Abstract
Objectives
The OPRL1 gene encodes the nociceptin/orphanin FQ receptor (NOP-R), which plays a role in regulating tolerance and behavioral responses to morphine. However, there is limited information on whether variants of OPRL1 are associated with vulnerability to develop opiate addiction. In this study, we examined five variants of OPRL1 and their role in determining vulnerability to develop opiate addiction.
Methods
We recruited 447 subjects: 271 former severe heroin addicts and 176 healthy controls. Using a 5′-fluorogenic exonuclease assay (TaqMan®), we genotyped subjects at five variants in OPRL1. It was then determined whether there was a significant association of allele, genotype, or haplotype frequency with vulnerability to develop opiate addiction.
Results
When the cohort was stratified by ethnicity, we found that, in Caucasians but not in African Americans or Hispanics, the allele frequency of rs6090041 and rs6090043 were significantly associated point-wise with opiate addiction (P = 0.03 and 0.04, respectively). Of the haplotypes formed by these two variants, one haplotype was found to be associated with protection from developing opiate addiction in both African Americans (point-wise P = 0.04) and Caucasians (point-wise P = 0.04), and another haplotype with vulnerability to develop opiate addiction in Caucasians only (P = 0.020).
Conclusions
This study provides evidence for an association of two variants of the OPRL1 gene, rs6090041 and rs6090043, with vulnerability to develop opiate addiction, suggesting a role for NOP-R in the development of opiate addiction.
Keywords: OPRL1, ORL1, nociceptin/orphanin FQ receptor, polymorphism, variant, opiate addiction, heroin
Introduction
The development of opiate addiction is a complex process that is influenced by a variety of factors (Kreek et al., 2004; Kreek et al., 2005a, Kreek et al., 2005b). It has been established that genetic, as well as social, psychological, and environmental factors, play a role in the development of opiate addiction. For example, family and twin studies have found that the genetic variance contributing to an individual’s vulnerability to develop heroin addiction is estimated to be up to 0.54 (Tsuang et al., 1998 and Kendler et al., 2003). However, many of the specific gene variants that are responsible for influencing an individual’s genetic vulnerability to develop addiction have yet to be identified.
The endogenous opioid system is involved in transducing the effects of opiates, such as heroin and morphine (Kreek, 1996). Opioids exert their effects, in part, by binding opioid receptors on the surface of GABA-ergic neurons in the mesocorticolimbic system. GABA-ergic neurons exert a tonic inhibition on the dopaminergic neurons, and the binding of opiates causes inhibition of the GABA-ergic neurons and disinhibition of the dopaminergic system, resulting in an increase in synaptic dopamine levels in the terminal fields of the mesocorticolimbic dopamine system (Johnson and North, 1992). The mesocorticolimbic system projects from the ventral tegmental area to the nucleus accumbens, olfactory tubercle, cingulate cortex, and amygdala. The increase in dopaminergic tone due to opiate inhibition contributes to the rewarding properties of opiates. Blocking opioid receptors in the nucleus accumbens or ventral tegmental area with opiate antagonists ablates the rewarding properties of opiates (reviewed in Kreek and Koob, 1998).
One component related to the endogenous opioid system is the nociceptin/orphanin FQ receptor (NOP-R), encoded by the OPRL1 gene. The ligand of NOP-R is nociceptin/orphanin FQ (N/OFQ). Both NOP-R and N/OFQ are expressed at various levels throughout brain tissues (Mollereau and Mouledous, 2000). NOP-R is a G protein-coupled receptor that, when activated by N/OFQ, signals via inhibition of adenylyl cyclase, decreases cAMP formation (Wu et al., 1997). The N/OFQ peptide is similar in structure to the endogenous opioid dynorphin (Houtani et al., 1996), and its processing has been reported to be similar to that of dynorphin (Yu et al., 1996). Functionally, N/OFQ has been reported to inhibit the rewarding effect of addictive drugs (Ciccocioppo et al., 2000; Cowen and Lawrence, 2006; Reinscheid et al., 2006). When N/OFQ is injected into mice, there is a dose-dependent inhibition of mesolimbic dopamine release (Murphy et al., 1996) with a concomitant loss of morphine conditioned place preference (Murphy et al., 1999). N/OFQ inhibits dopaminergic neurons in the ventral tegmental area, potentially via an increase in GABA levels, which in turn decreases dopamine transmission to the nucleus accumbens (Murphy and Maidment, 1999). It has been reported that there is conditioned place aversion to naloxone in mice following administration of N/OFQ (Sakoori and Murphy, 2004). In two studies, nociceptin was found to have no detectable effect on heroin self-administration in rats (Walker et al., 1998; Martin-Fardon et al., 2000). In most studies, however, the effects of N/OFQ are similar to those of dynorphin, which decreases basal and drug of abuse-induced increases in dopamine (Zhang et al., 2004). This has been further supported by a study in mice lacking the nociceptin receptor gene, in which a 50% reduction in the development of morphine tolerance was observed (Ueda et al., 1997). Thus, these studies indicate that N/OFQ and its receptor, NOP-R, block some of the rewarding properties of opiates and are involved in the development of opioid tolerance.
Given the role of NOP-R in the endogenous opioid system, we have evaluated whether variants of OPRL1 are associated with vulnerability to develop opiate addiction. In this study, we evaluated five variants in the coding, the intronic, or upstream regions of the human OPRL1 gene to examine whether there are significant associations of allele, genotype, or haplotype frequency with vulnerability to develop opiate addiction.
Methods
Study Subjects
Subjects were consecutive unrelated volunteers, who were a part of a study on the genetics of drug addiction at the Laboratory of the Biology of Addictive Diseases at The Rockefeller University. The subjects were recruited from February 7, 1995 to January 20, 2000 from methadone maintenance treatment programs in New York City and through advertisements in local media. There were 447 subjects who met the inclusion criteria defined below: former severe heroin addicts stabilized in methadone maintenance pharmacotherapy or healthy controls (Table 1).
