Abstract
Visual Rhodopsins (VR) are recognized members of the large and diverse family of G protein-coupled receptors (GPCRs), but their evolutionary origin and relationships to other proteins, are not known. In an earlier publication (Shlykov et al., 2012), we characterized the 4-Toulene Sulfonate Uptake Permease (TSUP) family of transmembrane proteins, showing that these 7 or 8 TMS proteins arose by intragenic duplication of a 4 TMS-encoding gene, sometimes followed by loss of a terminal TMS. In this study, we show that the TSUP, GPCR and Microbial Rhodopsin (MR) families are related to each other and to six other currently recognized transport protein families. We designate this superfamily the Transporter-Opsin-G protein-coupled receptor (TOG) Superfamily. Despite their 8 TMS origins, members of most constituent families exhibit 7 TMS topologies that are well conserved, and these arose by loss of either the N-terminal (more frequent) or the C-terminal (less frequent) TMS, depending on the family. Phylogenetic analyses revealed familial relationships within the superfamily and protein relationships within each of the nine families. The statistical analyses leading to the conclusion of homology were confirmed using HMMs, Pfam, and 3D superimpositions. Proteins functioning by dissimilar mechanisms (channels, primary active transporters, secondary active transporters, group translocators and receptors) are interspersed on a phylogenetic tree of the TOG superfamily, suggesting that changes in the transport and energy-coupling mechanisms occurred multiple times during the evolution of this superfamily.
Keywords: Transport proteins, channels, secondary carriers, receptors, rhodopsin
Introduction
Using functional and phylogenetic information derived from over 10,000 publications on transport systems, our laboratory has been able to classify virtually all recognized transport proteins into over 700 families [1–2]. The resultant system of classification is summarized in the IUBMB-approved Transporter Classification (TC) Database (TCDB; http://www.tcdb.org) [3–4]. Our current efforts serve to identify distant relationships, allowing the placement of these families into superfamilies. Since transport systems play crucial roles in virtually all processes associated with life, their importance cannot be overstated [5–6].
The present study reports the identification of a novel superfamily, a group of proteins showing a common evolutionary origin, that we have designated the Transporter-Opsin-G protein-coupled receptor (TOG) superfamily, based on the best-characterized families of proteins present in this superfamily. In addition to (1) ion-translocating Microbial Rhodopsins (MR; TC# 3.E.1) and (2) G protein-coupled receptors (GPCRs; TC# 9.A.14) including visual rhodopsins (VRs), we show that members of the following families (see Table 1) share a common origin with microbial, invertebrate and vertebrate rhodopsins: (3) Sweet sugar transporters (Sweet; TC# 9.A.58), (4) Nicotinamide Ribonucleoside Uptake Permeases (PnuC; TC# 4.B.1), (5) 4-Toluene Sulfonate Uptake Permeases (TSUP; TC# 2.A.102), (6) Ni2+-Co2+ Transporters (NiCoT; TC# 2.A.52), (7) Organic Solute Transporters (OST; TC# 2.A.82), (8) Phosphate:Na+ Symporters (PNaS; TC# 2.A.58), and (9) Lysosomal Cystine Transporters (LCT; TC# 2.A.43). Furthermore, our research indicates that the invertebrate Heteromeric Odorant Receptor Channel (HORC; TC# 1.A.69) family may also share a common origin with members of the TOG superfamily, although this could not be established using our standard statistical criteria.
Table 1. Summary of nine TOG Superfamily family members.
The family number, name, abbreviation, TC number, number of TMSs, normal protein size range in numbers of amino acyl residues, dominant topology, TMS gain or loss, organismal distribution and Pfam id designation are presented. Minor topologies, represented in a minority of family members, are listed in small numbers in parentheses in column 7. Putative topological transitions are described in footnotes 1–4.
