-type spleens, lanes 2 and 4 are extracts from Vav-Cre;B/A spleens. The identity of the phospho-protein in lanes 1–3 that migrates just above the 150 kDa marker is not known.
See also Figure S1.
Routine Bcr-Abl genotype analysis
The Bcr/Abl conditional knockin mice were genotyped from tail snips using real-time PCR assays designed by Transnetyx (Cordova, TN). The assays were designed to detect the wild-type and the mutant alleles in the presence or absence of the lox-Stop-lox cassette. A PCR assay for GFP was also used to confirm the presence of the targeted allele
Transgenic Mice
Double-transgenic Mx1-Cre, Vav-Cre or CMV-Cre;Bcr+/LSL-B/A (Vav-, Mx1- or CMV-Cre;B/A) mice were generated by crossing the Bcr+/LSL-B/A mice with mice transgenic for the Cre transgenes. Mx1-Cre was induced with pIpC as described (Oravecz-Wilson et al., 2009). Triple transgenic Vav-Cre; Bcr+/LSL-B/A;Aml+/LSL-A/E mice were generated by crossing Vav-Cre;Bcr+/LSL-B/A mice with Aml+/LSL-A/E mice (Higuchi et al., 2002). Mice were housed in the Unit for Laboratory Animal Medicine at the University of Texas Southwestern Medical Center under specific pathogen-free conditions and were monitored regularly for evidence of disease and abnormal peripheral blood cell counts. All mouse experiments were conducted after approval by the UT Southwestern Medical Center Committee on Use and Care of Animals.
Drug Treatments
Imatinib treatments were carried out for two weeks. Imatinib pills from the hospital pharmacy were dissolved in water and filtered through a 0.45-μm syringe filter. The stock concentration was 20 mg/ml (100 μl doses). Mice were given two doses a day by oral gavage.
Bone Marrow Transplantation and Long-Term Observation
Bone marrow from six-week old Creneg;B/A or Vav-Cre;B/A donor mice (CD45.2) was mixed with bone marrow from PepboyJ BL6 wild-type mice (CD45.1) prior to retroorbital reconstitution of lethally-irradiated recipients. Each recipient received a 50:50 ratio of CD45.1 to CD45.2 bone marrow cells to allow for subsequent quantitation of donor and recipient chimerism in transplanted mice. Small cohorts (n=3) of mice receiving Creneg;B/A and Vav.Cre;B/A bone marrow cells were sacrificed and analyzed at 6, 10, and 16 weeks post-transplant. Remaining recipients were preserved for long-term observation (6 months, 1 year and 1.5 years of observation).
Hematopoietic Analysis
Peripheral blood, spleen, and bone marrow cells were analyzed for lineage (CD3, B220, Mac-GR1), progenitor (lin−,Sca−,c−kit+, CD16/32+) and hematopoeitic stem cell (lin−,Sca+, CD150+) markers using the LSR Fortessa flow cytometer (BD). CBC analysis was performed on peripheral blood using the Hemavet 950 with MULTI-TROL Mouse as an equilibration control (Drew Scientific).
Western blot and Immunoprecipitation
Mouse tissues were solubilized by extraction in 1X RIPA lysis buffer (Cell Signaling Technology, Beverly, MA), electrophoresed on 6% SDS-PAGE gels, and transferred to a PVDF membrane. Proteins were detected using affinity-purified anti-BCR antiserum (Cell Signaling Technology) or anti c-Abl antiserum (Cell Signaling Technology) and visualized using Supersignal West Pico chemiluminescence substrate (Pierce, Rockford, IL). Immunoprecipitation of solubilized brain and spleen extracts was performed using anti c-Abl antiserum following pre-clearing of extracts with protein G-Sepharose. Immune complexes were washed with 1X RIPA lysis buffer, analyzed by SDS-PAGE, and blotted as indicated.
Results
To assess the role of a single copy of the BCR/ABL oncogene in disease pathogenesis and to avoid reported embryonic lethality (Castellanos et al., 1997; Heisterkamp et al., 1991), we generated a conditional knockin allele of BCR/ABL. A loxP-bracketed transcriptional stop cassette was engineered upstream of the human BCR/ABL p210-IRES-GFP cDNA (Pear et al., 1998) in exon 1 of the murine Bcr locus (BcrLSL-BCR/ABL, Figure 1A and S1A). This conditional knockin allele allowed for the use of different Cre transgenic mice to drive temporal and tissue-specific expression in concert with the endogenous mouse promoter and remaining regulatory elements of the locus. C57BL/6 ES cells were electroporated and screened for targeted knockin alleles and two ES cell lines generated mice carrying the BcrLSL-BCR/ABL mutation (Figure 1B and S1B). The C57BL/6 background has been used previously to induce disease in other BCR/ABL mouse models, and this background can be readily followed for donor chimerism following bone marrow transplantation using CD45.1 and CD45.2 variants. Furthermore, we sequenced the knockin allele from two different genomic Bcr+/LSL-BCR/ABL spleens to confirm the absence of de novo mutations in the BCR/ABL sequence. Bcr+/LSL-BCR/ABL mice were born at predicted Mendelian frequency and displayed normal growth. Lack of expression of wild-type Bcr from the knockin locus resulted in Bcr haploin sufficiency reflected in reduced Bcr message (Figure 1C) and protein (Figure 1D).