Table 1.
Primary ascertainment and classification of subjects
| Ethnicity
|
Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| African American | Caucasian | Hispanic | Asian | Mixed/Other | ||||||||
|
| ||||||||||||
| Classification | N | Male (%) | N | Male (%) | N | Male (%) | N | Male (%) | N | Male (%) | N | Male (%) |
|
|
|
|
|
|
|
|
||||||
| Former Severe | ||||||||||||
| Heroin Addicts | 59 | 30 (51) | 100 | 65 (65) | 94 | 51 (54) | 1 | 0 (0) | 17 | 6 (35) | 271 | 152 (56) |
| Controls | 44 | 26 (59) | 76 | 41 (54) | 29 | 11 (38) | 20 | 11 (55) | 7 | 6 (86) | 176 | 95 (54) |
|
|
|
|
|
|
|
|
||||||
| Total | 103 | 56 (54) | 176 | 106 (60) | 123 | 62 (50) | 21 | 11 (52) | 24 | 12 (50) | 447 | 247 (55) |
N, Total Number
%, Percentage of male subjects in that classification
Each subject was evaluated by a psychiatrist, clinical psychologist, or research nurse and signed an informed consent form approved by The Rockefeller University Hospital Institutional Review Board. This form gives consent specifically for genetic studies. The addiction history of each subject was evaluated using the Addiction Severity Index (ASI) (McLellen et al., 1992). Urine samples were obtained and screened for drugs of abuse, and blood samples were taken by venipuncture. Subjects were excluded if they had an ongoing psychotic illness.
All of the former severe heroin addicts met U.S. Federal criteria for methadone maintenance treatment, including demonstration of drug-seeking behavior, self-administration of multiple doses of opiate drugs daily for one year or more, and the acquisition of dependence and tolerance (Rettig and Yarmolinsky, 1995). Control subjects were excluded if they used an illicit drug (except cannabis) within the last 30 days, had any instance of alcohol intoxication in the last 30 days, or had a history of greater than six months of illicit drug or alcohol intoxication more than three times per week as defined by ASI criteria. Control subjects were excluded if they had used cannabis three or more times per week for four years or more.
Each subject provided information about place of birth and the ethnic/cultural background of him/herself, parents, grandparents, and great grandparents. Individuals were classified into five groups: African American, Asian, Caucasian, Hispanic, or Other/Mixed.
The Asian group was not included in the association studies since only one subject was a former severe heroin addict. The 24 subjects who identified themselves as in the mixed/other group were not included in the association studies, to avoid possible confounding factors due to population admixture.
Genotyping
We decided a priori to study five variants of OPRL1. Two variants, rs2229205 and rs2295448, were initially selected using the UCSC Genome Browser (http://genome.ucsc.edu, Build 36.1, May 2008). rs2229205 was chosen due to its location in the coding region of OPRL1. Using the Tagger software (http://www.broad.mit.edu/mpg/tagger), seven tagged variants were identified from the CEU (European) HapMap analysis panel (r2 = 1.00, minimum allele frequency ≥ 5%) (Bakker et al., 2005). The allele frequencies of these were examined using HapMap data found at dbSNP (http://www.ncbi.nlm.nih.gov/SNP) (Build 127), and three variants, rs6011280, rs6090041, and rs6090043, were chosen based on allele frequencies greater than 0.05 in the CEU, HCB (Asian), JPT (Asian), and YRI (Sub-Saharan African) populations. The variants in intron 1 (rs6090041 and rs6090043) were evaluated for transcription factor binding sites using TESS: Transcription Element Search System (Schug and Overton, 1998).
A 5′-fluorogenic exonuclease assay (TaqMan®, Applied Biosystems, Foster City, CA) was used to determine genotypes at the five variants. TaqMan® probes were designed for variants rs2229205 and rs2295448 using Primer Express software (Applied Biosystems) and synthesized by Applied Biosystems. We used the following oligonucleotides for genotyping variant rs2229205: 5′-TGTTCACCAGCACCTTCACC-3′ (forward), 5′-ACACCGACAACAGAGGCCAG-3′ (reverse), 5′-AGGCTGTCAATGTG-3′ (FAM-labeled), and 5′-AGGCTGTTAATGTGG-3′ (VIC-labeled), and for genotyping variant 2295448: 5′-GGCTCCCGGGTGGCT-3′ (forward), 5′-GCCGGATCATGAGGCTGTAG-3′ (reverse), 5′-AGGGCACGTGGGC-3′ (FAM-labeled), and 5′-AGGGCACATGGGC-3′ (VIC-labeled). Pre-designed TaqMan® primer-probe sets (Applied Biosystems) were used for genotyping variants rs6011280 (Assay ID C_69507_10), rs6090041 (Assay ID C_29614941_10), and rs6090043 (Assay ID C_26713862_10). PCR amplification was performed in duplicate using Platinum® quantitative PCR SuperMix-UDG (Invitrogen, Carlsbad, CA) on a GeneAmp® PCR system 9700 and the dual 384-well sample block module (Applied Biosystems). Samples were amplified at 50°C for 2 min, 95° C for 10 min, and then 50 cycles of 95°C for 15s and 60°C for 1 min. The amplification products were analyzed using an Applied Biosystems Prism® 7900 sequence detection system and SDS 2.2 software (Applied Biosystems) by an investigator who was unaware of the subjects’ diagnoses.
Statistical Analysis
The frequency of marker alleles and genotypes in cases and controls were compared using the two-sided Fisher’s exact test. Odds ratios (OR) with 95% confidence intervals were determined. A P ≤ 0.05 was considered significant point-wise prior to correcting for multiple comparisons.