| Family # | Family Name | Family Abb’n | TC # | # TMSs | Common Protein Size Range | Topology | TMS Gain or Loss (Primary) | Organismal Distribution | Pfam id |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Ion-Translocating Microbial Rhodopsin | MR | 3.E.1 | 7 | 250–350 | 3 + 1 + 3 | 7 arose from 8 by loss of the N-terminal TMS. | Archaea, Bacteria, Eukaryota | Bac_rhodopsin |
| 2 | Sweet1 | Sweet | 9.A.58 | 3 or 7 | 200–290 | 3 + 1 + 3 (3) | 7 arose from 8 by loss of the N-terminal TMS. | Archaea, Bacteria, Eukaryota | MtN3_slv |
| 3 | Nicotinamide Ribonucleotide Uptake Permease | PnuC | 4.B.1 | 7 or 8 | 210–270 | 3 + 1 + 3 (4 + 4) | 7 arose from 8 by loss of the N-terminal TMS. | Bacteria, Eukaryota | NMN_transporter, AAA_28 |
| 4 | 4-Toluene Sulfonate Uptake Permease2 | TSUP | 2.A.102 | 7–9 | 250–600 | 4 + 4 (3 + 1 + 3; 1 + 4 + 4) | 8 arose from internal duplication of 4 TMSs | Archaea, Bacteria, Eukaryota | TauE |
| 5 | Ni2+-Co2+ Transporter3 | NiCoT | 2.A.52 | 6–8 | 300–380 | 4 + 4 (3 + 1 + 3; 2 + 1 + 1 + 2) | 8 arose from internal duplication of 4 TMSs | Archaea, Bacteria, Eukaryota | NicO |
| 6 | Organic Solute Transporter | OST | 2.A.82 | 7 | 180–400 | 3 + 1 + 3 | 7 arose from 8 by loss of the N-terminal TMS. | Eukaryota | Solute_trans_a |
| 7 | Phosphate:Na+ Symporter4 | PNaS | 2.A.58 | 8 or 9 | 500–700 | 4 + 4 (4 + 4 + 1) | 8 arose from internal duplication of 4 TMSs | Bacteria, Eukaryota | Na_Pi_cotrans |
| 8 | Lysosomal Cystine Transporter | LCT | 2.A.43 | 7 | 300–400 | 3 + 1 + 3 | 7 arose from 8 by loss of the N-terminal TMS. | Eukaryota | PQ-loop |
| 9 | G-protein Coupled Receptor | GPCR | 9.A.14 | 7 | 300–1200 | 3 + 1 + 3 | 7 arose from 8 by loss of the N-terminal TMS 1. | Eukaryota | 7tm_1, 7tm_2, 7_tm3 |
3 arose from 4 TMSs by loss of one TMS.
7 arose from 8 by loss of the N-terminal TMS; 9 arose from 8 by gain of an N-terminal TMS.
7 arose from 8 by loss of the 8th (C-terminal) TMS; 6 arose from 7 by loss of the N-terminal TMS.
9 arose from 8 by gain of a C-terminal TMS.
Our evidence suggests that all of the diverse proteins included in the TOG superfamily derive from a common ancestor via similar pathways. It can therefore be anticipated that the structures of most of these proteins will exhibit common features [7–8]. Since rhodopsins are the transmembrane proteins with the highest resolution X-ray structures solved to date [9–11], we now have the capacity to apply this structural information to the other protein families included within this superfamily. The work reported here provides the groundwork for comparative studies that should lead to a more detailed understanding of how a single structural scaffold can vary to accommodate a wide diversity of functions. It should serve as a guide in future studies revealing how sequence divergence can lead to alterations in this scaffold.
Results
All protein families within TCDB belonging to subclass 2.A consist of electrochemical potential-driven uniporters, symporters and antiporters. Based on preliminary evidence reported by Shlykov et al. (2012), we used a modified SSearch program [12–13] to compare Toluene Sulfonate Uptake Permease (TSUP) homologues with all other secondary carriers and identified potential superfamily relationships. Subsequently, these analyses were extended to other TC classes. Comparisons to the TC subclasses 9.A, 3.E and 4.B proved fruitful.
The MR and LCT families had previously been shown to be related [14]. Analyses dealing with the MR, LCT, PnuC, PNaS, Sweet, and GPCR families provided sufficient evidence to include them in the TOG superfamily. Our results led to the formulation of the novel TOG superfamily for which trees were generated using the ClustalX, Superfamily Tree 1 (SFT1) and SFT2 programs [15–17].
