Abstract
The structural integrity and conformational stability of an IgG1 monoclonal antibody (mAb), after partial and complete enzymatic removal of the N-linked Fc glycan, was compared to the untreated mAb over a wide range of temperature (10° to 90°C) and solution pH (3 to 8) using circular dichroism, fluorescence spectroscopy, and static light scattering combined with data visualization employing empirical phase diagrams (EPDs). Subtle to larger stability differences between the different glycoforms were observed. Improved detection of physical stability differences was then demonstrated over narrower pH range (4.0-6.0) using smaller temperature increments, especially when combined with an alternative data visualization method (radar plots). Differential scanning calorimetry and differential scanning fluorimetry were then utilized and also showed an improved ability to detect differences in mAb glycoform physical stability. Based on these results, a two-step methodology was used in which mAb glycoform conformational stability is first screened with a wide variety of instruments and environmental stresses, followed by a second evaluation with optimally sensitive experimental conditions, analytical techniques and data visualization methods. With this approach, high-throughput biophysical analysis to assess relatively subtle conformational stability differences in protein glycoforms is demonstrated.
Keywords: Glycosylation, Monoclonal Antibody, Stability, Structure, Conformation, Biophysical, Formulation
Introduction
Monoclonal antibodies (mAbs) have emerged as a key category of therapeutic protein drugs with over 30 mAbs currently approved in the USA and Europe and many hundreds under clinical development.1, 2 The commonly used IgG mAb includes two light chains and two heavy chains forming a homo-dimeric, multidomain structure containing an N-linked glycosylation site in each of the two CH2 domains found in the Fc portion of the heavy chain.3, 4 The glycosylation pattern of the Fc region of IgG molecules plays a key role in IgG functionality and clearance, where the type and amount of glycan moieties control the ability and affinity of the Fc region to bind to the various Fc receptors in vivo.5-11 These Fc receptors are responsible for Fc effector function activities and regulating clearance of IgGs from circulation in vivo.12-18
The extent and type of glycosylation has been shown to influence the conformational stability of proteins in general and mAbs in particular.19 There are several studies examining the effect of deglycosylation on the structure and stability of the Fc region of IgGs.20, 21 These studies typically use a single measurement type (e.g., differential scanning calorimetry) over limited solution conditions (e.g., one or two pH values) to examine the effect of varying mAb glycosylation patterns.22-26, 30 In addition, the protease sensitivity of an IgG (e.g., papain digestion), has been used to examine mAb stability, in which more cleavage has been noted when the Fc was deglycosylated.34, 35 Recent studies have also examined the conformational stability of purified Fc domains as function of varying glycosylation.20, 21, 27-30, 32 Interactions between the glycan moieties and specific residues within the CH2 domains are responsible for stabilizing the structure of the CH2 domain, and disruption of these non-covalent interactions by partial or full deglycosylation leads to destabilization of the entire domain.20, 21, 30-33 The effect of deglycosylation on the structural integrity of the CH2 domain has been examined by a variety of structural analysis including X-ray crystallography, 56 SAXS 40 and HDX-MS 54, 55 as well as examined by molecular modeling. 6, 57
The pharmaceutical properties (e.g., storage stability and solubility) of mAbs are also affected by glycosylation, although not necessarily in predictable ways. For example, the solubility of an IgG1 was increased dramatically after the introduction of an additional glycosylation site on the Fab domain.36 In contrast, an isolated cryoimmunoglobulin species from human serum, known to have dramatically reduced cold solubility, was shown to contain an additional glycosylation moiety in the variable region of the antibody.37 The aggregation propensity of IgGs may increase upon deglycosylation, which has been attributed to the destabilization of the CH2 domain as well as exposure of an aggregation-prone regions within the CH2 domain that are masked in the native IgG by the glycan moiety.28, 38, 39, 59, 60
Due to the potential for changes in critical quality attributes for biotech drugs as a result of manufacturing and/or formulation modifications, comparability studies are performed in which the pre and post-change drug candidates are evaluated to ensure that these process and product changes do not affect the drug's structure, safety and function.41-44 Structural equivalence between pre and post-change protein drug candidates is evaluated in a step-wise fashion which may include analytical, biological and clinical evaluations.41 The effect of varying glycosylation profiles on the design of comparability assessments of protein therapeutics, including effects on Fc effector function activity of mAbs, has recently been reviewed.42, 45
Analytical characterization for comparability evaluations includes determination of primary and higher-order structural integrity using a combination of methods.43, 46 A combination of chromatographic (SE, RP and IE- HPLC) and electrophoretic (cIEF, cSDS) methods are typically utilized along with mass spectrometry (intact molecular weight, peptide and oligosaccharides maps) to characterize protein's primary structure and post-translational modifications (e.g., glycosylation patterns). In contrast, analytical methods available to examine higher-order structure are typically lower resolution in nature (e.g., CD, fluorescence), ultimately resulting in the requirement for functional potency assays to be performed to ensure biological activity. Since accelerated or long term stability studies are typically monitored as part of comparability assessments, 43, 46 there is a need for new analytical methods for determining the higher-order structure of protein drug candidates that would provide a more comprehensive picture of their structural integrity and stability.
In the past decade, multiple papers have appeared from our laboratories describing the ability of empirical phase diagrams (EPDs) to summarize physical stability data and use this information for formulation development of therapeutic proteins and vaccines.47-52 These EPDs are created through the utilization of large data sets from high throughput analysis of a protein's conformational stability as a function of environmental stresses (e.g., temperature and pH) by multiple lower-resolution biophysical techniques (e.g., CD, fluorescence, light scattering, etc.), followed by the application of multidimensional mathematical analysis techniques, to produce a colored diagram representing structural changes as a function of environmental stress. Furthermore, the ability of EPDs to detect major conformational stability changes in a series of site-directed mutants of acidic fibroblast growth factor (FGF-1) was recently demonstrated.50 Recent work has also established additional data visualization methods such as radar charts, Chernoff faces and comparative signature diagrams to better compare protein samples.51, 53
The purpose of this study is to test the feasibility of using high through-put biophysical analysis and data visualization methods to rapidly evaluate, over a wide range of solution conditions, differences in the structural integrity and conformational stability of an IgG1 mAb of varying glycosylation patterns. This work required, for the first time, incorporation of differential scanning calorimetry and differential scanning fluorimetry data into the EPD analysis. We also included radar chart methods to better visualize conformational stability data differences across the IgG1 glycoforms.
Materials and Methods
Materials
An IgG1 mAb solution was received from Janssen R&D/J&J at 40 mg/ml and frozen in aliquots at −80° C. Reagent chemicals were purchased from Sigma–Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburg, PA).
Deglycosylatation of IgG1 mAb
Fully deglycosylated mAb was prepared using PNGase F from Prozyme™ (San Leandro, CA). Samples of mAb were diluted to 10 mg/ml with reaction buffer (100 mM Tris, 100 mM NaCl, pH 7.5), and then 20 μl (200 μg) of the diluted protein sample was added to 172 μl of the reaction buffer and 8 μl of PNGase F to achieve a 1:100 (w/w) enzyme: protein ratio. The mixture was incubated at 37 °C for 15 h. Partially deglycosylated IgG was produced using Endoglycosydase F2 (Endo F2) from Prozyme™ (San Leandro, CA). Samples of mAb were diluted to 10 mg/ml using deionized water. Twenty μl (200 μg) of the diluted protein sample was then added to 38 μl of deionized water, followed by addition of 10 μl of 5X reaction buffer (250 mM sodium acetate, pH 4.5), and 2 μl of Endo F2 to the mixture. The mixture was incubated for 1h at 37 °C.