To account for any allelic correlation at the locus level, the exact test of Hardy-Weinberg proportions was performed as implemented in the genetics library of R v2.4.1 (Warnes, 2003). A P ≤ 0.05/15 (five markers on three ethnicities) was considered significant experiment-wise (correcting for multiple testing).
The G-test of independence was applied to the distribution of allele frequencies in the control samples across the different ethnicities to test for hidden structure (Sokal et al., 1994). This is an asymptotically distributed chi-square test with the same number of degrees of freedom as the corresponding chi-square test of independence.
Score tests of association with disease as well as frequencies were computed for haplotypes with a special software [haplo.score from the R package haplo.stats v1.4.3 (Sinnwell and Schaid, 2009)]. This program provides a global association test P value as well as P values resulting from evaluating each haplotype against the rest.
To adjust for multiple testing, single-marker and haplotype results were examined using a special software [R package qvalue v1.1 (Storey and Tibshirani, 2003)]. This package takes a given set of P values and estimates the minimum false discovery rate (FDR) (Benjamini and Hochberg, 1995) that is incurred when calling a particular test significant, which is the q-value of the test (Storey, 2003). An FDR of 0.05 was used as a cut-off for experiment-wise significance, and the proportion of true null hypotheses was estimated by the bootstrap method.
Pairwise Linkage Disequilibrium (LD)
The pattern of pairwise LD between variants was measured by the standardized disequilibrium value (D′) and r2 (Lewontin, 1964) as implemented in Haploview v3.32 (Barrett et al., 2005). D′ varies from −1 to 1. A value of |D′| of 1 implies complete LD, a value of 0 implies linkage equilibrium, and intermediate levels of D′ suggest recombination between the loci or recurrent mutation. Haplotype blocks were selected using the 4-gamete rule definition (Wang et al., 2002) and were reconstructed using the accelerated expectation-maximization (EM) algorithm implemented in Haploview (which is similar to the partition-ligation method) (Qin et al., 2002).
Results
Demographics
In this study, there were 447 subjects: 247 (55%) were male; 103 (23%) were African American, 176 (39%) Caucasian, 123 (28%) Hispanic, 21 (5%) Asian, and 24 (5%) Mixed/Other (Table 1). In total, 271 (61%) were methadone-stabilized former severe heroin addicts and 176 (39%) were healthy controls.
Variant Analysis
The 447 subjects were genotyped at five variants (rs6011280, rs6090041, rs6090043, rs2229205, and rs2295448), the first of which is found 6,348 nucleotides upstream of the OPRL1 transcription start site (in the 3′ untranslated region of RGS19, a gene that encodes a regulator of G-protein signaling (Ito et al., 2000)), and the remaining four are found in intron 1, intron 1, exon 4, and intron 4, respectively, of the NOP-R gene OPRL1 (Figure 1). Variant rs2229205 is located in the coding region of exon 4 but is a synonymous substitution that does not alter amino acid sequence. As is standard convention in most genetics papers, the variant nucleotide indicated is in the sense strand of OPRL1.
Figure 1.
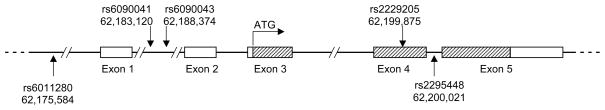
Organization of the human OPRL1 gene. The location of each variant used in this study is shown with its nucleotide position on chromosome 20 (NCBI Build 36.1). Coding region is shown as
 and non-coding region as white. ATG transcription start site is indicated.
and non-coding region as white. ATG transcription start site is indicated.
Genotype frequencies in the control groups were examined in order to determine whether they deviated significantly from Hardy-Weinberg equilibrium (HWE). None of the variants significantly deviated from HWE (P < 0.05) in any of the four major ethnic groups: African Americans, Caucasians, Hispanics, and Asians. The allele frequencies of the five variants were evaluated among ethnicities. Allele frequencies differed significantly among ethnicities for variant rs6090041 only (P = 0.0004, Table 2). Since one of the five variants was significantly different in allele frequencies among the ethnicities, case-control comparisons were done after stratification by ethnicity.
Table 2.
Allele frequencies in controls by ethnicity
| Variant | Allele | African American | Caucasian | Hispanic | Asian | LR χ2 a | P value |
|---|---|---|---|---|---|---|---|
|
| |||||||
| N (frequency) | N (frequency) | N (frequency) | N (frequency) | ||||
| rs6011280 | C | 32 (0.41) | 45 (0.36) | 18 (0.39) | 8 (0.20) | 5.91 | 0.116 |
| A | 46 (0.59) [0.62]c |
79 (0.64) [0.72] |
28 (0.61) | 32 (0.80) [0.73] |
|||
| rs6090041 | G | 37 (0.47) | 28 (0.23) | 14 (0.30) | 6 (0.15) | 18.33 | 0.0004b |
| A | 41 (0.53) [0.49] |
94 (0.77) [0.59] |
32 (0.70) | 34 (0.85) [0.69] |
|||
| rs6090043 | C | 34 (0.44) | 51 (0.42) | 21 (0.46) | 15 (0.38) | 0.66 | 0.883 |
| T | 44 (0.56) [0.52] |
71 (0.58) [0.51] |
25 (0.54) | 25 (0.62) [0.63] |
|||
| rs2229205 | T | 6 (0.07) | 28 (0.19) | 8 (0.14) | 4 (0.10) | 7.75 | 0.051 |
| C | 80 (0.93) [0.87] |
118 (0.81) [0.86] |
50 (0.86) | 36 (0.90) [0.87] |
|||
| rs2295448 | T | 12 (0.14) | 18 (0.12) | 4 (0.07) | 4 (0.10) | 2.17 | 0.537 |
| C | 72 (0.86) | 128 (0.88) | 54 (0.93) | 36 (0.90) | |||
Probabilities calculated with likelihood ratio (LR) χ2 test with 3 degrees of freedom (df) for the differences in allele frequencies of the control groups among the African American, Asian, Caucasian, and Hispanic groups.