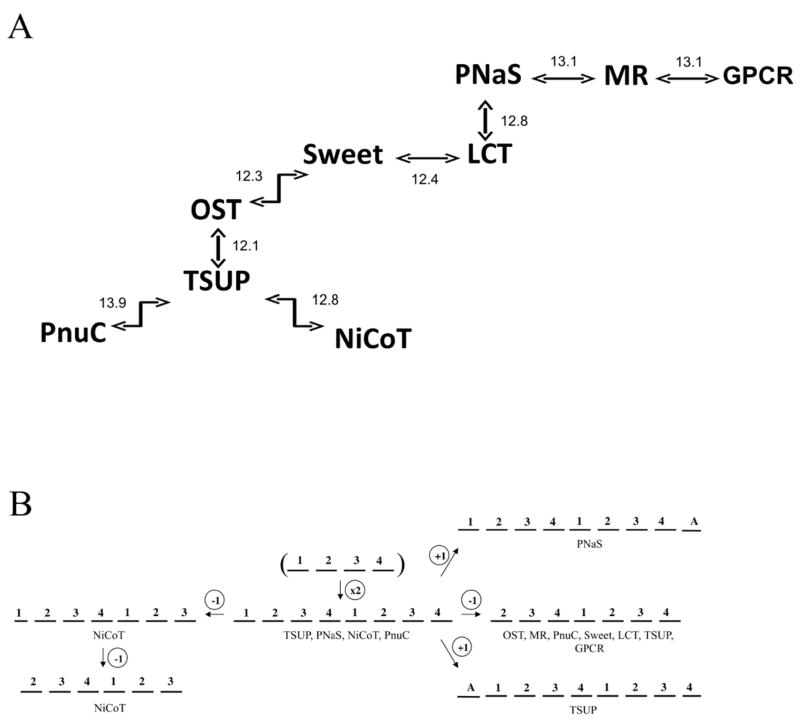
The TOG superfamily consists of nine, and possibly ten, currently recognized protein families with members of primarily 6, 7, 8 and 9 putative TMSs (Table 1). A summary of the comparisons performed is presented in Figure 1A and Table 2, while the proposed evolutionary pathway for the appearance of the different members of the TOG superfamily is presented in Figure 1B. The TSUP family has been characterized previously [18]. Therefore, unlike the other eight families, the AveHAS plots, phylogenetic trees and a table listing TSUP homologues have been excluded from this study and are reported by Shlykov et al. [18].
Figure 1.
(A): TOG superfamily homology established through the use of GSAT/GAP and the Superfamily Principle. TOG superfamily proteins from TCDB and their homologues were used to establish homology between all members of the nine families. GSAT/GAP comparison scores, adjacent to the arrows, are expressed in terms of standard deviations (S.D.). (B): Proposed evolutionary pathway for the appearance of nine recognized families within the TOG superfamily. The TOG superfamily is believed to have arisen from a 4 TMS precursor that duplicated to an 8 TMS precursor, common to the superfamily constituents, before diverging in topology via the loss or gain of specific TMSs.
Table 2. The highest comparison scores between TOG superfamily members.
Protein representatives of families used in these comparisons are found in tables S1–S9. Representative alignments for highlighted squares are usually provided in figures S1–S7 and S9–S10, but in some cases better scores are reported here than in the figures, based on other alignments. Values above 12.0 S.D., which are considered sufficient to establish homology and interconnect families, are shaded. These values are sufficient to establish homology based on the criteria discussed in the methods section1,2.