Electrospray Ionization Mass Spectrometry (ESI-MS)
To confirm deglycosylation, antibody samples with and without reduction, using 10 mM dithiotreitol (DTT), were diluted to 0.25 mg/ml with 0.1% formic acid. Ten μL of this solution (approximately 16 pmoles of intact mAb or 50 pmoles of Fc monomer) was injected into the sample loop of the LC (Agilent Technologies 1200 Series) with a two pump system. An isocratic loading pump carried the sample from the loop to a Protein Concentration and Desalting Micro Trap (Bruker-Michrom, Auburn, CA, USA) for 5 minutes desalting with 0.1% formic acid at 200 μL/min. A gradient pump eluted the protein from the trap at 50 μL/min flow rate using a 5-60 % linear acetonitrile gradient in 3 min followed by a 60-95% gradient in 1 min for cleaning the trap onto the electrospray ionization (4 kV capillary) source of a time of flight mass spectrometer (model 6220, Agilent Technologies). The mobile phases used for gradient elution were 0.1% formic acid and 90% acetonitrile /10% water /0.1% formic acid. The mass spectrometer was operated in 2 GHz extended dynamic range mode with fragmentor voltage of 150V, desolvation gas flow rate of 10 L/min at 325 °C and nebulizer pressure of 20 psig. Mass spectra were acquired over an m/z range of 300-3200 with an acquisition rate of 1 spectrum per second with reference mass correction. The raw mass spectral data was processed using Agilent Mass Hunter Qualitative Analysis (version B.04). The mass spectra were deconvoluted at specific mass ranges to search for intact or reduced mAb species.
Sample Preparation for Stability Assessments
Samples were dialyzed overnight at 4° C using a 10 kDa molecular-weight cutoff membrane (Pierce, Rockford, IL) against 20 mM citrate-phosphate buffer to achieve the targeted pH range (3-8 with one pH unit increment or 3.5-6.0 with 0.5 pH increments). The ionic strength in all buffers was adjusted to 0.15 using NaCl. Protein concentration was determined at each pH using Agilent 8453 spectrophotometer (Palo Alto, California) and adjusted to 1 mg/ml. For physical measurements, two different sets of experimental parameters were evaluated: (1) for the pH range of 3-8, the temperature was raised from 10 °C to 90 °C at 2.5 °C intervals using a 180 sec equilibration time, or (2) for the pH range of 3.5-6.0, 1.25 °C intervals were used as a temperature ramp with a 30 sec equilibration time.
Biophysical Measurements
Far UV circular dichroism (CD), intrinsic tryptophan and extrinsic (using 1-analino-8-naphthalene ,ANS, dye) fluorescence spectroscopy, static light scattering and differential scanning calorimetry (DSC) studies were performed as described earlier.58 For more detailed description of these methods, please refer to the supplemental section.
DSF measurements were performed using MX3005P QPCR system (Agilent Technologies), with a protein concentration of 0.2 mg/ml and total sample volume of 100 μl. SYPRO™ orange purchased from Invitrogen, Inc. (Carlsbad, CA) supplied in a concentrated form (5000x) dissolved in DMSO. The dye was diluted to 40x and then added to the protein samples to achieve 1x dye concentration for measurements. Using FAM filter sets, the mixture was excited at 492 nm and the emission intensity change with temperature at 516 nm was followed. Temperature was raised from 25 °C to 90 °C using 60 °C/hr as a heating rate and 1 °C as a step size. Data were transferred to Excel software (Microsoft, Redmond, WA) for data analysis. A pH range of 4-6 with 0.5 pH unit increment was used, and buffers were run and subtracted from all samples. DSF curves were integrated, before being utilized in the construction of the EPD, to get a sigmoidal curve, in a step to get a better representation of the data in the EPD.
Construction of Empirical Phase Diagrams (EPDs) and Radar Charts
EPDs are constructed to summarize and visualize the conformational stability of IgG1 mAbs using data sets from selected experimental techniques as a function of pH and temperature. The experimental measurements are organized in the form of a multi-dimensional vector matrix and analyzed by Singular value decomposition (SVD) as described in detail elsewhere.51, 52 Results are mapped to a RGB color scheme and visualized as changes in color which indicate changes in the physical states of the protein. Additional data visualization scheme with radar charts was also used to analyze the data as described in detail elsewhere.51 A radar chart can have any number of polar axes each of which is mapped to an experimental technique. A polygon drawn by connecting all points in the polar axes represents changes in the physical states of the protein. Similar to an EPD, difference in the shapes of polygons indicate changes in the conformational state of a protein. Apparent boundaries between states can be assessed visually or by use of computational aid such as k-Means clustering. A detailed explanation of radar charts and clustering methods can be found elsewhere.50
Results
Deglycosylation of the IgG1 mAb
The IgG1 mAb used in this study had a glycosylation pattern typically observed for recombinant mAbs produced from SP2/0 cells with ~75% consisting of G1F, G2F and G0F structures, and the remaining ~25% being spread over various charged species (e.g., mono and disialylated glycoforms) as described in detail elsewhere.43 The ability of the different enzymes to remove the two major glycans (G0F, G1F) was monitored by mass spectrometry analysis of the heavy chain Fc from reduced mAb samples (Table 1). Mass spectrometric analysis of the reduced mAb samples showed no significant changes to the light chain (data not shown) while the heavy chain manifested a molecular weight consistent with the specific enzymatic treatments (Table 1). For example, partial deglycosylation was achieved by the treatment with endoglycosydase F2 (Endo F2) which cleaves between two GlcNAc residues and leaves Asn 297 in the protein backbone attached to GlcNAc-Fuc. Full deglycosylation was achieved using N-Glycanase (PNGase F) which fully removes the glycan (see Table 1) and deamidates the Asn to Asp, adding a negative charge to both CH2 domains. Additional confirmation was obtained by SDS-PAGE and capillary isoelectric focusing analysis which qualitatively showed the expected shifts in migration of fully deglycosylated mAb in terms of molecular weight and charge heterogeneity due to glycan removal (data not shown).
Table 1.
Mass spectrometry results of the heavy chain region of reduced samples of the IgG1 mAb. First column shows the average masses of the PNGase F and Endo F2 treated IgG1 mAb obtained from MS analysis along with the masses measured for the two most abundant glycoforms (G0F and G1F) of the untreated mAb. The second, third and fourth columns show observed change in mass, predicted change in mass, and modification expected due to enzymatic treatment of the IgG1 mAb, respectively.
| IgG1 Glycoform | Heavy Chain (Da) | Δ mass observed (Da) | Δ mass predicted (Da) | Modification of Asn 297 |
|---|---|---|---|---|
| PNGase F treated | 49168.3 * | - | - | Deglycosylation, N → D |
| Endo F2 treated | 49516.8 | +348.5 | +348.3 | +GlcNAc-Fucose |
| Untreated: ** | ||||
| G0F glycosylated | 50612.5 | +1445 | +1444.6 | +G0F |
| G1F glycosylated | 50774.7 | +1607 | +1606.5 | +G1F |
The theoretical mass of the aglycosylated mAb heavy chain is 49167.3 Da.
G0F, G1F are two main peaks used to follow deglycosylation (Supplemental figure 4)
Abbreviations: N: Asparagine, D: Aspartic acid, GlcNAc: N-acetylglucosamine, F: Fucose, G: Galactose, G0F: GlcNAc2Man3GlcNAc2Fuc, G1F: GalGlcNAc2Man3GlcNAc2Fuc.
Initial biophysical characterization of a native and fully deglycosylated IgG1
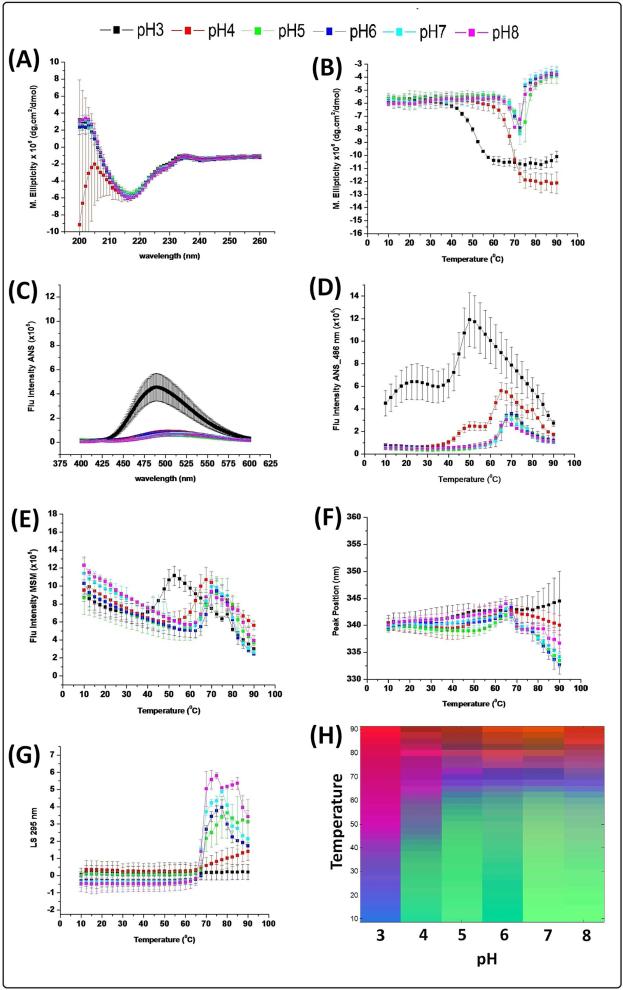
The secondary structure of the native and fully deglycosylated IgG1 mAb was evaluated by far-UV CD analysis from 260 nm to 200 nm at 10 °C (Figure 1A, 2A). Both samples share the same structural features across all pH values in the form of a broad negative peak at 217 nm, indicating the expected β-sheet rich structure. By following the CD intensity change at 217 nm with increasing temperature (Figures 1B, 2B), the two IgG1 glycoforms show similarity in their secondary structure stability behavior at higher pH values (pH 6-8) with an onset temperature of ~60 °C. A lower pH, however, (pH range 3-5), the native IgG1 shows a trend toward enhanced stability compared to its deglysosylated form, with ~1 °C difference in the onset temperature.