To express BCR/ABL from this knockin allele, we crossed the Bcr+/LSL-BCR/ABL mice with Vav-Cre and Mx1-Cre transgenic mice. The Vav and interferon (pIpC) induced Mx1 promoters are specifically activated in embryonic (Georgiades et al., 2002) and adult hematopoietic tissues including HSCs (Georgiades et al., 2002; Rajewsky et al., 1996). Mx1-Cre and Vav-Cre-mediated recombination and BCR/ABL expression was predicted to emulate BCR/ABL expression in CML patients as Cre expression is not only restricted to hematopoietic tissues but the knocked in BCR/ABL expression is further constrained by the endogenous regulatory elements of the Bcr locus. Expression of GFP was detected by flow cytometry of peripheral blood in white blood cells (WBCs) using either Vav-Cre or Mx1-Cre mediated recombination (Figure 1E and data not shown). Peripheral blood GFP levels were highest in the Mac1+Gr1+ population (Figure S1C) and GFP expression in bone marrow was highest in the stem/progenitor LSK (Lin−Sca+;c-Kit+) population (Figure 1F and Figure S1D). Since Bcr is expressed primarily in hematopoietic stem and progenitor cells (Wetzler et al., 1993), high GFP expression in hematopoietic stem cells was expected. These BCR/ABL expression patterns are also consistent with previous findings of enriched BCR/ABL expression in human bone marrow progenitors from CML patients (Marega et al., 2010). Additionally, GFP and constitutively phosphorylated BCR/ABL were detected by western blot of whole spleen extracts (Figure 1E; insert and Figure 1G). We tested for constitutive phosphorylation of CRKL and STAT5 in isolated LSK cells, but did not observe phosphorylation (data not shown). These data indicate that the knockin allele is expressed in a cellular pattern similar to the Bcr protein and the expressed oncogene has the expected constitutive tyrosine kinase activity. However, the amount of signaling is not strong enough for us to detect phosphorylation of its downstream targets.
To determine if the levels of BCR/ABL were similar to the human situation where BCR/ABL is expressed from the human BCR locus, we compared the copy number of the knocked-in BCR/ABL message to the copy number of the mouse Bcr message in Vav-Cre;Bcr+/BCR-ABL mice. Using qPCR and a huBCR/ABL-mBcr chimeric standard, we measured the absolute levels of the splenic huBCR/ABL message (8 copies/ng RNA; range 2–15; n= 13) and the mouse Bcr message (13 copies/ng RNA; range 3 to 15; n=10) in Vav-Cre;BCR/ABL knockin mice. The numbers of BCR/ABL and Bcr copies were in the same order of magnitude, although as expected, Bcr was higher. We did not expect the BCR/ABL:Bcr ratio to be 1:1 as Vav-Cre does not induce 100% recombination and is not expressed in every cell of the spleen. These data, together with specific GFP expression in stem and progenitor cells (Figure 1F), indicate that regulated expression of the BCR/ABL knockin allele is similar to the endogenous Bcr.
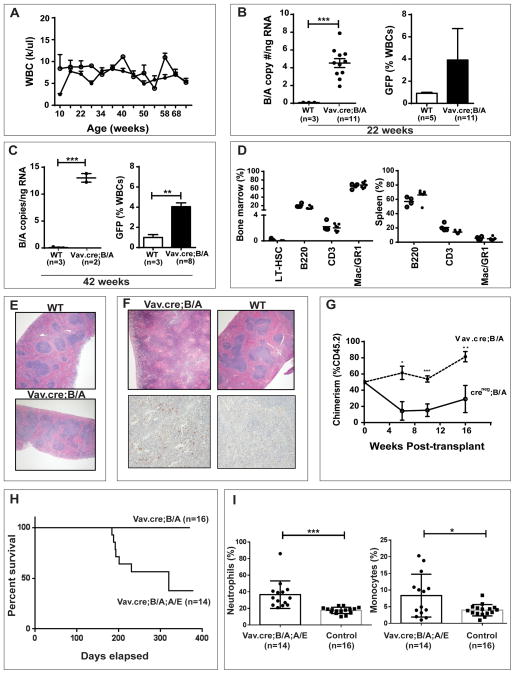
During the first year of observation of cohorts that were of either Creneg;Bcr+/LSL-BCR/ABL, Vav-Cre;Bcr+/LSL-BCR/ABL (n=14) or pIpC induced Mx1-Cre; Bcr+/LSL-BCR/ABL (n=13) genotypes, we saw no signs or symptoms of a hematopoietic illness. Mice expressing BCR/ABL (Cre;Bcr+/LSL-BCR/ABL ) had normal WBC counts (Figure 2A; Figure S2A), and their WBC differentials displayed normal myeloid and monocytic frequencies with only a slight trend of decreased lymphocyte frequencies (data not shown). Over the 1.5-years of observation, Vav-Cre;Bcr+/LSL-BCR/ABL mice were sacrificed and necropsied at 3 month intervals (n=3 per time point). During this period, none of the Vav-Cre;Bcr+/LSL-BCR/ABL mice or Mx1-Cre; Bcr+/LSL-BCR/ABL mice displayed abnormalities at necropsy, and flow cytometry of bone marrow showed normal stem and progenitor frequencies (Figure S2B and S2C). Furthermore, no changes in BCR/ABL expression were observed with age, as evidenced by steady peripheral blood GFP and spleen BCR/ABL message levels (Figure 2B and C). The bone marrow, spleen and peripheral blood of a final set of Vav-Cre; Bcr+/LSL-BCR/ABL mice sacrificed at 75 weeks of age had normal HSC and early progenitor frequencies as well as normal myeloid and lymphoid progenitor frequencies (Figure 2D). Spleen structure and histology as well as frequency of myeloid methylcellulose colonies (CFU-GEMM and CFU-GM) from the final groups of Vav-Cre;B/A and Mx1-Cre;B/A mice were normal (Figure 2E and data not shown). We observed seven homozygous Vav-Cre;Bcr LSL-BCR/ABL;LSL-BCR/ABL mice (have twice as much GFP as the heterozygous Vav-Cre;Bcr +/LSL-BCR/ABL mice) for 10–12 months. All of these homozygous mice remained disease free indicating that two copies of BCR/ABL are also not sufficient for disease development.