| 9.A.14 GPCR |
2.A.102 TSUP |
2.A.82 OST |
3.E.1 MR |
9.A.58 Sweet |
2.A.52 NiCoT |
2.A.58 PNaS |
4.B.1 PnuC |
|
|---|---|---|---|---|---|---|---|---|
| 9.A.14 GPCR |
||||||||
| 2.A.102 TSUP |
11.4 S.D. (2 TMSs) |
|||||||
| 2.A.82 OST |
10.8 S.D. (2 TMSs) |
12.1 S.D. (2 TMSs) |
||||||
| 3.E.1 MR |
13.1 S.D. (2 TMSs) |
11.0 S.D. (2 TMSs) |
12.4 S.D. (3 TMSs) |
|||||
| 9.A.58 Sweet |
12.3 S.D. (2 TMSs) |
10.1 S.D. (3 TMSs) |
12.3 S.D. (2 TMSs) |
11.2 S.D. (2 TMSs) |
||||
| 2.A.52 NiCoT |
10.4 S.D. (2 TMSs) |
12.8 S.D. (3 TMSs) |
11.9 S.D. (2 TMSs) |
10.7 S.D. (2 TMSs) |
11.8 S.D. (3 TMSs) |
|||
| 2.A.58 PNaS |
11.2 S.D. (2 TMSs) |
10.6 S.D. (2 TMSs) |
11.5 S.D. (4 TMSs) |
13.1 S.D. (3 TMSs) |
11.6 S.D. (2 TMSs) |
12.2 S.D. (3 TMSs) |
||
| 4.B.1 PnuC |
10.2 S.D. (3 TMSs) |
13.9 S.D. (2 TMSs) |
11.4 S.D. (2 TMSs) |
10.9 S.D. (2 TMSs) |
13.1 S.D. (3 TMSs) |
11.2 S.D. (2 TMSs) |
9.8 S.D. (2 TMSs) |
|
| 2.A.43 LCT |
9.4 S.D. (3 TMSs) |
11.3 S.D. (2 TMSs) |
11.1 S.D. (2 TMSs) |
11.0 S.D. (2 TMSs) |
13.6 S.D. (2 TMSs) |
11.5 S.D. (2 TMSs) |
12.8 S.D. (3 TMSs) |
11.3 S.D. (3 TMSs) |
Average comparison score and average number of TMSs in all TOG superfamily alignments: 2.3 TMSs; 11.5 S.D.
Average comparison score and average number of TMSs in alignments used to establish homology and interconnect all families within the TOG superfamily: 2.5 TMSs; 12.8
The Lysosomal Cystine Transporter (LCT) and the Ion-Translocating Microbial Rhodopsin (MR) Families (TC# 2.A.43 and 3.E.1, respectively)
The evolutionary pathway of the 7 TMS LCT family has been elucidated [14], and LCT family members were found to be homologous to members of the MR family, including putative fungal chaperone proteins present in the Microbial Rhodopsin family (see Table 1 and TC entries under TC# 3.E.1). Most of the known MR transporters are light-driven ion pumps or light-activated ion channels.
LCT family members range in size from 300 to 400 amino acyl residues (aas) and are generally larger than MR proteins, which have ~220 to 300 residues. Eukaryotic homologues within a single transporter family tend to be ~40% larger than their bacterial homologues [19]. Whereas the LCT family is found exclusively in the eukaryotic domain, the MR family is present in all three domains of life (Table 1). Despite these differences, both families possess a 7 TMS topology (Tables 1, S1, S2 and Figs. S1A–B, S2A–B).
TMSs 1–3 in LCT family members duplicated to give rise to TMSs 5–7, with TMS 4 showing insignificant sequence similarity to any one of the other six TMSs [14]. The precursor could have been an 8 TMS protein that generated the present-day 7 TMS proteins by loss of TMS 1 or 8, and strong evidence for this possibility is presented in [18] and here.
The 7 TMS Bba2 protein of the LCT family is homologous to the 7 TMS Aae2 protein of the Sweet family. The alignment between these two proteins using GSAT yielded a comparison score of 12.4 S.D. (Fig. S1C). These comparisons establish homology between the LCT and Sweet families. TMSs 3–7 of Aae2 aligned with TMSs 3–7 of Bba2, demonstrating that the two families both arose via the same evolutionary pathway (Fig. S1C). Loss of TMS 1 in an 8 TMS predecessor yielded the 7 TMS topology found in members of the LCT and Sweet families.
Expansion of the TOG superfamily resulted from comparisons between the LCT and PNaS families. Comparing TMSs 2–4 of LCT Ago1 (7 putative TMSs) with TMSs 6–8 of PNaS Cre1 (11 putative TMSs) yielded a comparison score of 12.8 S.D. (Fig. S1D). This comparison establishes homology between regions of proteins in the LCT and PNaS families and further supports the proposed evolutionary pathway for the LCT family, as TMSs 2–4 of Ago1 and TMSs 6–8 of Cre1 correspond to the last three TMSs in the proposed 4 TMS predecessor. PSI-BLAST searches of Cre1 yielded two separate conserved PNaS domains within the protein. The extended 11 TMS topology in Cre1 likely arose from the fusion of a 7 TMS protein with another 4 TMS repeat unit.