Figure 1.
Biophysical characterization of the untreated (control) IgG1 mAb as a function of temperature and pH. (A) CD spectra at 10 °C, (B) CD intensity change at 217 nm with temperature, (C) ANS spectra at 10 °C, (D) ANS melting curve at 486 nm, (E) Fluorescence intensity vs. temperature, (F) Fluorescence peak position changes with temperatures, (G) Static light scattering intensity change with temperature, and (H) empirical phase diagram analysis of data. Data shown for n=3 measurements.
Figure 2.
Biophysical characterization of PNGase treated, fully deglycosylated IgG1 mAb as a function of temperature and pH. (A) CD spectra at 10 °C, (B) CD intensity change at 217 nm with temperature, (C) ANS spectra at 10 °C, (D) ANS melting curve at 486 nm, (E) Fluorescence intensity vs. temperature, (F) Fluorescence peak position changes with temperatures, (G) Static light scattering intensity change with temperature, and (H) empirical phase diagram analysis of data. Data shown for n=3 measurements.
At 10 °C, the native IgG1 produced a higher ANS intensity at pH 3 than other pH values, suggesting increased exposure of apolar regions at low pH (Figure 1C). For the fully deglycosyalted IgG1, higher ANS intensity was observed at all pH values compared to native IgG1 (Figure 2C). Thermal stability was studied by following the ANS intensity change at 486 nm with increasing temperature as shown in Figures 1D and 2D. The native and deglycosylated forms of the mAb show a similar transition at high pH values (pH 6-8) with one, major structural transition observed starting at ~60 °C. Differences between the two IgG1 forms are observed, however, in more acidic environments (pH 3-5), including an additional structural transition at lower temperatures. At pH 3, the first transition begins at approximately 10 °C for the native IgG1, while the protein is already structurally altered at 10 °C for the fully deglycosylated form, suggesting that the fully deglycosylated IgG1 is experiencing a higher degree structural disruption at pH 3. In contrast, the second transition starts at ~35 °C for both samples. Two structural transitions are seen at pH 4 for both IgG1s, with a similar onset temperature for the second transition (~57 °C), but a different onset temperatures in the first transition (~35 °C for the native IgG vs. ~25 °C for the fully deglycosylated IgG). The native IgG1 at pH 5 shows only one structural transition starting at ~60 °C while the deglycosylated form manifest two transitions at ~42 and ~60 °C.
Intrinsic (Trp) fluorescence spectroscopy was used to probe the overall tertiary structure stability of the native and fully deglycosylated mAb. Comparing the two spectra at 10 °C, no major differences were observed at all pH values (data not shown). By following the intensity change with increasing temperature for the IgG1 in the native (Figure 1E) and fully deglycosylated states (Figure 2E), similar structural transitions are seen including an initial decrease in fluorescence intensity (due to the intrinsic temperature dependent decrease in quantum yield of the tryptophan's indol ring), a second sudden increase in fluorescence intensity (indicating the start of a tertiary structure unfolding event), and a third transition marked by a sudden decrease in fluorescence intensity consistent with aggregation/precipitation. The major unfolding event (second transition) for both IgG1 forms starts earlier at pH 3 (~38 °C) and pH 4 (~55 °C) compared to other pH values. Approximate 2-3 °C stability difference for the IgG1 in the native state (~62.5 °C) was observed for pH (5-8) compared to the fully deglycosylated IgG (~60 °C). Peak position changes as a function of increasing temperature were also analyzed. The peak position for the native IgG1 (Figure 1F) was 340 nm (shifted from an actual value of 330 nm due to the use of “spectral central of mass” method) for all pH values at 10°C representative of native/folded tertiary structure. The peak position gradually increased with increasing temperature followed by a decrease at around 65 °C (except at pH 3) suggesting aggregation/precipitation. The fully deglycosylated IgG1 (Figure 2F) at 10 °C was in a partially unfolded state as indicated by the red shifted peak position (345 nm at pH 3 and 342 nm for the pH range 4-8 using MSM analysis). With increasing temperature, both proteins showed a gradual increase in their peak position as a result of gradual unfolding followed by aggregation/ precipitation and concomitant loss of signal.
Figures 1G and 2G show the temperature induced aggregation behavior of the native and fully deglycosylated IgG1, respectively, as measured by static light scattering. Comparing the two IgG1 forms, pH 3 shows a similar trend with no increase in light scattering intensity even up to 90 °C, whereas, the native and deglycosylated IgG1 at pH 4 starts aggregating at 64.5 °C and 62 °C, respectively, with a greater intensity upon aggregation for the deglycosylated form. At pH 5 and 6, native IgG1 starts aggregating at ~62.5 °C and ~65 °C compared to ~60 °C and ~62.5 °C for the deglycosylated IgG1, implying a destabilizing influence of removal of the sugar moiety.
The thermal melting curves from CD, ANS, fluorescence peak intensity, fluorescence peak position, and SLS analysis were used to generate EPDs for the native (Figure 1H) and fully deglycosylated mAbs (Figure 2H). Comparing the two EPDs, three major regions are identified that signify different conformational states. The green region represents the IgG1 in its stable native state and the blue region in a structurally perturbed, partially unfolded state. The purple/red region represents an aggregated state. Both IgG1 forms have similar transition temperatures in the native to the unfolded state events (blue region) in the pH range of 6-8, suggesting no detectable differences in conformational stability between the native and fully deglycosylated IgGs in this pH range. In contrast, notable differences in conformational stability between the two IgG1 forms were seen under more acidic conditions of pH 3-5. Comparing the native (green) region of the EPDs at pH 5 for the two IgG1 forms, a large stability difference is observed, in which the native IgG1 starts transitioning to the unfolded state at ~57.5 °C and the fully deglycosylated form at ~47.5 °C. At pH 4, conformational stability differences are even more dramatic, with transitions to the unfolded state appearing at ~47.5 °C and ~35 °C for the native and fully deglycosylated IgG1, respectively. For both IgG1 forms at pH 3, the structurally perturbed state initiates at 10 °C with this unfolded state undergoing aggregation at ~50 °C.