Figure 2. Phenotype of Vav-Cre;Bcr+/BCR/ABL Mice.
A. Complete blood counts (CBCs) from wild-type (WT open circles) and BCR/ABL knockin mice (closed circles) were normal. Members of a cohort of WT (n=9) and Vav-Cre;Bcr+/BCR/ABL (n=15) mice were observed for at least one year. Averages and standard deviations of WBCs counts of all mice are reported. No significant differences were observed between the two groups over the entire time course. Upon necropsy of a subset of the mice (n=5 of each group) at 6 months and1 year, no gross or histologic diseases were uncovered. Error bars denote SEM.
B. BCR/ABL message in the spleen (left) and expression of GFP in the peripheral blood (right) of WT and Vav-Cre;B/A mice at 5 months of age. BCR/ABL message was measured by absolute quantitative PCR, and GFP fluorescence was measured by flow cytometry as described.
C. BCR/ABL message levels in spleens (left) and GFP in peripheral blood (right) of WT and BCR/ABL mice at 11 months of age.
D. Final bone marrow (left) and spleen (right) progenitor frequencies in the members of the 1.5-year old aged mouse cohort. All mice were without evidence of disease at necropsy. Open circles represent aged control mice and closed circles represent aged Vav-Cre;B/A mice. Hematopoietic stem (LT-HSCs), B (B220), T (CD3) and myeloid (Mac+Gr1+) cell frequencies were not altered in the presence of the BCR/ABL oncogene.
E. Representative H&E stains of spleens from 1.5-year old aged control and Vav-Cre;B/A mice at 4x magnification.
F. Displayed are spleen H&E stains (top) of the one Vav-Cre;B/A mouse that succumbed to a myeloproliferative neoplasm at 58 weeks of age and a WT control mouse (4x magnification). A myeloid infiltration was confirmed in the Vav-Cre;B/A spleen with myeloperoxidase staining (bottom; 10x magnification). Spleen from the wild type (WT) mouse was myeloperoxidase negative.
G. Competitive bone marrow transplantation indicated that BCR/ABL enhances engraftment. Bone marrow from Cre-negative B/A (n=3) or Vav-Cre-positive donor mice (n=3) (CD45.2) was mixed with an equal number of WT competitor bone marrow (CD45.1) cells, and then 1 × 106 cells were transplanted into 15 lethally-irradiated recipients per donor (n=45 recipients per genotype) in a CD45.1:CD45.2 ratio of 1:1. At the indicated time points, peripheral blood and bone marrow was assessed for donor chimerism by flow cytometry. The dotted line represents percent of CD45.2 chimerism in mice that received Vav-Cre;Bcr+/BCR/ABL donor cells, and the solid line represents chimerism analysis in mice that received Cre-negative Bcr+/BCR/ABL donor cells. Percent CD45.2 on the Y-axis was expected to be 50% throughout the experiment. Results represent average and SEM of the chimerism data from mice in each cohort. The engraftment of the Vav-Cre;B/A cells were significantly than the Creneg;B/A cells at point; *p<0.05; **p<0.01; ***p<0.001.
H. Kaplan-Meier survival curve of Vav-Cre;B/A;A/E and Vav-Cre;B/A control mice. Vav-Cre;B/A;A/E were found dead or were sacrificed when moribund. P<0.01 (Logrank test).
I. Complete blood counts (CBCs) from Vav-Cre;B/A;A/E knockin mice showed significant monocytosis and neutrophilia. Members of a cohort of controls (n=16) and Vav-Cre;Bcr+/BCR/ABL;Aml+/AML1/ETO (n=14) mice were observed for one year. If a mouse died before the year endpoint, the last available CBC data were included. Averages and standard deviations of are reported. Error bars denote SEM. *P<0.05; ***P<0.001.
See also Figure S2.
To determine if the expression of BCR/ABL was necessary for the survival of the cells expressing it, we treated Vav-Cre;B/A mice with imatinib for two weeks and measured GFP in peripheral blood. We detected no changes in GFP after imatinib treatment indicating that the majority of the BCR/ABL expressing cells are not addicted to BCR/ABL (Figure S2D). This supports prior data that indicates leukemia cells expressing low levels of BCR/ABL do not undergo apoptosis when treated with imatinib (Modi et al., 2007). These data are also consistent with the hypothesis that low levels of BCR/ABL expression are a reason for persistence of CML in patients treated with imatinib (Kumari et al., 2012).