Point-wise significant P values are in bold.
Allele frequency from HapMap of the allele in line 2 is provided in brackets under each variant except rs2295448, for which there is no data in HapMap. The allele frequency for the African Americans are from the HapMap-YRI (Sub-Saharan African), for the Caucasian from the HapMap-CEU (European), and for the Asian from the HapMap-HCB (Asian) (dbSNP, Build 130).
Association of Allele, Genotype, and Haplotype Frequency with Opiate Addiction
The five OPRL1 variants were analyzed for association with vulnerability to develop opiate addiction separately in each of the three major ethnicities in which there were adequate sample sizes (African Americans, Caucasians, and Hispanics, Table 3). After stratifying by ethnicity, the allele frequencies of rs6090041 and of rs6090043 were found to be associated significantly point-wise with opiate addiction in Caucasians (P = 0.03, Table 3B; P = 0.04, Table 3C, respectively). No experiment-wise significance was found after correction for multiple testing. At the genotype level (Table 3), no significant association of OPRL1 variants with opiate addiction was found in any of the ethnic groups tested.
Table 3.
Association of the genotypes of variants of the OPRL1 gene with opiate addiction
| A. Variant rs6011280 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Genotype | Allele Frequency | ||||||||
|
| |||||||||
| Ethnicity | Group | AA | CA | CC | P value | A | C | ORa (95% CI) | P value |
| African American | Opiate | 21 | 25 | 7 | 0.84 | 0.63 | 0.37 | 0.84 (0.44, 1.59) | 0.65 |
| Control | 13 | 20 | 6 | 0.59 | 0.41 | ||||
| Caucasian | Opiate | 38 | 35 | 9 | 0.66 | 0.68 | 0.32 | 0.84 (0.50, 1.41) | 0.54 |
| Control | 24 | 31 | 7 | 0.64 | 0.36 | ||||
| Hispanic | Opiate | 28 | 43 | 14 | 1.00 | 0.59 | 0.41 | 1.11 (0.55, 2.32) | 0.87 |
| Control | 8 | 12 | 3 | 0.61 | 0.39 | ||||
| B. Variant rs6090041 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Genotype | Allele Frequency | ||||||||
|
| |||||||||
| Ethnicity | Group | AA | AG | GG | P value | A | G | OR (95% CI) | P value |
| African American | Opiate | 8 | 25 | 20 | 0.17 | 0.39 | 0.61 | 1.75 (0.93, 3.31) | 0.07 |
| Control | 10 | 21 | 8 | 0.53 | 0.47 | ||||
| Caucasian | Opiate | 36 | 34 | 12 | 0.08 | 0.65 | 0.35 | 1.83 (1.05, 3.25) | 0.03b |
| Control | 36 | 22 | 3 | 0.77 | 0.23 | ||||
| Hispanic | Opiate | 33 | 44 | 8 | 0.83 | 0.65 | 0.35 | 1.25 (0.59, 2.73) | 0.60 |
| Control | 10 | 12 | 1 | 0.70 | 0.30 | ||||
| C. Variant rs6090043 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Genotype | Allele Frequency | ||||||||
|
| |||||||||
| Ethnicity | Group | CC | CT | TT | P value | C | T | OR (95% CI) | P value |
| African American | Opiate | 15 | 30 | 8 | 0.13 | 0.57 | 0.43 | 0.59 (0.31, 1.11) | 0.10 |
| Control | 8 | 18 | 13 | 0.44 | 0.56 | ||||
| Caucasian | Opiate | 24 | 41 | 17 | 0.09 | 0.54 | 0.46 | 0.61 (0.37, 1.00) | 0.04b |
| Control | 13 | 25 | 23 | 0.42 | 0.58 | ||||
| Hispanic | Opiate | 28 | 43 | 14 | 0.27 | 0.58 | 0.42 | 0.60 (2.96, 1.22) | 0.14 |
| Control | 5 | 11 | 7 | 0.46 | 0.54 | ||||
| D. Variant rs2229205 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Genotype | Allele Frequency | ||||||||
|
| |||||||||
| Ethnicity | Group | CC | CT | TT | P value | C | T | OR (95% CI) | P value |
| African American | Opiate | 47 | 8 | 0 | 1.00 | 0.93 | 0.07 | 0.95 (0.26, 3.33) | 1.00 |
| Control | 37 | 6 | 0 | 0.93 | 0.07 | ||||
| Caucasian | Opiate | 65 | 27 | 5 | 0.96 | 0.81 | 0.19 | 0.99 (0.56, 1.79) | 1.00 |
| Control | 48 | 22 | 3 | 0.81 | 0.19 | ||||
| Hispanic | Opiate | 75 | 14 | 1 | 0.42 | 0.91 | 0.09 | 0.61 (0.23, 1.75) | 0.32 |
| Control | 22 | 6 | 1 | 0.86 | 0.14 | ||||
| E. Variant rs2295448 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Genotype | Allele Frequency | ||||||||
|
| |||||||||
| Ethnicity | Group | CC | CT | TT | P value | C | T | OR (95% CI) | P value |
| African American | Opiate | 41 | 11 | 3 | 0.25 | 0.85 | 0.15 | 1.10 (0.46, 2.69) | 0.84 |
| Control | 30 | 12 | 0 | 0.86 | 0.14 | ||||
| Caucasian | Opiate | 69 | 23 | 5 | 0.55 | 0.83 | 0.17 | 1.46 (0.76, 2.88) | 0.28 |
| Control | 57 | 14 | 2 | 0.88 | 0.12 | ||||
| Hispanic | Opiate | 63 | 23 | 1 | 0.41 | 0.86 | 0.14 | 2.26 (0.73, 9.34) | 0.17 |
| Control | 25 | 4 | 0 | 0.93 | 0.07 | ||||
Odds ratio (OR) > 1 indicates vulnerability to develop opioid addiction; OR < 1 indicates protective for the minor allele in controls.