Optimization of experimental parameters to better compare conformational stability of Native and Fully Deglycosylated mAb glycoforms
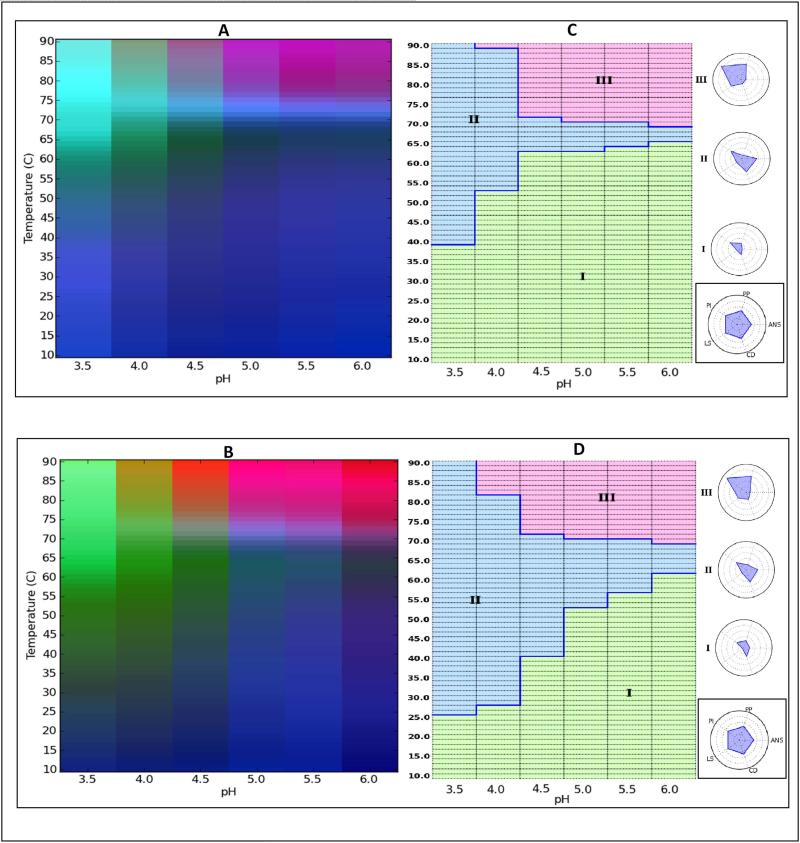
Based on the studies above, EPD data analysis permitted a rapid, visual assessment of conformational thermal stability differences between the native and fully deglycosylated IgG1 in the pH range of 3 to 5. Given the size of the temperature steps (2.5 °C) and pH steps (one pH unit), however, it was difficult to analyze these stability differences in more detail. Thus, a narrower pH range (3.5 to 6 in 0.5 pH increments) with smaller temperature increments (1.25°C) was used to generate a new set of EPDs for both IgG1 mAb forms. Using the same techniques, a more detailed EPD was generated for native protein (Figure 3A) and deglycosylated protein (Figure 3C). The data from these experiments are provided in Supplementary Figures S1 and S2. These improved EPDs resulted in observation of four structural regions including a blue region, where both IgG1 forms are in their stable-native like states, a dark black region, immediately above the blue region probably representing the IgG's in a molten globule-like state. The third region (green) represents a highly structurally altered, extensively unfolded state, while the purple/red region comprising protein that is aggregating and/or precipitating. Comparing the blue regions of the two IgG1 forms to each other, we see this region covers most of the native state, but covers a much smaller area in the fully deglycosylated form. This more clearly illustrates conformational stability differences between the two IgG1 forms. For example, in the pH range of 3.5-4.5, a noticeable difference in the structural transition temperatures from the native to the molten globular and unfolded states are observed. Transition temperature differences were less substantial in the pH range of 5-6 in the EPD with an ~2-3 °C stability difference between the two IgG1 forms.
Figure 3.
Empirical Phase Diagram (EPD) and Radar Chart analysis of the conformational stability of IgG1 mAb samples. (A) EPD of untreated (control) IgG1, (C) radar chart of untreated (control) IgG1, (B) EPD of fully deglycosylated IgG1, and (D) radar chart of fully deglycosylated IgG1. The temperature ramp was in 1.25°C increments.
We used the same data set to generate radar charts, a newly developed data visualization method for protein biophysical data.51 Radar charts for the native and fully deglycosylated IgG1 mAb forms are shown in Figures 3C and 3D, respectively. The reference radar chart guide to the right of the figure shows the position of the five analytical techniques that are being evaluated. Consequently, five radii are projecting out from the center of the chart creating one larger pentagonal image. According to the clustering analysis (K=3), for both IgG forms, the radar charts are divided into three regions, regions I, II, and III. These three regions are similar to the three regions describe above with EPD analysis (i.e., native-like state, structurally altered state, and more extensively altered form with aggregation). Region I is of small quadrilateral shape and corresponds to minimal change in the signals, thus representing the more stable-native like state. Region I covers 60% (Figure 3C) vs. 42% (Figure 3D) of the total area of the radar charts for the untreated and fully deglycosylated IgG mAbs, respectively.
Conformational Stability of Three Different IgG1 mAb Glycoforms Analyzed with Optimized Analytical and Data Visualization Methods
As a final set of experiments, three forms of the IgG1 mAb of varying glyscoylation were analyzed: the untreated control, the fully deglycoyslated PNGase F treated protein, and a partially deglycosylated form generated by treatment with Endo F2 (see Table 1). In addition, as noted above, since various biophysical techniques (Figures 1-3) differed in their ability to detect the destabilizing effect of deglycosylation, we selected the most sensitive methods described above (extrinsic fluorescence spectroscopy with ANS and static light scattering) for the additional experiments described below. In addition, we also employed differential scanning calorimetry (DSC) and differential scanning fluorimetry (DSF) which have previously been shown to be capable of detecting conformational stability differences in deglycosylated IgG1 mAbs.21, 24, 27, 28, 30-33 Finally, the pH range of analysis was also narrowed to pH 4.0-6.0 to better focus on the structural transition regions and due to complex, irreproducible behavior of DSC and DSF results when heated at pH 3.5 and below (data not shown). The ANS, DSC and DSF data are discussed below and the SLS data for the three mAb samples are provided in Supplementary Figure S3.
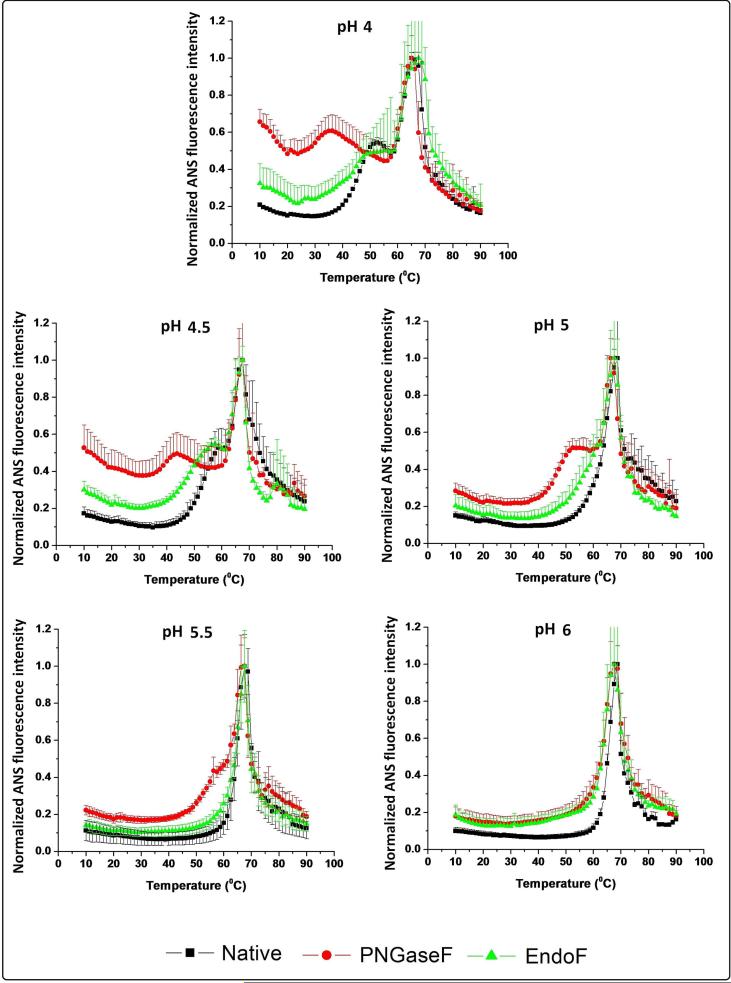
The thermal melting curves for the native (black), partially (green), and fully deglycosylated (red) mAbs in the presence of ANS are shown in Figure 4. At 10 °C, at all pH values tested, the fully deglycosylated IgG1 has the highest ANS intensity value, followed by the partially deglycosylated IgG1. The ANS intensity differences between the three IgG1 glycoforms at 10 °C decreases as the pH increases. As the temperature is increased, two transitions are evident for the three IgG glycoforms at lower pH with only a single transition observed at higher pH values. The loss of the first transition occurs at pH 5.0, 5.5, and 6.0 for the native, partially deglycosylated and the fully deglycosylated IgG1 mAbs, respectively. The second thermal transition temperature seems to be less affected by the glycosylation state of the mAbs, since the three samples display similar transition temperatures (~68°C at pH 4.5-6.0 and ~65°C at pH 4.0).