Despite an initial year in which no hematopoietic abnormalities or addiction to BCR/ABL expression were observed, a single Vav-Cre;Bcr+/LSL-BCR/ABL knockin mouse spontaneously died at 58 weeks of age. At necropsy, this mouse was found to have an effaced spleen with infiltration of neutrophils, mimicking what is seen in the chronic phases of CML (Figure 2F). In addition to this mouse, upon sacrifice of the remaining members of this cohort at 1.5-years of age, we observed a slight effacement of the normal white/red pulp spleen structure in two out of the remaining seven mice. However, the rest of the hematopoietic analysis (bone marrow, peripheral blood and spleen flow cytometry) of these two mice was normal. This single case of a myeloproliferative disease could be due to either expression of the BCR/ABL oncogene or to the normal increased frequency of myeloproliferation neoplasms that occurs with age in mice. In fact, the presence of a single myeloproliferative neoplasm in the BCR/ABL cohort was not significantly different from the controls (p<0.2).
Although expression of BCR/ABL from its endogenous locus did not lead to disease, bone marrow cells from Vav-Cre; Bcr+/LSL-BCR/ABL mice did exhibit an engraftment advantage compared to wild type bone marrow when the two were co-transplanted into lethally-irradiated mice (Figure 2G, dotted line). It is possible that CD44 molecules on the BCR/ABL-expressing stem cells in the knockin model are more functional than CD44 of the co-transplanted wild type cells. This possibility was considered due to a recent report indicating that BCR/ABL-expressing stem cells have a greater dependence on CD44 for bone marrow engraftment compared to normal stem cells (Krause et al., 2006). However, we did not observe increased expression of CD44 in the BCR/ABL-expressing stem and progenitor populations (data not shown). This enhanced reconstitution could be a result of accumulation genetic mutations over time. Contrasting with the Cre-positive cells, Cre-negative Bcr+/LSL-BCR/ABL haploinsufficient bone marrow cells displayed a competitive disadvantage compared to co-transplanted wild-type cells (Figure 2E, solid line). The decreased engraftment by the Cre-negative Bcr+/LSL-BCR/ABL bone marrow may be due to Bcr haploinsufficiency (Figure 1D). Indeed, we found that the haploinsufficiency of Bcr in young Cre negative Bcr+/LSL-BCR/ABL mice (1–3 months of age) did lead to a decrease in the number of hematopoietic stem cells (Figure S2E). The fact that expression of BCR/ABL overrode this negative engraftment effect and provided an additional engraftment advantage (Figure 2G) may simply be due to the well-known constitutive proliferative and survival signals from BCR/ABL that sustain CML in humans(Cilloni and Saglio, 2012). Because we did not observe increased frequencies of LT-HSCs in any of the Vav-Cre;BCR/ABL mice, an engraftment advantage was likely not due to BCR/ABL leading to increased self-renewal (Figure S2B). Instead, BCR/ABL expression may provide a better environment for cells to survive the normally harsh cytokine-poor state of an irradiated bone marrow niche.
The infrequent occurrence of myeloproliferation after a long latency in the mice bearing the BCR/ABL knockin allele suggests that additional genetic events are required for transformation of hematopoietic cells. We previously observed this for the HIP1/PDGFβR knockin allele (Oravecz-Wilson et al., 2009). To begin to test this, we crossed the BCR/ABL mice with mice bearing a conditional AML1/ETO knockin allele (Higuchi et al., 2002). This AML1/ETO knockin allele on its own, like HIP1/PDGFβR and BCR/ABL, is insufficient for leukemogenesis. Dual conditional knockin mice with the Vav-Cre transgene were born at the expected Mendelian ratio and appeared healthy until spontaneous deaths occurred in over 50% of the mice between 6 and 12 months of age (Figure 2H). Upon necropsy, we observed splenic architecture effacement in those mice whose spleens were intact at death (Figure S2F). Prior to death lymphopenia, thrombocytopenia, neutrophilia and/or monocytosis were present in the double knockin mice (Figure 2I and data not shown). In the mice that remained in this dual knockin cohort, peripheral blood GFP levels of the double knockin mice (Vav-Cre;BCR/ABL;AML1/ETO) were similar to those of the Vav-Cre;BCR/ABL mice (Figure S2G, top panel). By the time of this last analysis of GFP levels, all of the mice had aged enough to develop a monocytosis (Figure 2SG, bottom panel), which is the harbinger to a lethal myeloproliferation (Figure 2H). There was one “outlier” mouse with a higher GFP level than the rest of the mice (14% vs. average of 5%). However, this level is similar to that found in homozygous BCR/ABL mice (two copies of BCR/ABL) that do not develop disease. These data indicate that a second hit (e.g. AML1/ETO), rather than an increased BCR/ABL level, is key to the development of myeloproliferation in this mouse (Figure S2G). These data also indicate that even though AML1/ETO has been found in many tyrosine kinase-driven leukemias (Golub et al., 1994; Miyoshi et al., 1991; Schessl et al., 2005) and cooperates with BCR/ABL in these mice, the 5 month latency to disease suggests even more than the two genetic changes are needed for the development of BCR/ABL associated leukemia.