Point-wise significant P values are in bold.
Since the OPRL1 variants rs6090041 and rs6090043 showed point-wise significant association with opiate addiction, association analyses of the haplotypes of these variants with opiate addiction were conducted. Point-wise significant differences were found in the African Americans and in the Caucasians (Table 4). The AT haplotype was found to be associated with protection from developing opiate addiction in both African Americans and Caucasians (point-wise P = 0.041 for each). In the Caucasians only, the GC haplotype was found to be associated with vulnerability to develop opiate addiction (point-wise P = 0.020).
Table 4.
Association of OPRL1 haplotypes of rs6090041 and rs6090043 with opiate addiction
| Haplotype | Estimate of haplotype frequency
|
Haplotype P valuea | Global P valueb | |
|---|---|---|---|---|
| Opiate | Control | |||
| A. African American | ||||
| A T | 0.288 | 0.421 | 0.041c | 0.215 |
| A C | 0.099 | 0.104 | 0.895 | |
| G C | 0.467 | 0.332 | 0.061 | |
| G T | 0.146 | 0.143 | 0.939 | |
| B. Caucasian | ||||
| A T | 0.417 | 0.541 | 0.041c | 0.105 |
| A C | 0.229 | 0.229 | 1.000 | |
| G C | 0.313 | 0.189 | 0.020c | |
| G T | 0.040 | 0.041 | 0.984 | |
| C. Hispanic | ||||
| A T | 0.373 | 0.464 | 0.231 | 0.406 |
| A C | 0.274 | 0.231 | 0.530 | |
| G C | 0.308 | 0.225 | 0.228 | |
| G T | 0.045 | 0.079 | 0.256 | |
Haplotype-specific P value, obtained from analyzing a particular haplotype against the rest.
Global P value, obtained from estimating the significance of the overall set of haplotypes.
Point-wise significant P values are in bold.
Linkage Disequilibrium
Linkage disequilibrium among the five variants was evaluated in the controls of the four primary ethnic groups. As a service to the reader, haplotype blocks were determined, but were not used in the statistical analyses. A single haplotype block was found at the 3′ end of the gene in the African American, Caucasian, and Hispanic groups that encompasses rs6090043 in intron 1, rs2229205 in exon 4, and rs2295448 in intron 4, and spans 11,547 nucleotides (Table 5a, 5b, 5c). In the Asian group, in addition to this block, a second haplotype block was found encompassing rs6011280, which is upstream of the OPRL1 gene (in the 3′ untranslated region of RGS19), and rs6090041 in intron 1, and this block spans 7,536 nucleotides (Table 5d). Thus, variants rs6090043, rs2229205, and rs2295448 are in high linkage disequilibrium in all four ethnic groups, while the variants rs6011280 and rs6090041 are in high linkage disequilibrium in Asians only.
Table 5.
Pairwise linkage disequilibrium among the five OPRL1 variants in the four primary ethnic groupsa
| A. African American
| |||||
|---|---|---|---|---|---|
| Variant | rs6011280 | rs6090041 | rs6090043 | rs2229205 | rs2295448 |
| rs6011280 | --- | 0.48 | 0.83 | 1.00 | 0.33 |
| rs6090041 | 0.12 | --- | 0.56 | 1.00 | 0.53 |
| rs6090043 | 0.41 | 0.26 | --- | 1.00 | 1.00 |
| rs2229205 | 0.05 | 0.09 | 0.08 | --- | 1.00 |
| rs2295448 | 0.03 | 0.04 | 0.17 | 0.01 | --- |
| B. Caucasian
| |||||
|---|---|---|---|---|---|
| Variant | rs6011280 | rs6090041 | rs6090043 | rs2229205 | rs2295448 |
| rs6011280 | --- | 0.06 | 0.46 | 0.83 | 0.77 |
| rs6090041 | 0.001 | --- | 0.74 | 0.78 | 0.92 |
| rs6090043 | 0.11 | 0.24 | --- | 1.00 | 1.00 |
| rs2229205 | 0.09 | 0.06 | 0.23 | --- | 1.00 |
| rs2295448 | 0.05 | 0.33 | 0.17 | 0.04 | --- |
| C. Hispanic
| |||||
|---|---|---|---|---|---|
| Variant | rs6011280 | rs6090041 | rs6090043 | rs2229205 | rs2295448 |
| rs6011280 | --- | 0.08 | 0.68 | 0.77 | 0.68 |
| rs6090041 | 0.005 | --- | 0.66 | 1.00 | 0.85 |
| rs6090043 | 0.26 | 0.18 | --- | 0.88 | 1.00 |
| rs2229205 | 0.04 | 0.05 | 0.10 | --- | 1.00 |
| rs2295448 | 0.05 | 0.22 | 0.13 | 0.02 | --- |
| D. Asian
| |||||
|---|---|---|---|---|---|
| Variant | rs6011280 | rs6090041 | rs6090043 | rs2229205 | rs2295448 |
| rs6011280 | --- | 1.00 | 1.00 | 1.00 | 1.00 |
| rs6090041 | 0.04 | --- | 0.66 | 1.00 | 1.00 |
| rs6090043 | 0.42 | 0.13 | --- | 1.00 | 1.00 |
| rs2229205 | 0.03 | 0.02 | 0.08 | --- | 1.00 |
| rs2295448 | 0.03 | 0.63 | 0.19 | 0.01 | --- |
The upper diagonal displays the absolute D′ (linkage disequilibrium) value. The lower diagonal displays the r2.