The Ni2+-Co2+ Transporter (NiCoT) Family (TC# 2.A.52)
Members of subfamily 1 within the ubiquitous NiCoT family are typically 300 to 380 aas in size and possess 6–8 putative TMSs [20] (Table S3 and Figs. S3A–B). NiCoT subfamily 2 is comprised of distant homologues of great size, sequence and topological variation. NiCoT transporters catalyze the uptake of Ni2+ and Co2+ using a pmf-dependent mechanism; however, a Ni2+ and Co2+ resistance protein that is believed to export the two metals to the external environment has been reported [21–22]. Smaller members of subfamily 2 exhibit 6–8 putative TMSs.
Figure 6. Structural superimposition of bovine and bacteriorhodopsin.
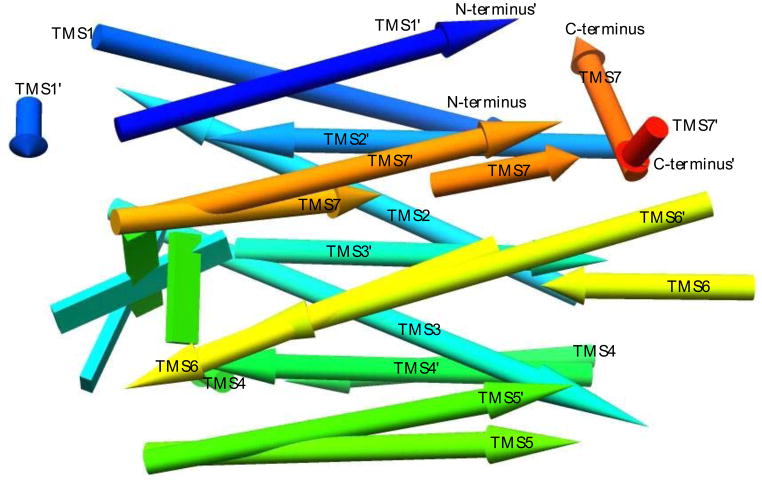
A superimposition of a bovine (Visual) rhodopsin (VR) structure (PDB:1F88, chain A) and a bacteriorhodopsin (MR) structure (PDB:3NS0), both containing 7 TMSs, was performed in Chimera 1.7. The prime (‘) notation indicates TMS numbering of the MR structure. The overall RMSD is 7.176. While the protein is shown as a set of helices, these can deviate from perfect helices. When such data are used as templates to switch to the “Pipes and Planks” (α-helices and β-sheets) representation mode in Chimera, idealized “Pipes” (helices) are placed in a way that best represents the current separate stretches of helix-annotated sequences. Several idealized helices are presented, each representing its own discontinuous helix segment. The result is that the idealized TMS arrow is placed in a way that shows a compromise of how that TMS has traversed the membrane. In the figure, all helices point towards the N-termini, except constituent helices of the C-terminal TMSs which point towards the C-termini. The coloring is a scale starting from the N-termini (blue) and ending in the C-termini (red), going through an intermediate color scale from blue to light blue to turquoise to spring green to light green to yellow, orange, and finally red. The color scale is placed on the sequence considering the entire sequence including the non-TMS sequence which is not shown in the figure.
Comparing TMSs 1–3 of TSUP Pla1 (8 putative TMSs) with TMSs 4–6 of NiCoT Bja1 (6 putative TMSs) yielded a comparison score of 12.8 S.D. (Fig. S3C). This comparison establishes homology between members of these two families and serves to confirm our proposed evolutionary pathway for the appearance of the NiCoT family as a member of the TOG superfamily (Figs. 1A and 1B). Based on alignments, it is likely that the 6 TMS NiCoT proteins arose by the loss of both TMSs 1 and 8 after the 4 TMS intragenic duplication event took place.