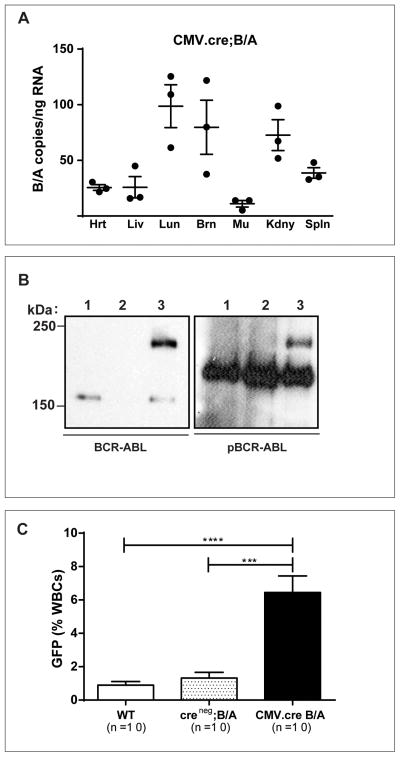
Germline expression of transgenic BCR/ABL has been reported to result in embryonic lethality (Heisterkamp et al., 1991), and prior attempts to generate a germline knockin of p190 BCR/ABL allele were not successful (Castellanos et al., 1997). To determine when in embryogenesis lethality occurs and understand why, the Bcr+/LSL-BCR/ABL mice were crossed with transgenic CMV-Cre mice to allow Cre recombination of the LoxP sites in the germline (Schwenk et al., 1995). To our surprise, germline expression of BCR/ABL in our mice was not embryonic lethal. CMV-Cre; Bcr+/LSL-BCR/ABL mice were born at the predicted Mendelian frequency and displayed no gross abnormalities. Members of a cohort of these mice are now a year of age and without evidence of cancer or other abnormalities. Although CMV-Cre recombination leads to LoxP site recombination in all cells of the body, the expression of the BCR/ABL knockin allele also depends on many of the natural regulatory elements of the Bcr locus. Thus, we predicted that the expression of BCR/ABL would closely mimic that of Bcr (Heisterkamp et al., 1993). Indeed, the BCR/ABL message (Figure 3A) was especially prevalent in the brain, in agreement with earlier reports that expression of Bcr protein is highest in the CNS (Heisterkamp et al., 1993). Despite normal brain histology and size, the brains from these mice had easily detectable BCR/ABL protein that exhibited constitutively active tyrosine kinase activity (Figure 3B). Expression of the knockin allele was also maintained in the peripheral blood of these CMV-Cre; Bcr+/LSL-BCR/ABL mice at frequencies similar to that in Vav-Cre; Bcr+/LSL-BCR/ABL mice (Figure 3C). This CMV-Cre experiment further demonstrates that this knockin allele mimics the tissue specific expression pattern of the endogenous Bcr locus.
Figure 3. Germline BCR/ABL expression mimics that of BCR and does not alter the health of embryos, newborns, or mature mice.
A. Quantitative PCR analysis for BCR/ABL RNA expression in CMV-Cre;B/A mouse organs. Hrt, heart; Lvr, liver; Lng, lung; Brn, brain; Mu,muscle; Kdny, kidney; Spln, spleen. Each assay was performed in triplicate and three mice were evaluated (total assays n=9).
B. Immunoprecipitation and western blot analysis for BCR/ABL expression and constitutive phosphorylation (4G10) in brain extracts from CMV-Cre;B/A mice. The identity of the phosphoprotein that migrates just above the 150 kDa marker in the right hand panel is not known but serves as an internal loading control. Lane 1 =wild-type brain; Lane 2 = Creneg;B/A brain; Lane 3 = CMV-Cre;B/A brain.
C. Peripheral blood cells of adult CMV-Cre;B/A mice have GFP positive white blood cells similar to that observed for the Vav-Cre;B/A mice. Error bars are SEM; ***p<0.001; ****p<0.0001.
Discussion
Although CML has been considered exemplary of a malignancy caused by a single genetic event, three main lines of evidence prior to this report led us to question whether BCR/ABL alone is responsible for to induce disease. First, blood cells from up to 30% of healthy individuals repeatedly test positive for the Philadelphia chromosome (Bayraktar and Goodman, 2010; Biernaux et al., 1995; Bose et al., 1998). Second, treatment of patients with potent BCR/ABL inhibitors infrequently cures their CML (Savona and Talpaz, 2008). And third, based on the low copy number of BCR/ABL messages in residual diseased progenitors from imatinib-treated CML patients, Kumari and colleagues recently proposed that chronically low levels of BCR/ABL in imatinib-refractory CML-initiating cells may be independent of BCR/ABL signals for survival and explain their persistence in the presence of imatinib (Kumari et al., 2012). These data suggest that in addition to the presence of the BCR/ABL oncogene, distinct molecular changes may be required for disease onset, progression and maintenance.
To address the challenges associated with current CML treatments and to determine whether single-gene-copy expression of BCR/ABL is sufficient for CML development, we generated a mouse model in which BCR/ABL p210 is expressed from the endogenous Bcr locus. This allele allows for the expression of the oncogene from its endogenous locus and mimics heterozygous BCR knockout that occurs as a result of the translocation. Although heterozygous Bcr knockout mice are grossly normal (Voncken et al., 1995), Bcr haploinsufficiency has been proposed by Lin and colleagues to influence myeloid leukemogenesis by the BCR/ABL oncogene (Lin et al., 2001). In the latter report, the acute phase of CML was evaluated by injecting K562 cells into NOD/SCID mice under conditions where endogenous BCR expression was low, resulting in rapid development of disease in 100% of the recipient mice. This 100% chance of disease development was compared to a 20% disease rate in mice that were transplanted with K562 cells that were modified to have high levels of BCR expression (Lin et al., 2001). However, our haploinsufficiency data do not support an inhibitory role for Bcr expression in the chronic phase of CML development as the lower levels of the Bcr protein together with expression of BCR/ABL from the knockin allele did not lead to disease (Figures 1D and 2A).