N = 423
Discussion
In this study, we evaluated the association of five variants in the OPRL1 gene with vulnerability to develop opiate addiction in the three ethnicities with adequate sample sizes (African Americans, Caucasians, and Hispanics). Variant rs6011280 is located 6,348 nucleotides upstream of the OPRL1 transcription start site, variants rs6090041 and rs6090043 are in intron 1, variant rs2229205 is in exon 4, and variant rs22295448 is in intron 4. The variant rs2229205, located in the coding region of exon 4, does not result in an amino acid change.
In the analysis of the 447 subjects stratified by ethnicity, significant point-wise associations were found between the allele frequency of the variants rs6090041 and rs6090043 with vulnerability to develop opiate addiction in Caucasians. One haplotype (AT) formed by these two variants was significantly associated point-wise with protection from developing opiate addiction in both African Americans and Caucasians. Since we had similar results with both ethnicities, this provides further support for a role of this haplotype in protection from opiate addiction. In Caucasians only, another haplotype (GC) was found to be associated with vulnerability to develop opiate addiction.
The two variants that were point-wise significantly associated with opiate addiction, rs6090041 and rs6090043, are located in the first intron of the OPRL1 gene. These variants, which are in low linkage disequilibrium (r2 = 0.26, 0.24, and 0.13 in African Americans, Caucasians, and Hispanics, respectively), may themselves alter function, or they may be markers for nearby functional variants that have a role in determining an individual’s vulnerability to develop opiate addiction. An evaluation of predicted transcription factor binding sites, using TESS (Schung and Overton, 1998), suggests that the sequence surrounding the G allele of rs6090041 predicts a putative AP-2 transcription factor binding site. The presence of the A allele would disrupt this AP-2 binding site. The AP-2 binding site containing the G allele is GCGGTGGG, which conforms to the AP-2 consensus motif CCCMNSSS (TESS - TRANSFAC Site Record R02121) in reverse orientation (SSSNKGGG), while the site is altered to GCGGTGAG, which no longer conforms to the consensus motif and therefore would not bind AP-2. AP-2 has been found to alter the transcription of genes impacting the morphogenesis of the peripheral nervous system during embryogenesis (Mitchell et al., 1991). Similarly, analyses using TESS suggest that the C allele of rs6090043 predicts several transcription factor binding sites that the T allele would disrupt, including binding sites for CREB and AP-1. The CREB binding motif containing the C allele is TGACG, the same sequence as the CREB binding motif that is found in the insulin gene (TESS - TRANSFAC Site Record R02710; Boam et al., 1990); when this motif is altered to TGATG, it would not bind CREB. CREB is activated in neurons by a wide range of extracellular stimuli and is involved in the function, development, and plasticity of the nervous system (Lonze et al., 2002). The AP-1 binding motif containing the C allele is TGAC, the same sequence as the AP-1 binding motif found in adenovirus 5 (TESS - TRANSFAC Site Record R00368; Merino et al., 1989); when the motif is changed to TGAT, it should not bind AP-1. AP-1 has been shown to be required for efficient axonal regeneration (Raivich, et al., 2004). Thus, both variants may alter transcriptional regulation.
In this study in Caucasians, the G allele at rs6090041 and the C allele at rs609004, which are both associated with an increased risk for developing opiate addiction (see Table 3), each form potential additional transcription factor binding sites that may increase NOP-R expression. Since NOP-R has anti-opioid effects, it is possible that individuals with increased NOP-R expression may require a greater amount of an opiate to experience drug reward and therefore may be more likely to become addicted to opiates.
There are three published genetic studies of the nociceptin system in humans. The first study found that there was no association between the number of copies of a 39-base pair repeat in OPRL1 and panic disorder in a case-control study in Japanese subjects (Kobayashi et al., 2007). The second study examined the association of OPRL1 and N/OFQ variants with alcohol, illicit drug, or opiate dependence (Xuei et al. 2007). This study examined 10 variants of the OPRL1 gene, and an adjacent regulatory gene (RGS19), and 15 variants of the gene that encodes N/OFQ (PNOC). Among the variants of OPRL1 were rs6011280, rs6090041, rs6090043, and rs2229205, which were examined in our study. rs2295448 was not studied by Xuei et al. No experiment-wise significant associations with alcohol or illicit drug dependence were found in that study. When opiate addiction was examined, they found a “marginally significant” association of variants rs6512305 and rs6090043 with opiate addiction (P = 0.05). However, they did not state which allele of rs6512305 or rs6090043 was associated with opiate dependence. In our study, we found an association between rs6090043 and opiate addiction that had point-wise significance (P = 0.04). The Xuei et al. study found that variants rs6512305 and rs6090043 were in high linkage disequilibrium (D′ = 0.99) in European Americans. It may be that rs6512305, or another nearby variant in linkage disequilibrium with rs6090043, may be the functional marker. The convergence of our results with that of Xuei et al. may be due the alteration of the overlapping CREB/AP-1 binding sites, resulting in an altered expression of OPRL1. We also found that variant rs6090041, which Xuei et al. studied, had a significant association with opiate addiction (P = 0.03). The lack of significance found in the Xuei et al. study may be due to their small number of cases (83 cases). The third genetic study examined the association between OPRL1 variants and alcohol dependence in a Swedish population (Huang et al., 2008). They studied 15 variants of OPRL1, including four of the five variants that we examined (rs6090041, rs6090043, rs2229205, and rs2295448). They found variant rs6010718 (a variant we did not examine) in OPRL1 and two haplotypes of the OPRL1 gene to be significantly associated with alcohol dependence. A rare haplotype (5.9% in cases, 0.6% in controls) containing the T allele of rs6090043 was found to be associated with alcohol dependence. We found the common (46% in cases, 58% in controls) T allele of rs6090043 to be protective from opiate addiction in Caucasians. It may be that alterations in function caused by the rs6090043 allele (or a nearby linked functional allele) may influence vulnerability to develop alcohol dependence and opiate addiction in different ways.