The Organic Solute Transporter (OST) Family TC# 2.A.82
Members of the OST family are almost exclusive to animals and are known to transport organic anions including estrone-3-sulfate, bile acids, taurocholate, digoxin and prostaglandins [23–25]. Distant homologues of the α-subunits in plants, fungi and bacteria were retrieved in NCBI searches, but their scores usually bordered or fell below our threshold cutoff for establishing homology. Furthermore, each well characterized transporter within this family functions as part of a two-component system utilizing the α-(280–400 aas) and β-subunits (180–290 aas). The α-subunits generally contain seven TMSs, whereas the β-subunits contain only one. To date, neither subunit has been found to function without the other (Table S4 and Figs. S4A–B) [23].
Comparing TMSs 2–3 of TSUP Tsp1 (8 putative TMSs) with TMSs 1–2 of OST Cre2 (7 putative TMSs) yielded a comparison score of 12.1 S.D. (Fig. S4C). This comparison demonstrates the loss of TMS 1 in OST transporters and establishes homology between the two families; the loss of TMS 1 from an 8 TMS precursor generated the 7 TMS topology of the OST family. Another alignment between TMSs 2–4 of TSUP Gfo1 (7 putative TMSs) with TMSs 2–4 of OST Dre1 (7 TMSs) also supports homology between the TSUP and OST families and the proposed evolutionary pathway. It yielded a comparison score of 11.3 S.D. (Fig. S4D).
The Sweet (PQ-loop; Saliva; MtN3) (Sweet) Family TC# 9.A.58
Eukaryotic Sweet family channels or carriers catalyze facilitated diffusion (uptake or efflux) of sugars across the ER and plasma membranes of plants and animals [26]. Bacterial pathogens upregulate specific plant Sweet transporters, allowing them to utilize the sugar efflux function of these proteins to meet their energy needs [27]. Eukaryotic homologues possess 7 TMSs in a 3 + 1 + 3 repeat arrangement and are 200–290 aas in size (Table S5 and Figs. S5A–B). Although 7 TMS bacterial homologues exist, most bacterial putative Sweet channels possess 3 TMSs and are about half the size of their eukaryotic and larger bacterial relatives. The 3 TMS proteins show greatest sequence similarity to the first (N-terminal) repeat in the 7 TMS proteins. It is unclear whether the eukaryotic or prokaryotic proteins function as channels or carriers. However, no well-documented examples of carriers with fewer than 4 TMSs per polypeptide chain have been reported, suggesting that the 3 TMS proteins may function as oligomeric channels [28].
Comparing TMSs 6–7 of Sweet Rco4 (7 putative TMSs) with TMSs 6–7 of OST Ath8 (7 putative TMSs) yielded a comparison score of 12.3 S.D. (Fig. S5C). This result indicates that as for the MR and OST families (as well as several other TOG superfamily members), the N-terminal TMS was lost from the 8 TMS topology to generate the 7 TMS Sweet proteins. A second alignment between TMSs 4–6 of Sweet Asu3 (7 putative TMSs) and TMSs 4–6 of OST Ncr1 (7 putative TMSs) also yielded a comparison score of 10.3 S.D. (Fig. S5D), further confirming the establishment of homology between Sweet and OST families.
The Phosphate:Na+Symporter (PNaS) Family TC# 2.A.58
Both bacterial and eukaryotic PNaS homologues usually range in size between 500 and 700 residues, but the bacterial homologues can be as small as 350 residues. Most members of this family possess 8 or 9 TMSs in a 4 + 4 or a 4 + 4 + 1 TMS arrangement, which has been demonstrated previously (Saier, 2003; Table S6 and Fig. S6A–B). However, some proteins such as NPT2 of Rattus norvegicus can have as many as 12 TMSs, with the extra ones appearing at the N-termini [29]. Mammalian PNaS porters may catalyze the electroneutral cotransport of 3 Na+ with inorganic phosphate (Pi). Their activities can be regulated by parathyroid hormone and dietary Pi.
Comparing TMSs 4–6 of PNaS Odi8 (8 putative TMSs) with TMSs 3–5 of MR Hwa1 (7 putative TMSs) yielded a comparison score of 13.1 S.D. (Fig. S6C). This comparison demonstrates homology between the MR and PNaS families and further supports the conclusion that TMS loss in PNaS family members occurred at their N-termini.