The BCR/ABL oncogene in the human disease is expressed not only from the BCR promoter but also from a derivative t(9;22) Philadelphia chromosome. Epigenetic effects or new distal enhancers that result from the translocated arm of chromosome 9 may change expression of the oncogene beyond what is expected from the BCR locus alone. It is therefore possible that the level of BCR/ABL in this knockin mouse model does not perfectly mimic the human disease, due to a lack of these translocation effects. Measurements of the levels of BCR/ABL compared to wild type BCR in CML cells from patients with early phases of the disease will address the hypothesis that the translocated arm of chromosome 9 leads to altered expression of the BCR locus.
The lack of hematopoietic neoplasia in the BCR/ABL knockin mice up to one and a half years of age contrasts to previous retroviral and transgenic models using the same human BCR/ABL cDNA. Prior models demonstrated high penetrance of malignancies between one and four months of age(Daley et al., 1990; Elefanty et al., 1990; Honda et al., 1998; Huettner et al., 2003; Jaiswal et al., 2003; Kelliher et al., 1990; Koschmieder et al., 2005; Voncken et al., 1995; Wong and Witte, 2001). There was one report that described a transgenic model with somewhat longer disease latency than the other models. In this model with delayed disease, p210 was driven by the promoter of the tec gene, which is expressed specifically in early hematopoietic progenitors and a total of 10 mice were afflicted with hematopoietic neoplasms between 4 and 12 months of age (Honda and Hirai, 2001). On the other hand, the data from our knockin mice are consistent with the fact that low levels of BCR/ABL do not confer factor-independent growth to hematopoietic cell lines (Barnes et al., 2005), and these findings suggest 15 that the malignancies induced in prior models may be due, in part, to achievement of very high levels of BCR/ABL expression. Although there may also be secondary mutations induced by the prior models (retroviral or transgene insertional mutagenesis, genome instability), it has also been reported that oncogene levels in retroviral mouse models are much higher than single copy oncogene knockin models (Chen et al., 2008). BCR/ABL levels have, in fact, been reported to increase upon progression from the chronic phase of the disease to the blast crisis phase (Barnes et al., 2005; Gaiger et al., 1995). However, Vav-Cre;BCR/ABL;AML1/ETO double knockin mice that did develop a myeloproliferative disease, did not show a large increase in GFP levels. These data support the assertion that BCR/ABL expression from the Bcr locus is insufficient on its own for development of disease and requires additional genetic hits that achieve more than just an increased amount of BCR/ABL.
Unfortunately, genome sequencing experiments to identify BCR/ABL cooperators have primarily analyzed CML blast crisis cells rather than cells from the earlier chronic phase of the disease. Recently, pilot exon sequencing and single nucleotide polymorphism analyses of the earliest stages of CML have been initiated by a few investigators (Grossmann et al., 2011; Huh et al., 2011). No frequent genetic aberrations have yet been reported from these studies. WT1 and IDH1 mutations have been identified in a few chronic phase patients and, thus, are certainly genes of interest for examination in the BCR/ABL knockin mouse background. Because the AML1/ETO mutation has been found in tyrosine kinase associated leukemias (Golub et al., 1994; Miyoshi et al., 1991; Schessl et al., 2005), we generated a group of mice that were double mutant BCR/ABL and AML1/ETO knockin alleles and did observe a myeloproliferative phenotype. These data support the idea that additional genetic aberrations are needed to cooperate with BCR/ABL as a single copy oncogene to induce CML.
It remains to be determined if the BCR/ABL knockin mice will develop CML and progress to accelerated or frank blast crisis when combined with additional known genetic stressors such as expression of Nup98/HoxA9 (Neering et al., 2007), abnormal IDH1 (Mardis et al., 2009), or deficiencies of TET2 (Moran-Crusio et al., 2011), p53 and/or Ink4a/Arf. Alternatively, a non-biased discovery of spontaneous or ENU induced cooperating mutations and other conditions that promote myeloid leukemogenesis in this mouse model such as those that induce the Bcr promoter will likely shed new light on mechanisms of CML development and progression. The unique result of no disease in the single BCR/ABL knockin mouse despite expression of an active oncogene suggests that additional mouse models of other leukemias may provide new insights about the role(s) of well-known oncogenes to leukemogenesis. We predict that this conditional knockin model will be a powerful tool for investigators to use to improve CML prevention in those “previvors” with “normal” bone marrow bearing BCR/ABL(Bayraktar and Goodman, 2010; Biernaux et al., 1995; Bose et al., 1998) and treatment for individuals with the earliest phases of CML.
Supplementary Material
Highlights.
A BCR/ABL knockin allele that expresses active BCR/ABL was generated.
BCR/ABL from the endogenous Bcr locus in insufficient for leukemogenesis.
The BCR/ABL germline knockin is not embryonic lethal.
BCR/ABL and AML1/ETO knockin alleles cooperate to induce myeloproliferation.
Acknowledgments
We are grateful to members of the Ross lab for continuous and critical review of the data. We especially thank Dr. Corbin Meacham for her skilled assistance in hematopoietic analysis of the 1.5 year old cohort of Vav-Cre;BCR/ABL mice. This work was funded in part by the Burroughs Wellcome Fund (Clinical Scientist Award; TSR) and the National Cancer Institute (R01 CA82363-03, R01 CA098730-01;TSR) and TSR holds the Jeanne Ann Plitt Professorship in Breast Cancer Research and the H. Ben and Isabelle T. Decherd Chair in Internal Medicine at UT Southwestern Medical Center. TSR was a Leukemia and Lymphoma Society Scholar when these experiments were initiated.