Figure 4.
Normalized ANS fluorescence intensity change as a function of temperature in the presence of untreated (control) IgG1 (black line), partially deglycosylated IgG1 (green line), and fully deglycosylated IgG1 (red line) from pH 4 to 6. Normalized results were generated by fitting the data to be equal to 1 at the maxima and to 0 at the minima for incorporation into the EPDs and radar charts. Curves shown here are averages of three runs.
Figure 5 shows DSC thermograms of the three IgG1s at pH 4-6. The second and third endothermic peaks observed by DSC are very similar in terms of melting temperatures (74 °C and 82.5 °C, respectively). The first endothermic peak onset temperature, however, shows both a solution pH and glycosylation pattern dependence. In the pH range of 4 to 5, the transition onset temperature initiates earlier in the case of the deglycosylated IgG, followed by the partially deglycosylated and then the native IgG1. The latter two samples show similarity in their onset temperature, except at pH 4.5 where the partially deglycosylated IgG1 seems to be less stable than the native molecule. At pH 5, the first peak starts merging with the second peak for both the native and partially deglycosylated IgGs, ultimately forming a shoulder. The first peak for the fully deglycosylated mAb, however, remains completely separated from the second. At higher pH values of 5.5 and 6, the first transition for both the native and the partially deglycosylated IgG1 forms are completely merged into the second peak, while the first transition in the fully deglycosylated IgG form remains separated from the other thermal transition peaks at pH 5.5.
Figure 5.
Differential scanning calorimetry (DSC) analysis of untreated (control) IgG1 (black line), partially deglycosylated IgG1 (green line), and fully deglycosylated IgG1 (red line) from pH 4 to 6. Normalized heat capacity changes were generated by fitting the data to be equal to 1 at the maxima and to 0 at the minima for incorporation into the EPDs and radar charts. Curves shown here are averages of three runs.
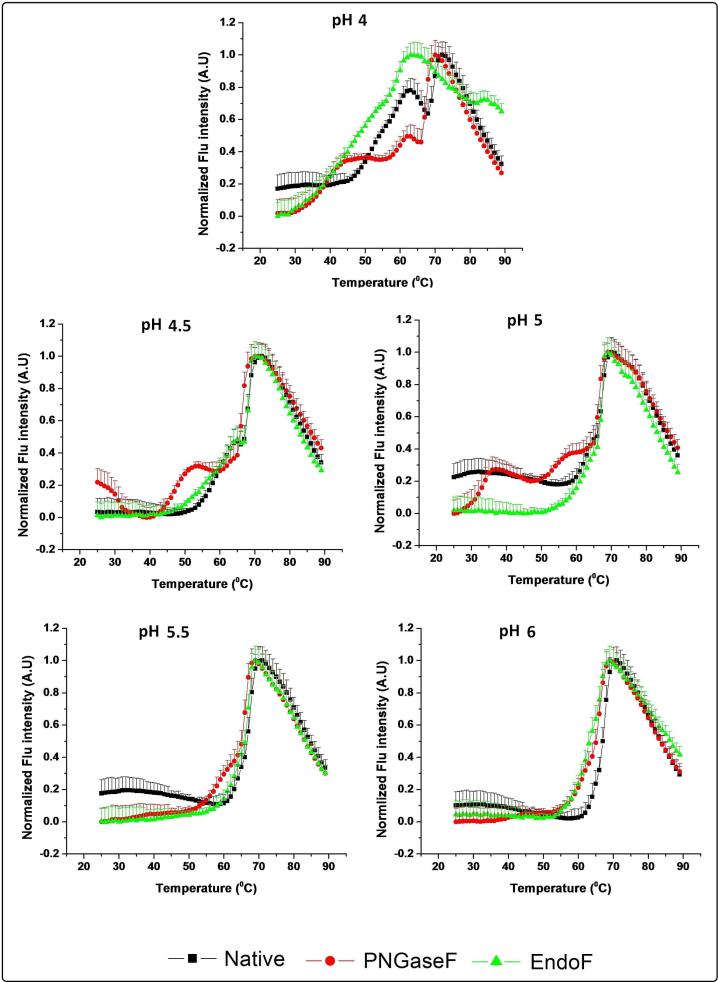
Using SYPRO orange as an extrinsic dye which shows increased fluorescence upon exposure to more apolar environments such as those associated with structural alterations in proteins, the thermal stability of the different IgG1 samples as a function of pH and temperature was followed (Figure 6). Among the three IgG1 mAb samples and pH values examined, a major transition at 70 °C was consistently observed except for the partially deglycosylated IgG1 at pH 4, in which the observed transition is very broad and is seen at a lower temperature (62 °C). Multiple additional transitions (prior to the main transition) are evident for the three IgG1 samples at pH 4.0. A destabilization effect due to carbohydrate removal is evident since both the partially and fully deglycosylated IgG1 forms start unfolding at ~28 °C compared to the native IgG at ~46 °C. At pH 4.5 and 5.0, an initial transition is noted at a lower temperature for the partially deglycosylated compared to the native IgG1 although they peak at about the same temperature. The presence of the multiple transitions for the fully deglycosylated IgG1 at pH 4.5 and 5.0, indicates more structurally disrupted states than the other mAb samples at lower temperatures. At pH 5.5 and 6.0, two of the IgG1 forms show a single major structural transition, with the fully deglycosylated form also manifesting a pre-transition at pH 5.5.
Figure 6.
Differential scanning fluorimetry (DSF) analysis of untreated (control) IgG1 (black line), the partially deglycosylated IgG (green line), and the fully deglycosylated IgG1 (red line) from pH 4 to 6. Normalized results were generated by fitting the data to be equal to 1 at the maxima and to 0 at the minima for incorporation into the EPDs and radar charts. Curves shown here are averages of three runs.
Using the results obtained from the four techniques described above, EPDs and radar charts were generated for the three IgG1 samples (Figure 7). Comparing the EPDs of the native (Figure 7A), the partially deglycosylated (Figure 7B), and the fully deglycosylated proteins (Figure 7C), the three EPDs share a common blue and green region. The blue region (based on separate evaluation of the data) signifies the region where the protein is in its stable, native-like state. The green color defines the region where the protein is in a structurally altered state. A third region appears in all the three EPDs, but with different colors and different intensities, corresponds to aggregation/precipitation of the protein. Comparing the blue (native structure) region across the three IgG1 glycoforms at pH 5.5 and 6.0, the native and the partially deglycosylated IgG1 samples have the same transition temperatures near 64 °C. The fully deglycosylated IgG, however, starts its transition at lower a temperature (59 °C). At pH 5.0, the native and the partially deglycosylated IgG forms show a transition at 63 °C and 61 °C, respectively. For the fully deglycosylated IgG at pH 5.0, additional color elements are observed indicating the existence of a partially unfolded, conformationally disrupted state at this pH (see the ANS and DSF data in Figures 4 and 6, respectively). At pH 4.0 and 4.5, the native IgG1 is less stable, with structural transitions at 50 and 55 °C, respectively. The partially deglycosylated IgG1 at pH 4 and 4.5 develops an additional light blue region, consistent with the existence of a partially unfolded state. (See the ANS data in Figure 4). The fully deglycosylated IgG at pH 4.5 shows a more structurally disrupted state compared to the other IgG forms, while an additional green region is observed for the fully deglycosylated IgG1 form at pH 4.0, indicating the existence of the extensively structurally altered state even at lower temperatures.
Figure 7.
Empirical Phase Diagram (EPD) and Radar Chart analysis of the conformational stability of IgG1 mAb samples. Panel A shows the EPD (left) and the radar chart (right) for the untreated (control) IgG1. Panel B shows the EPD (left) and the radar chart (right) for the partially deglycosylated mAb due to Endo F2 treatment. Panel C shows the EPD (left) and the radar chart (right) for the fully deglycosylated mAb due to PNGase F treatment.