The Nicotinamide Ribonucleotide Uptake Permease (PnuC) Family TC# 4.B.1
PnuC family proteins are restricted to bacteria and archaea as well as several bacteriophage. These proteins possess 8 or 7 TMSs in a 4 + 4 or 3 + 1 + 3 repeat arrangement, respectively. The 7 TMS proteins arose by the loss of the N-terminal TMS in the 8 TMS homologues. Some members may be energized by multifunctional NadR homologues, which perform the required step of phosphorylating nicotinamide ribonucleoside (NR), thus allowing its transport in a “group translocation” or “metabolic trapping” process [30–32]. The ribonucleoside kinase domains of NadR homologues are responsible for the transfer of a phosphoryl group from ATP onto NR [33–34]. Therefore, ATP appears to be required for NR accumulation. Proteins of the PnuC family are typically 210 to 270 aas in size (Table S7 and Figs. S7A–B).
Comparing TMSs 2–3 of PnuC Spr1 (7 putative TMSs) with TMSs 3–4 of TSUP Cba4 (8 putative TMSs) yielded a comparison score of 12.4 S.D. (Fig. S7C). This comparison demonstrates homology between the PnuC and TSUP families. An alignment between TMSs 3–6 of PnuC Sde2 (7 putative TMSs) and TMSs 4–6 of TSUP Ere1 (8 putative TMSs) provides additional evidence of homology and supports the PnuC evolutionary pathway (Fig S8D). Our results and the placement of the PnuC family into the TOG superfamily supports the proposal that a 4 TMS precursor duplicated to give 8 TMS proteins, and that the N-terminal TMS was then deleted.
The G protein-coupled Receptor (GPCR) Family TC# 9.A.14
Members of the GPCR family [35–41] encompass an extremely diverse range of cellular membrane proteins and constitute the largest family of transmembrane proteins found in humans [39, 42]. While all share a general signaling mechanism wherein extracellular signals are transduced into intracellular effectors via ligand binding, members vary tremendously in both ligand type and function. GPCR family members each consists of a 7 TMS α-helical bundle, connected by three extracellular and three intracellular loops. This 7 TMS bundle displays distinctive hydrophobic patterns (Fig. S8A and S8E) and is commonly recognized as the most conserved element of GPCRs [43]. Because the GPCR family includes receptors for a wide variety of hormones, neurotransmitters, chemokines, calcium ions and photons (see Tables S8A, S8B and TCDB), they are among the most targeted therapeutic proteins for drugs, and their analyses have tremendous implications for future pharmacological developments [44].
Comparing TMSs 5–6 of the GPCR, Dre1 (7 putative TMSs) with TMSs 5–6 of Mos1 (7 putative TMSs) of the Microbial Rhodopsin (MR) family yielded a comparison score of 13.1 S.D. (Fig. S8C). This comparison establishes homology between the GPCR family and the MR family; the topology of members of the GPCR family, like the MR family, likely arose from the loss of the N-terminal TMS from the proposed 8 TMS predecessor. TMSs 1–4 of GPCR Dre 1 (7 putative TMSs) also aligned with TMSs 1–4 of MR Cga1 (7 putative TMSs) and yielded a comparison score of 10.6 S.D., further supporting establishment of homology (Fig S7D) between GPCRs and MRs.
The Heteromeric Odorant Receptor Channel (HORC) Family TC# 1.A.69
Olfactory sensory neurons in insects express between one to three members of the channel-forming olfactory receptor (OR) gene family as well as the highly conserved Or83b co-receptor TC# 1.A.69.1.1. Functional odorant receptors consist of a heteromeric complex comprised of at least one odorant-binding subunit and the aforementioned Or83b co-receptor [45]. Immunocytochemical experiments demonstrated that insect odorant receptors possess a 7 transmembrane topology, but in contrast to members of the GPCR family, they exhibit a cytoplasmic N-terminus and extracellular C-terminus. Several authors [45–47] suggested that heteromeric insect ORs comprise a new class of ligand-activated non-selective cation channels. We obtained preliminary evidence that insect ORs and GPCRs are homologous. However, based on our criteria, we could not establish homology because comparison scores were insufficient (10.3 S.D.). Nevertheless, the intriguing possibility of homology will provide the basis for future investigations.