Footnotes
Authorship
S.B.F., Z.L.H. and A.A.S. performed research, analyzed data and edited the paper. Y.W., J.A.M and K.I.O.W. performed research and analyzed data. T.S.R designed and performed the research, analyzed the data and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barnes DJ, Schultheis B, Adedeji S, Melo JV. Dose-dependent effects of Bcr-Abl in cell line models of different stages of chronic myeloid leukemia. Oncogene. 2005;24:6432–6440. doi: 10.1038/sj.onc.1208796. [DOI] [PubMed] [Google Scholar]
- Bayraktar S, Goodman M. Detection of BCR-ABL Positive Cells in an Asymptomatic Patient: A Case Report and Literature Review. Case Report Med. 2010;2010:939706. doi: 10.1155/2010/939706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernaux C, Loos M, Sels A, Huez G, Stryckmans P. Detection of major bcr-abl gene expression at a very low level in blood cells of some healthy individuals. Blood. 1995;86:3118–3122. [PubMed] [Google Scholar]
- Bose S, Deininger M, Gora-Tybor J, Goldman JM, Melo JV. The presence of typical and atypical BCR-ABL fusion genes in leukocytes of normal individuals: biologic significance and implications for the assessment of minimal residual disease. Blood. 1998;92:3362–3367. [PubMed] [Google Scholar]
- Castellanos A, Pintado B, Weruaga E, Arevalo R, Lopez A, Orfao A, Sanchez-Garcia I. A BCR-ABL(p190) fusion gene made by homologous recombination causes B-cell acute lymphoblastic leukemias in chimeric mice with independence of the endogenous bcr product. Blood. 1997;90:2168–2174. [PubMed] [Google Scholar]
- Chen W, Kumar AR, Hudson WA, Li Q, Wu B, Staggs RA, Lund EA, Sam TN, Kersey JH. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13:432–440. doi: 10.1016/j.ccr.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilloni D, Saglio G. Molecular pathways: BCR-ABL. Clin Cancer Res. 2012;18:930–937. doi: 10.1158/1078-0432.CCR-10-1613. [DOI] [PubMed] [Google Scholar]
- Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, Capdeville R, Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001a;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001b;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- Elefanty AG, Hariharan IK, Cory S. bcr-abl, the hallmark of chronic myeloid leukaemia in man, induces multiple haemopoietic neoplasms in mice. EMBO J. 1990;9:1069–1078. doi: 10.1002/j.1460-2075.1990.tb08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Hochhaus A. Chronic myeloid leukemia: clinical impact of BCR-ABL1 mutations and other lesions associated with disease progression. Semin Oncol. 2012;39:58–66. doi: 10.1053/j.seminoncol.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Gaiger A, Henn T, Horth E, Geissler K, Mitterbauer G, Maier-Dobersberger T, Greinix H, Mannhalter C, Haas OA, Lechner K, et al. Increase of bcr-abl chimeric mRNA expression in tumor cells of patients with chronic myeloid leukemia precedes disease progression. Blood. 1995;86:2371–2378. [PubMed] [Google Scholar]
- Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, Print CG. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis. 2002;34:251–256. doi: 10.1002/gene.10161. [DOI] [PubMed] [Google Scholar]
- Golub TR, Barker GF, Lovett M, Gilliland DG. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- Grossmann V, Kohlmann A, Zenger M, Schindela S, Eder C, Weissmann S, Schnittger S, Kern W, Muller MC, Hochhaus A, et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011;25:557–560. doi: 10.1038/leu.2010.298. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N, Jenster G, Kioussis D, Pattengale PK, Groffen J. Human bcr-abl gene has a lethal effect on embryogenesis. Transgenic Res. 1991;1:45–53. doi: 10.1007/BF02512996. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N, Kaartinen V, van Soest S, Bokoch GM, Groffen J. Human ABR encodes a protein with GAPrac activity and homology to the DBL nucleotide exchange factor domain. J Biol Chem. 1993;268:16903–16906. [PubMed] [Google Scholar]
- Higuchi M, O’Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- Honda H, Hirai H. Model mice for BCR/ABL-positive leukemias. Blood Cells Mol Dis. 2001;27:265–278. doi: 10.1006/bcmd.2000.0374. [DOI] [PubMed] [Google Scholar]
- Honda H, Oda H, Suzuki T, Takahashi T, Witte ON, Ozawa K, Ishikawa T, Yazaki Y, Hirai H. Development of acute lymphoblastic leukemia and myeloproliferative disorder in transgenic mice expressing p210bcr/abl: a novel transgenic model for human Ph1-positive leukemias. Blood. 1998;91:2067–2075. [PubMed] [Google Scholar]
- Huettner CS, Koschmieder S, Iwasaki H, Iwasaki-Arai J, Radomska HS, Akashi K, Tenen DG. Inducible expression of BCR/ABL using human CD34 regulatory elements results in a megakaryocytic myeloproliferative syndrome. Blood. 2003;102:3363–3370. doi: 10.1182/blood-2003-03-0768. [DOI] [PubMed] [Google Scholar]
- Huh J, Jung CW, Kim JW, Kim HJ, Kim SH, Shin MG, Kim YK, Suh JS, Moon JH, Sohn SK, et al. Genome-wide high density single-nucleotide polymorphism array-based karyotyping improves detection of clonal aberrations including der(9) deletion, but does not predict treatment outcomes after imatinib therapy in chronic myeloid leukemia. Ann Hematol. 2011;90:1255–1264. doi: 10.1007/s00277-011-1195-2. [DOI] [PubMed] [Google Scholar]