We found point-wise significance of an association of OPRL1 variants with the vulnerability to develop opiate addiction. A future study with a larger sample size should have more power to detect an association. However, our study demonstrates a moderate association of OPRL1 variants with vulnerability to develop opiate addiction specifically and thereby contributes to the growing evidence that the OPRL1 gene may contribute to the development of illicit drug and alcohol dependence.
Conclusions
In this study, we found that two variants of the OPRL1 gene, rs6090041 and rs6090043, are significantly associated point-wise with vulnerability to develop opiate addiction, and also form a haplotype that was associated with vulnerability to develop opiate addiction and a haplotype that was protective. These two variants, located in the first intron of the gene, may alter transcription factor binding sites, thus altering OPRL1 gene transcription and, finally, the development of opiate addiction. These two variants, which are not in linkage disequilibrium, may be markers for nearby functional variants that themselves may have a role in an individual’s vulnerability to develop opiate addiction. Our study provides evidence for an association between OPRL1 variants and vulnerability to develop opiate addiction, which may ultimately lead to better prevention strategies or new therapeutic approaches.
Acknowledgments
Sources of support: National Institutes of Health (NIH) grants P60-DA005130 (M.J.K.), R01-MH79880, RR UL1RR024143 (B.C.), NIH-MH R01-44292 (J.O.), R01-DA12848 (M.J.K.), NSFC grants 30730057 and 30700442 from the Chinese Government (J.O.), and American Federation for Aging Research Medical Student Training in Aging Research grant (J.A.B.).
We thank all of our clinical staff for recruiting, screening, and assessing study subjects. We also thank Matthew Randesi for technical assistance and Sara Hamon for statistical assistance.
References
- De Bakker R, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nature Genetics. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Boam DSW, Clark AR, Docherty K. Positive and negative regulation of the human insulin gene by multiple Trans-acting Factors. J Biol Chem. 1990;265:8285–8296. [PubMed] [Google Scholar]
- Ciccocioppo R, Angeletti S, Panocka I, Massi M. Nociceptin/orphanin FQ and drugs of abuse. Peptides. 2000;21:1071–1080. doi: 10.1016/s0196-9781(00)00245-x. [DOI] [PubMed] [Google Scholar]
- Cowen MS, Lawrence AJ. Alcoholism and neuropeptides: an update. CNS Neurol Disord Drug Targets. 2006;5:233–239. doi: 10.2174/187152706776359646. [DOI] [PubMed] [Google Scholar]
- Houtani T, Nishi M, Takeshima H, Nukada T, Sugimoto T. Structure and regional distribution of nociceptin/orphanin FQ precursor. Biochem Biophys Res Commun. 1996;219:714–719. doi: 10.1006/bbrc.1996.0300. [DOI] [PubMed] [Google Scholar]
- Huang J, Young B, Pletcher MT, Heilig M, Wahlestedt C. Assocation between the nociceptin receptor gene (OPRL1) single nucleotide polymorphisms and alcohol dependence. Addict Biol. 2008;13:88–94. doi: 10.1111/j.1369-1600.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- Ito E, Xie G, Maruyama K, Palmer PP. A core-promoter region functions bi-directionally for human opioid-receptor-like gene ORL1 and its 5′-adjacent gene GAIP. J Mol Biol. 2000;304:259–270. doi: 10.1006/jmbi.2000.4212. [DOI] [PubMed] [Google Scholar]
- Johnson SW, North RA. Opioids excite dopamine neurons by hyperpolarization of local interneurons. J Neurosci. 1992;12:483–488. doi: 10.1523/JNEUROSCI.12-02-00483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003;160:687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Akiyoshi J, Kanehisa M, Ichioka S, Tanaka Y, Tsuru J, et al. Lack of polymorphism in genes encoding mGluR 7, mGluR 8, GABA(A) receptor alfa-6 subunit and nociceptin/orphanin FQ receptor and panic disorder. Psychiatric Genetics. 2007;17:9. doi: 10.1097/YPG.0b013e32801118bc. [DOI] [PubMed] [Google Scholar]
- Kreek MJ. Opiates, opioids and addiction. Mol Psychiatry. 1996;1:232–254. [PubMed] [Google Scholar]
- Kreek MJ, Bart G, Lilly C, LaForge KS, Nielsen DA. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev. 2005;57:1–26. doi: 10.1124/pr.57.1.1. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Koob GF. Drug dependence: stress and dysregulation of brain reward pathways. Drug Alcohol Depend. 1998;51:23–47. doi: 10.1016/s0376-8716(98)00064-7. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Nielsen DA, LaForge KS. Genes associated with addiction: alcoholism, opiate, and cocaine addiction. Neuromolecular Med. 2004;5:85–108. doi: 10.1385/NMM:5:1:085. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. Genetic influences on impulsivity, risk taking, stress responsitivity and vulnerability to drug abuse and addiction. Nat Neurosci. 2005;8:1450–7. doi: 10.1038/nn1583. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The interaction of selection and linkage I. General considerations. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze BC, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35(4):605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Martin-Fardon R, Ciccocioppo R, Massi M, Weiss F. Nociceptin prevents stress-induced ethanol- but not cocaine-seeking behavior in rats. Neuroreport. 2000;11:1939–1943. doi: 10.1097/00001756-200006260-00026. [DOI] [PubMed] [Google Scholar]