Radar charts were generated from the same data as shown on the right side of Figure 7. Spanning the same pH and temperature range examined with colored EPDs, similar regions were identified for the different IgG1 glycoforms indicating the existence of different conformational states (in the radar plots, structural transitions were identified by clustering analysis with k=3 as described in the methods section). The native-like, stable region for the untreated, control mAb (Figure 7A), the partially deglycosylated mab (Figure 7B), and the fully deglycosylated protein (Figure 7C) are depicted as dot-like entities (minimal structural transitions) and occupy 67, 52 and 40% of the radar chart's total area, respectively. Based on this simple analysis of the areas in the radar plots representative of native-like, stable form of the mAb, the effect of glycosylation on IgG1 conformational stability is readily evident, with the decreased area qualitatively proportional to the enzymatic truncation of sugar moieties.
Discussion
In this study, a direct comparison of the structure and conformational stability of an IgG1 mAb and its truncated glycoforms was performed with a variety of biophysical techniques, over a wide range of solution conditions. Data was also analyzed with two vector based technologies. The different glycoforms were prepared by enzymatic treatments. Mass spectrometric analysis of the reduced IgG1 mAbs (Table 1) directly demonstrated that the partial and full deglycosylation was successful since the measured molecular weights of the heavy chain of the mAb glycoforms were in close proximity to the expected values. The native mAb contained the expected mixture of glycan structures for SP20 cells, in which the G2F/G1F, G1F/G1F and G2F/G0F glycosylation patterns are the dominant glycosylated forms as described elsewhere.43 The partially deglycosylated mAb, produced by enzymatic treatment with Endo F2, contained a Fucose-GlcNac as a major glycan species structures consistent with the theoretical (average) mass. The presence of the fully deglycosylated protein, achieved by digesting with PNGase F, was evident from MS analysis (Table 1), where complete removal of the glycan and subsequent deamidation of the backbone Asn to Asp, was achieved as a result of the enzymatic reaction.
The destabilizing effects of removing the N-linked glycan in the CH2 domain of the Fc region of IgGs has previously been demonstrated by selected techniques under certain conditions. In this regard, the ability of EPDs and radar charts to detect changes in conformational stability due to differences in post-translational modifications of a protein was examined in this work in a “model system” using three different glycoforms of an IgG1 mAb. Although this work covered a wider range of solution conditions, experimental techniques and data visualization approaches, some of the conditions examined here overlap with previous studies. The results with the IgG1 mAb of this work are consistent with previous reports with other IgG1 antibodies. For example, as a result of complete or partial removal of the CH2 domain N linked glycan, destabilization of the CH2 domain of an IgG1 has been shown by DSC. 22, 26, 30, 31, 33 In addition, an increase in fluorescence intensity spectra using intrinsic (Trp) and extrinsic (SYPRO orange dye) fluorescence spectroscopy was seen with full deglycosylation.33 Differential scanning fluorimetry (DSF) has been used to evaluate conformational stability differences of an IgG1 in different formulation buffers.23 The effect upon complete or partial removal of the N-linked glycan on the conformational stability of purified Fc proteins has also recently been evaluated using DSC,21, 27, 28 as well as CD and extrinsic (ANS) fluorescence spectroscopy.27 Results from DSC indicated a destabilization effect on the CH2 domain of the Fc protein upon complete or partial deglycosylation, while results from CD and extrinsic fluorescence indicated a significant destabilization effect under acidic conditions and less of an effect at neutral to basic pH.
The results of this work demonstrate a two-step methodology to evaluate differences in the conformational stability between several mAb glycoforms. First, “standard” EPD data analysis was used to screen mAb conformational stability using a variety of methods that probe different aspects of structure over a wide range of an environmental stresses (e.g., temperature and pH). Second, based on these initial results, an additional evaluation was conducted using the most sensitive experimental methods, with a focus on a narrower range of environmental conditions (near values associated with structural changes in the protein), using a combination of EPDs and radar charts for data visualization. For example, when comparing the untreated to the fully deglycosylated IgG1, a noticeable destabilization effect was first evaluated over a wide range of pH and temperature conditions by a variety of biophysical techniques (Figures 1 and 2). This initial evaluation showed the destabilization effects under mildly acidic conditions with some variability between the different analytical techniques in terms of their ability to detect structural changes as a result of deglycosylation. Based on these results, data were collected under “zoomed-in” conditions focusing on an optimal pH range (4.0-6.0 in 0.5 increments), where notable structural changes are detected, combined with 1.25 °C temperature steps to improve resolution (over the initially used 2.5 °C increments). We also implemented the most sensitive analytical methods including ANS fluorescence spectroscopy, DSC, DSF and light scattering (LS). In addition, we added a partially deglycosylated mAb glycoform to better assess the sensitivity of this methodology. The “zoomed-in” EPDs and radar charts from these three IgG1 glycoforms (Figure 7) clearly reflect differences in the major structural variations between the three samples. Radar charts have the advantage of reflecting the individual technique(s) which reflect the structural change at an identifiable site in the diagram. For the three mAb glycoforms studied, radar charts clearly point to an ANS-DSF influence in detected structural changes associated with region II, with LS becoming a major tool for monitoring aggregation (Region III) at higher temperatures.
Although this work demonstrates the ability to compare conformational stability trends across mAb samples of known glycosylation content, the generation of EPDs and Radar charts for more formal evaluations such as regulatory comparability studies currently has practical limitations. For example, although visualization and rank ordering of structural transitions between samples is clearly useful for semi-qualitative comparisons of conformational stability, it still requires expert analysis by individual scientists of the individual biophysical data sets to confirm (and if necessary adjust) readouts from the initial clustering analysis. This is due to a combination of potential effects including noise in raw data, propagation of error when combining data sets across different instruments and experiments, and limitations of clustering analysis. 51 Ongoing work in our laboratory is evaluating and addressing these topics including more advanced mathematical treatments to potentially allow for statistical comparisons and independence from a separate expert review. For example, comparative signature diagrams are an alternative approach recently described 53 to statistically compare differences in spectral readouts from different instruments when evaluating two protein samples across different temperatures and pH values.
In summary, this work combines data sets from multiple biophysical techniques that monitor different aspects of a protein's higher-order structural stability as function of environmental stress (e.g., secondary structure by CD, tertiary structure by fluorescence spectroscopy, quaternary structure and aggregation by light scattering). Data visualization by EPDs and Radar charts allows for convenient and rapid analysis of these large biophysical stability data sets. We also incorporated conformational stability data from DSC and DSF in EPD and Radar charts construction, for the first time, due to the sensitivity of these two methods in detecting more subtle structural stability differences between the different mAb glycoforms across different solution conditions. By assessing conformational stability as a function of environmental stress (pH, temperature), subtle differences in structural integrity may potentially be detected when these differences are not readily apparent when monitored using lower resolution methods under non-stressed conditions (analysis at low temperature at neutral pH). Thus, evaluation of conformational stability differences may not only be an effective surrogate to monitor subtle differences in higher order structure between protein samples as part of formulation development, but also a useful complement to traditional accelerated stability data often used in analytical comparability studies.
Supplementary Material
Acknowledgments
The authors wish to thank and acknowledge Janssen R&D for providing an IgG1 mAb for this study, King Saud University for the financial support of M.A. Alsenaidy, as well as the financial support for TT from NIH grant NIGMS RO1 GM090080.