- Jaiswal S, Traver D, Miyamoto T, Akashi K, Lagasse E, Weissman IL. Expression of BCR/ABL and BCL-2 in myeloid progenitors leads to myeloid leukemias. Proc Natl Acad Sci U S A. 2003;100:10002–10007. doi: 10.1073/pnas.1633833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H, Giles F, Wunderle L, Bhalla K, O’Brien S, Wassmann B, Tanaka C, Manley P, Rae P, Mietlowski W, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Sawyers C, Hochhaus A, Guilhot F, Schiffer C, Gambacorti-Passerini C, Niederwieser D, Resta D, Capdeville R, Zoellner U, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Shah NP, Hochhaus A, Cortes J, Shah S, Ayala M, Moiraghi B, Shen Z, Mayer J, Pasquini R, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2010;362:2260–2270. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- Kelliher MA, McLaughlin J, Witte ON, Rosenberg N. Induction of a chronic myelogenous leukemia-like syndrome in mice with v-abl and BCR/ABL. Proc Natl Acad Sci U S A. 1990;87:6649–6653. doi: 10.1073/pnas.87.17.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschmieder S, Gottgens B, Zhang P, Iwasaki-Arai J, Akashi K, Kutok JL, Dayaram T, Geary K, Green AR, Tenen DG, et al. Inducible chronic phase of myeloid leukemia with expansion of hematopoietic stem cells in a transgenic model of BCR-ABL leukemogenesis. Blood. 2005;105:324–334. doi: 10.1182/blood-2003-12-4369. [DOI] [PubMed] [Google Scholar]
- Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med. 2006;12:1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- Kumari A, Brendel C, Hochhaus A, Neubauer A, Burchert A. Low BCR-ABL expression levels in hematopoietic precursor cells enable persistence of chronic myeloid leukemia under imatinib. Blood. 2012;119:530–539. doi: 10.1182/blood-2010-08-303495. [DOI] [PubMed] [Google Scholar]
- Lin F, Monaco G, Sun T, Liu J, Lin H, Stephens C, Belmont J, Arlinghaus RB. BCR gene expression blocks Bcr-Abl induced pathogenicity in a mouse model. Oncogene. 2001;20:1873–1881. doi: 10.1038/sj.onc.1204409. [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marega M, Piazza RG, Pirola A, Redaelli S, Mogavero A, Iacobucci I, Meneghetti I, Parma M, Pogliani EM, Gambacorti-Passerini C. BCR and BCR-ABL regulation during myeloid differentiation in healthy donors and in chronic phase/blast crisis CML patients. Leukemia. 2010;24:1445–1449. doi: 10.1038/leu.2010.101. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci U S A. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi H, McDonald T, Chu S, Yee JK, Forman SJ, Bhatia R. Role of BCR/ABL gene-expression levels in determining the phenotype and imatinib sensitivity of transformed human hematopoietic cells. Blood. 2007;109:5411–5421. doi: 10.1182/blood-2006-06-032490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neering SJ, Bushnell T, Sozer S, Ashton J, Rossi RM, Wang PY, Bell DR, Heinrich D, Bottaro A, Jordan CT. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 2007;110:2578–2585. doi: 10.1182/blood-2007-02-073031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz-Wilson KI, Philips ST, Yilmaz OH, Ames HM, Li L, Crawford BD, Gauvin AM, Lucas PC, Sitwala K, Downing JR, et al. Persistence of leukemia-initiating cells in a conditional knockin model of an imatinib-responsive myeloproliferative disorder. Cancer Cell. 2009;16:137–148. doi: 10.1016/j.ccr.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- Rajewsky K, Gu H, Kuhn R, Betz UA, Muller W, Roes J, Schwenk F. Conditional gene targeting. J Clin Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Rudkin CT, Hungerford DA, Nowell PC. DNA Contents of Chromosome Ph1 and Chromosome 21 in Human Chronic Granulocytic Leukemia. Science. 1964;144:1229–1231. doi: 10.1126/science.144.3623.1229. [DOI] [PubMed] [Google Scholar]
- Savona M, Talpaz M. Getting to the stem of chronic myeloid leukaemia. Nat Rev Cancer. 2008;8:341–350. doi: 10.1038/nrc2368. [DOI] [PubMed] [Google Scholar]
- Schessl C, Rawat VP, Cusan M, Deshpande A, Kohl TM, Rosten PM, Spiekermann K, Humphries RK, Schnittger S, Kern W, et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J Clin Invest. 2005;115:2159–2168. doi: 10.1172/JCI24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O’Brien S, Nicaise C, Bleickardt E, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- Voncken JW, Kaartinen V, Pattengale PK, Germeraad WT, Groffen J, Heisterkamp N. BCR/ABL P210 and P190 cause distinct leukemia in transgenic mice. Blood. 1995;86:4603–4611. [PubMed] [Google Scholar]
- Wetzler M, Talpaz M, Van Etten RA, Hirsh-Ginsberg C, Beran M, Kurzrock R. Subcellular localization of Bcr, Abl, and Bcr-Abl proteins in normal and leukemic cells and correlation of expression with myeloid differentiation. J Clin Invest. 1993;92:1925–1939. doi: 10.1172/JCI116786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Witte ON. Modeling Philadelphia chromosome positive leukemias. Oncogene. 2001;20:5644–5659. doi: 10.1038/sj.onc.1204638. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rowley JD. Chronic myeloid leukemia: current perspectives. Clin Lab Med. 2011;31:687–698. x. doi: 10.1016/j.cll.2011.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.