- McLellen AT, Kushner H, Metzger D, Peters R, Smith I, Grisson G, et al. The fifth edition of the Addiction Severity Index. J Subst Abuse Treat. 1992;9:199–213. doi: 10.1016/0740-5472(92)90062-s. [DOI] [PubMed] [Google Scholar]
- Merino A, Buckbinder L, Mermelstein FH, Reinberg D. Phosphorylation of cellular proteins regulates their binding to the cAMP response element. J Biol Chem. 1989;264:21266–21276. [PubMed] [Google Scholar]
- Mitchell PJ, Timmons PM, Hebert JM, Rigby PW, Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- Mollereau C, Mouledous L. Tissue distribution of the opioid receptor-like (ORL1) receptor. Peptides. 2000;21:907–917. doi: 10.1016/s0196-9781(00)00227-8. [DOI] [PubMed] [Google Scholar]
- Mollereau C, Parmentier M, Mailleux P, Butour J-L, Moisand C, Chalon P, et al. ORL1, a novel member of the opioid receptor family: cloning, functional expression and localization. FEBS Lett. 1994;341:33–38. doi: 10.1016/0014-5793(94)80235-1. [DOI] [PubMed] [Google Scholar]
- Murphy NP, Lee Y, Maidment NT. Orphanin FQ/nociceptin blocks acquisition of morphine place preference. Brain Res. 1999;832:168–170. doi: 10.1016/s0006-8993(99)01425-0. [DOI] [PubMed] [Google Scholar]
- Murphy NP, Ly HT, Maidment NT. Intracerebroventricular orphanin FQ/nociceptin suppresses dopamine release in the nucleus accumbens of anaesthetized rats. Neuroscience. 1996;75:1–4. doi: 10.1016/0306-4522(96)00322-3. [DOI] [PubMed] [Google Scholar]
- Murphy NP, Maidment NT. Orphanin FQ/nociceptin modulation of mesolimbic dopamine transmission determined by microdialysis. J Neurochem. 1999;73:179–186. doi: 10.1046/j.1471-4159.1999.0730179.x. [DOI] [PubMed] [Google Scholar]
- Qin ZS, Niu T, Liu JS. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet. 2002;71(5):1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivich G, Bohatschek M, Da Costa C, Iwata O, Galiano M, Hristova M, et al. The AP-1 transcription factor c-Jun Is required for efficient axonal regeneration. Neuron. 2004;43(1):57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Reinscheid RK. The orphanin FQ/nociceptin receptor as a novel drug target in psychiatric disorders. CNS Neurol Disord Drug Targets. 2006;5:219–224. doi: 10.2174/187152706776359628. [DOI] [PubMed] [Google Scholar]
- Rettig RA, Yarmolinsky A, editors. Federal regulation of methadone treatment. Washington, DC: National Academy Press; 1995. [PubMed] [Google Scholar]
- Sakoori K, Murphy NP. Central administration of nociceptin/orphanin FQ blocks the acquisition of conditioned place preference to morphine and cocaine, but not conditioned place aversion to naloxone in mice. Psychopharmacology (Berl) 2004;172:129–136. doi: 10.1007/s00213-003-1643-3. [DOI] [PubMed] [Google Scholar]
- Schug J, Overton GC. TESS: Transcription element search software on the WWW Technical Report CBIL-TR-1997-1001-v00. Computational Biology and Informatics Laboratory. School of Medicine. University of Pennsylvania; 1998. URL: http://www.cbil.upenn.edu/tess. [Google Scholar]
- Sinnwell JP, Schaid DJ. haplo.stats: Statistical analysis of haplotypes with traits and covariates when linkage phase is ambiguous. R package version 1.4.3. 2009 URL: http://mayoresearch.mayo.edu/mayo/research/schaid_lab/software.cfm.
- Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. 3 New York: 1994. [Google Scholar]
- Storey JD, Tibshirani . Statistical significance for genome-wide experiments. 2003. [Google Scholar]
- Storey JD. The positive false discovery rate: A bayesian interpretation and the q-value. The Annals of Statistics. 2003;31:2013–2035. [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Ueda H, Yamaguchi T, Tokuyama S, Inoue M, Nishi M, Takeshima H. Partial loss of tolerance liability to morphine analgesia in mice lacking the nociceptin receptor gene. Neuroscience Lett. 1997;237:136–138. doi: 10.1016/s0304-3940(97)00832-x. [DOI] [PubMed] [Google Scholar]
- Walker JR, Spina M, Terenius L, Koob GF. Nociceptin fails to affect heroin self-administration in the rat. Neuroreport. 1998;9:2243–2247. doi: 10.1097/00001756-199807130-00017. [DOI] [PubMed] [Google Scholar]
- Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. American Journal of Human Genetics. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR. R News. 2003. The genetics package; p. 3. [Google Scholar]
- Wu X, Zhu K, Wang Z, Chen S, Yang L. GATA-1, -2, -3 genes expression in bone marrow microenvironment with chronic aplastic anemia. Hematology. 2007;12:331–335. doi: 10.1080/10245330701255288. [DOI] [PubMed] [Google Scholar]
- Wu YL, Pu L, Ling K, Zhao J, Cheng ZJ, Ma L, Pei G. Molecular characterization and functional expression of opioid receptor-like1 receptor. Cell Res. 1997;7:69–77. doi: 10.1038/cr.1997.8. [DOI] [PubMed] [Google Scholar]
- Xuei X, Flury-Wetherill L, Almasy L, Beirut L, Tischfield J, Schuckit M, et al. Assocation analysis of genes encoding the nociceptin receptor (OPRL1) and its endogenous ligand (PNOC) with alcohol or illicit drug dependence. Addict Biol. 2008;13:80–87. doi: 10.1111/j.1369-1600.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- Yu J, Chait BT, Toll L, Kreek MJ. Nociceptin in vitro biotransformation in human blood. Peptides. 1996;17:873–876. doi: 10.1016/0196-9781(96)00079-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelman ER, Schlussman SD, Ho A, Kreek MJ. Effect of the endogenous kappa opioid agonist dynorphin A(1–17) on cocaine-evoked increases in striatal dopamine levels and cocaine-induced place preference in C57BL/6J mice. Psychopharmacology. 2004;172:422–429. doi: 10.1007/s00213-003-1688-3. [DOI] [PubMed] [Google Scholar]