References
- 1.Reichert JM. Marketed therapeutic antibodies compendium. mAbs. 2012;4(3):413–415. doi: 10.4161/mabs.19931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aaron L. Nelson, E. D., Janice M. Reichert. Development trends for human monoclonal antibody therapeutics. Nature Reviews Drug Discovery. 2010;9:767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 3.Jefferis R. The antibody paradigm: present and future development as a scaffold for biopharmaceutical drugs. Biotechnology and Genetic Engineering Reviews. 2009;26(1):1–41. doi: 10.5661/bger-26-1. [DOI] [PubMed] [Google Scholar]
- 4.Wang W, Singh S, Zeng DL, King K, Nema S. Antibody structure, instability, and formulation. Journal of Pharmaceutical Sciences. 2007;96(1):1–26. doi: 10.1002/jps.20727. [DOI] [PubMed] [Google Scholar]
- 5.Jefferis R. Isotype and glycoform selection for antibody therapeutics. Archives of Biochemistry and Biophysics. 2012;526(2):159–166. doi: 10.1016/j.abb.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The Impact of Glycosylation on the Biological Function and Structure of Human Immunoglobulins. Annual Review of Immunology. 2007;25(1):21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 7.Kanda Y, Yamada T, Mori K, Okazaki A, Inoue M, Kitajima-Miyama K, Kuni-Kamochi R, Nakano R, Yano K, Kakita S, Shitara K, Satoh M. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: the high-mannose, hybrid, and complex types. Glycobiology. 2007;17(1):104–118. doi: 10.1093/glycob/cwl057. [DOI] [PubMed] [Google Scholar]
- 8.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8(3):226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 9.Jefferis R, Lund J. Interaction sites on human IgG-Fc for FcγR: current models. Immunology Letters. 2002;82(1–2):57–65. doi: 10.1016/s0165-2478(02)00019-6. [DOI] [PubMed] [Google Scholar]
- 10.Boyd PN, Lines AC, Patel AK. The effect of the removal of sialic acid, galactose and total carbohydrate on the functional activity of Campath-1H. Molecular Immunology. 1995;32(17-18):1311–1318. doi: 10.1016/0161-5890(95)00118-2. [DOI] [PubMed] [Google Scholar]
- 11.Radaev S, Sun PD. Recognition of IgG by Fcγ Receptor. Journal of Biological Chemistry. 2001;276(19):16478–16483. doi: 10.1074/jbc.M100351200. [DOI] [PubMed] [Google Scholar]
- 12.Yu M, Brown D, Reed C, Chung S, Lutman J, Stefanich E, Wong A, Stephan J-P, Bayer R. Production, characterization and pharmacokinetic properties of antibodies with N-linked Mannose-5 glycans. mAbs. 2012;4(4):475–487. doi: 10.4161/mabs.20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Putnam WS, Prabhu S, Zheng Y, Subramanyam M, Wang Y-MC. Pharmacokinetic, pharmacodynamic and immunogenicity comparability assessment strategies for monoclonal antibodies. Trends in Biotechnology. 2010;28(10):509–516. doi: 10.1016/j.tibtech.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Stadheim A, Hamuro L, Pittman T, Wang W, Zha D, Hochman J, Prueksaritanont T. Pharmacokinetics of IgG1 monoclonal antibodies produced in humanized Pichia pastoris with specific glycoforms: A comparative study with CHO produced materials. Biologicals. 2011;39(4):205–210. doi: 10.1016/j.biologicals.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Alessandri L, Ouellette D, Acquah A, Rieser M, LeBlond D, Saltarelli M, Radziejewski C, Fujimori T, Correia I. Increased serum clearance of oligomannose species present on a human IgG1 molecule. mAbs. 2012;4(4):509–520. doi: 10.4161/mabs.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goetze AM, Liu YD, Zhang Z, Shah B, Lee E, Bondarenko PV, Flynn GC. High-mannose glycans on the Fc region of therapeutic IgG antibodies increase serum clearance in humans. Glycobiology. 2011;21(7):949–959. doi: 10.1093/glycob/cwr027. [DOI] [PubMed] [Google Scholar]
- 17.Bumbaca D, Boswell C, Fielder P, Khawli L. Physiochemical and Biochemical Factors Influencing the Pharmacokinetics of Antibody Therapeutics. The AAPS Journal. 2012;14(3):554–558. doi: 10.1208/s12248-012-9369-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Liu YD, Flynn GC. The effect of Fc glycan forms on human IgG2 antibody clearance in humans. Glycobiology. 2009;19(3):240–249. doi: 10.1093/glycob/cwn120. [DOI] [PubMed] [Google Scholar]
- 19.Solá RJ, Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. Journal of Pharmaceutical Sciences. 2009;98(4):1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural Analysis of Human IgG-Fc Glycoforms Reveals a Correlation Between Glycosylation and Structural Integrity. Journal of Molecular Biology. 2003;325(5):979–989. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 21.Mimura Y, Church S, Ghirlando R, Ashton PR, Dong S, Goodall M, Lund J, Jefferis R. The influence of glycosylation on the thermal stability and effector function expression of human IgG1-Fc: properties of a series of truncated glycoforms. Molecular Immunology. 2000;37(12-13):697–706. doi: 10.1016/s0161-5890(00)00105-x. [DOI] [PubMed] [Google Scholar]
- 22.Ha S, Ou Y, Vlasak J, Li Y, Wang S, Vo K, Du Y, Mach A, Fang Y, Zhang N. Isolation and characterization of IgG1 with asymmetrical Fc glycosylation. Glycobiology. 2011;21(8):1087–1096. doi: 10.1093/glycob/cwr047. [DOI] [PubMed] [Google Scholar]
- 23.He F, Hogan S, Latypov RF, Narhi LO, Razinkov VI. High throughput thermostability screening of monoclonal antibody formulations. Journal of Pharmaceutical Sciences. 2010;99(4):1707–1720. doi: 10.1002/jps.21955. [DOI] [PubMed] [Google Scholar]
- 24.Wen J, Jiang Y, Narhi L. Effect of carbohydrate on thermal stability of antibodies. Am Pharm Rev. 2008;11:98–104. [Google Scholar]
- 25.Ha S, Wang Y, Rustandi RR. Biochemical and biophysical characterization of humanized IgG1 produced in Pichia pastoris. mAbs. 2011;3(5):453–460. doi: 10.4161/mabs.3.5.16891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hari SB, Lau H, Razinkov VI, Chen S, Latypov RF. Acid-Induced Aggregation of Human Monoclonal IgG1 and IgG2: Molecular Mechanism and the Effect of Solution Composition. Biochemistry. 2010;49(43):9328–9338. doi: 10.1021/bi100841u. [DOI] [PubMed] [Google Scholar]
- 27.Li CH, Narhi LO, Wen J, Dimitrova M, Wen Z.-q., Li J, Pollastrini J, Nguyen X, Tsuruda T, Jiang Y. Effect of pH, Temperature, and Salt on the Stability of Escherichia coli- and Chinese Hamster Ovary Cell-Derived IgG1 Fc. Biochemistry. 2012;51(50):10056–10065. doi: 10.1021/bi300702e. [DOI] [PubMed] [Google Scholar]
- 28.Latypov RF, Hogan S, Lau H, Gadgil H, Liu D. Elucidation of Acid- induced Unfolding and Aggregation of Human Immunoglobulin IgG1 and IgG2 Fc. Journal of Biological Chemistry. 2012;287(2):1381–1396. doi: 10.1074/jbc.M111.297697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, Westland K, Ma Y.-h., Gadgil H. Evaluation of effects of Fc domain high-mannose glycan on antibody stability. Journal of Pharmaceutical Sciences. 2012;101(11):4107–4117. doi: 10.1002/jps.23284. [DOI] [PubMed] [Google Scholar]
- 30.Liu H, Bulseco G-G, Sun J. Effect of posttranslational modifications on the thermal stability of a recombinant monoclonal antibody. Immunology Letters. 2006;106(2):144–153. doi: 10.1016/j.imlet.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Hristodorov D, Fischer R, Joerissen H, Müller-Tiemann B, Apeler H, Linden L. Generation and Comparative Characterization of Glycosylated and Aglycosylated Human IgG1 Antibodies. Molecular Biotechnology. 2013;53(3):326–335. doi: 10.1007/s12033-012-9531-x. [DOI] [PubMed] [Google Scholar]
- 32.Ghirlando R, Lund J, Goodall M, Jefferis R. Glycosylation of human IgG-Fc: influences on structure revealed by differential scanning micro-calorimetry. Immunology Letters. 1999;68(1):47–52. doi: 10.1016/s0165-2478(99)00029-2. [DOI] [PubMed] [Google Scholar]
- 33.Kai Zheng C. B. a. R. B. The impact of glycosylation on monoclonal antibody conformation and stability. mAbs. 2011;3(6):568–576. doi: 10.4161/mabs.3.6.17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raju TS, Scallon BJ. Glycosylation in the Fc domain of IgG increases resistance to proteolytic cleavage by papain. Biochemical and Biophysical Research Communications. 2006;341(3):797–803. doi: 10.1016/j.bbrc.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 35.Raju TS, Scallon B. Fc Glycans Terminated with N-Acetylglucosamine Residues Increase Antibody Resistance to Papain. Biotechnology Progress. 2007;23(4):964–971. doi: 10.1021/bp070118k. [DOI] [PubMed] [Google Scholar]
- 36.Wu S-J, Luo J, O'Neil KT, Kang J, Lacy ER, Canziani G, Baker A, Huang M, Tang QM, Raju TS, Jacobs SA, Teplyakov A, Gilliland GL, Feng Y. Structure-based engineering of a monoclonal antibody for improved solubility. Protein Engineering Design and Selection. 2010;23(8):643–651. doi: 10.1093/protein/gzq037. [DOI] [PubMed] [Google Scholar]
- 37.Middaugh CR, Litman GW. Atypical glycosylation of an IgG monoclonal cryoimmunoglobulin. Journal of Biological Chemistry. 1987;262(8):3671–3. [PubMed] [Google Scholar]
- 38.Kayser V, Chennamsetty N, Voynov V, Forrer K, Helk B, Trout BL. Glycosylation influences on the aggregation propensity of therapeutic monoclonal antibodies. Biotechnology Journal. 2011;6(1):38–44. doi: 10.1002/biot.201000091. [DOI] [PubMed] [Google Scholar]
- 39.Chennamsetty N, Helk B, Voynov V, Kayser V, Trout BL. Aggregation-Prone Motifs in Human Immunoglobulin G. Journal of Molecular Biology. 2009;391(2):404–413. doi: 10.1016/j.jmb.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 40.Borrok MJ, Jung ST, Kang TH, Monzingo AF, Georgiou G. Revisiting the Role of Glycosylation in the Structure of Human IgG Fc. ACS Chemical Biology. 2012;7(9):1596–1602. doi: 10.1021/cb300130k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chirino AJ, Mire-Sluis A. Characterizing biological products and assessing comparability following manufacturing changes. Nat Biotech. 2004;22(11):1383–1391. doi: 10.1038/nbt1030. [DOI] [PubMed] [Google Scholar]
- 42.Schiestl M, Stangler T, Torella C, Cepeljnik T, Toll H, Grau R. Acceptable changes in quality attributes of glycosylated biopharmaceuticals. Nat Biotech. 2011;29(4):310–312. doi: 10.1038/nbt.1839. [DOI] [PubMed] [Google Scholar]
- 43.Lubiniecki A, Volkin DB, Federici M, Bond MD, Nedved ML, Hendricks L, Mehndiratta P, Bruner M, Burman S, DalMonte P, Kline J, Ni A, Panek ME, Pikounis B, Powers G, Vafa O, Siegel R. Comparability assessments of process and product changes made during development of two different monoclonal antibodies. Biologicals. 2011;39(1):9–22. doi: 10.1016/j.biologicals.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Schenerman MA, Hope JN, Kletke C, Singh JK, Kimura R, Tsao EI, Folena-Wasserman G. Comparability Testing of a Humanized Monoclonal Antibody (Synagis®) to Support Cell Line Stability, Process Validation, and Scale-Up for Manufacturing. Biologicals. 1999;27(3):203–215. doi: 10.1006/biol.1999.0179. [DOI] [PubMed] [Google Scholar]
- 45.Jiang X-R, Song A, Bergelson S, Arroll T, Parekh B, May K, Chung S, Strouse R, Mire-Sluis A, Schenerman M. Advances in the assessment and control of the effector functions of therapeutic antibodies. Nat Rev Drug Discov. 2011;10(2):101–111. doi: 10.1038/nrd3365. [DOI] [PubMed] [Google Scholar]
- 46.Federici M, Lubiniecki A, Manikwar P, Volkin DB. Analytical lessons learned from selected therapeutic protein drug comparability studies. Biologicals. 2013;41(3):131–147. doi: 10.1016/j.biologicals.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Bhambhani A, Kissmann JM, Joshi SB, Volkin DB, Kashi RS, Middaugh CR. Formulation design and high-throughput excipient selection based on structural integrity and conformational stability of dilute and highly concentrated IgG1 monoclonal antibody solutions. Journal of Pharmaceutical Sciences. 2012;101(3):1120–1135. doi: 10.1002/jps.23008. [DOI] [PubMed] [Google Scholar]
- 48.Cheng W, Joshi SB, He F, Brems DN, He B, Kerwin BA, Volkin DB, Middaugh CR. Comparison of high-throughput biophysical methods to identify stabilizing excipients for a model IgG2 monoclonal antibody: Conformational stability and kinetic aggregation measurements. Journal of Pharmaceutical Sciences. 2012;101(5):1701–1720. doi: 10.1002/jps.23076. [DOI] [PubMed] [Google Scholar]
- 49.Hu L, Joshi SB, Andra KK, Thakkar SV, Volkin DB, Bann JG, Middaugh CR. Comparison of the structural stability and dynamic properties of recombinant anthrax protective antigen and its 2-fluorohistidine-labeled analogue. Journal of Pharmaceutical Sciences. 2012;101(11):4118–4128. doi: 10.1002/jps.23294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alsenaidy MA, Wang T, Kim JH, Joshi SB, Lee J, Blaber M, Volkin DB, Middaugh CR. An empirical phase diagram approach to investigate conformational stability of “second-generation” functional mutants of acidic fibroblast growth factor-1. Protein Science. 2012;21(3):418–432. doi: 10.1002/pro.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JH, Iyer V, Joshi SB, Volkin DB, Middaugh CR. Improved data visualization techniques for analyzing macromolecule structural changes. Protein Science. 2012;21(10):1540–1553. doi: 10.1002/pro.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maddux NR, Rosen IT, Hu L, Olsen CM, Volkin DB, Middaugh CR. An improved methodology for multidimensional high-throughput preformulation characterization of protein conformational stability. Journal of Pharmaceutical Sciences. 2012;101(6):2017–2024. doi: 10.1002/jps.23132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iyer V, Maddux N, Hu L, Cheng W, Youssef AK, Winter G, Joshi SB, Volkin DB, Middaugh CR. Comparative signature diagrams to evaluate biophysical data for differences in protein structure across various formulations. Journal of Pharmaceutical Sciences. 2013;102(1):43–51. doi: 10.1002/jps.23367. [DOI] [PubMed] [Google Scholar]
- 54.Houde D, Arndt J, Domeier W, Berkowitz S, Engen JR. Characterization of IgG1 Conformation and Conformational Dynamics by Hydrogen/Deuterium Exchange Mass Spectrometry. Analytical Chemistry. 2009;81(7):2644–2651. doi: 10.1021/ac802575y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational Modifications Differentially Affect IgG1 Conformation and Receptor Binding. Molecular & Cellular Proteomics. 2010;9(8):1716–1728. doi: 10.1074/mcp.M900540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gong R, Vu BK, Feng Y, Prieto DA, Dyba MA, Walsh JD, Prabakaran P, Veenstra TD, Tarasov SG, Ishima R, Dimitrov DS. Engineered Human Antibody Constant Domains with Increased Stability. Journal of Biological Chemistry. 2009;284(21):14203–14210. doi: 10.1074/jbc.M900769200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Kumar S, Buck PM, Singh SK. Impact of deglycosylation and thermal stress on conformational stability of a full length murine igG2a monoclonal antibody: Observations from molecular dynamics simulations. Proteins: Structure, Function, and Bioinformatics. 2013;81(3):443–460. doi: 10.1002/prot.24202. [DOI] [PubMed] [Google Scholar]
- 58.Shi S, Liu J, Joshi SB, Krasnoperov V, Gill P, Middaugh CR, Volkin DB. Biophysical characterization and stabilization of the recombinant albumin fusion protein sEphB4–HSA. Journal of Pharmaceutical Sciences. 2012;101(6):1969–1984. doi: 10.1002/jps.23096. [DOI] [PubMed] [Google Scholar]
- 59.Manikwar P, Majumdar R, Hickey JM, Thakkar SV, Samra HS, Sathish HA, Bishop SM, Middaugh CR, Weis DD, Volkin DB. Correlating excipient effects on conformational and storage stability of an IgG1 monoclonal antibody with local dynamics as measured by hydrogen/deuterium- exchange mass spectrometry. Journal of Pharmaceutical Sciences. 2013 doi: 10.1002/jps.23543. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 60.Majumdar R, Manikwar P, Hickey JM, Samra HS, Sathish HA, Bishop SM, Middaugh CR, Volkin DB, Weis DD. Effects of Salts from the Hofmeister Series on the Conformational Stability, Aggregation Propensity, and Local Flexibility of an IgG1 Monoclonal Antibody. Biochemistry. 2013;52(19):3376–3389. doi: 10.1021/bi400232p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.