Controls: the Major Intrinsic Protein (MIP) Family (TC# 1.A.8) and the Mitochondrial Carrier (MC) Family (TC# 2.A.29)
Members of the Major Intrinsic Protein (MIP) family are channel proteins that function in water, small carbohydrate, urea, NH3, CO2, H2O2 and ion transport by energy-independent mechanisms. The observed topology of the MIP family arose from the intragenic duplication of a 3 TMS predecessor [48]. Members of the Mitochondrial Carrier (MC) family are transporters involved in transporting keto acids, amino acids, nucleotides, inorganic ions and co-factors across the mitochondrial inner membrane. Proteins from the MC family arose from tandem intragenic triplication of a 2 TMS element, giving rise to a 6 TMS topology [49–50]. These two large 6 TMS protein families thus arose via different pathways and are not homologous. They provide an excellent control for homology.
The best comparison scores between the MC and MIP families and TOG superfamily members were 9.5 S.D., and 10.5 S.D., respectively (Table S9). Comparisons of the MC family against the NiCoT and PNaS families yielded a maximal comparison score of 9.5 S.D. A comparison of the MIP family against the PNaS family yielded a maximal comparison score of 10.5 S.D. The average score for all of the best comparisons between TOG superfamily members and the MC family was 8.8 S.D., and the average score for comparisons between TOG superfamily members and the MIP family was 8.9 S.D. When compared to each other, the MIP and MC families yielded maximal comparison scores of 9.2 S.D. By contrast, the average score for all of the best comparisons for the nine TOG superfamily families with each other was 11.5 S.D., and 12.6 S.D. between families used to establish homology. Based on these results, we suggest that 12.0 S.D. (Fig. S9) combined with the proper alignment of at least 2 transmembrane domains that fit a proposed evolutionary pathway, is sufficient to provide strong evidence for homology.
As a negative control, we searched for similarities between the MC (TC# 2.A.29) and MIP (TC# 1.A.8) families using Pfam-A. We found that even considering weak similarities, using the default cutoff of 10, these families showed links only through Pfam family PF12822 (DUF3816), an uncharacterized 5 TMS protein family. The edge linking 1.A.8.1.1 and DUF3816 scored only 2.8 (the best edge between MIP and DUF3816), considerably worse than any of the similarities we have reported to substantiate our conclusions about homology between members of the TOG superfamily. These results further establish the common origin of the family members of the TOG superfamily.
Integration of Topological Data
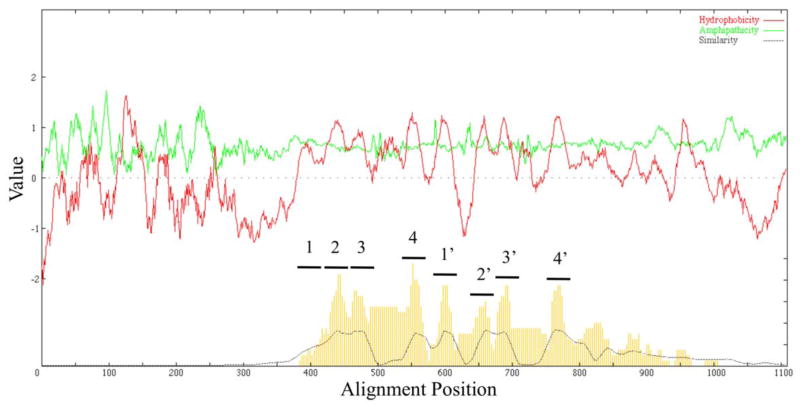
Using a phylogenetic tree that includes members of the nine established families of the TOG superfamily, proteins from each phylogenetic cluster were chosen and combined into a single file. The proteins were then aligned using ClustalX [51], and AveHAS plots [52] were generated for all families except the GPCR family (Fig. 2), as well as one for all families (Fig. S10). The large GPCR homologues rendered the AveHAS plot in Fig. S10 too large for easy viewing.
Figure 2. Average hydropathy, amphipathicity and similarity (AveHAS) plots based on a ClustalX multiple alignment.
All members of all TOG Superfamily families from TCDB were included except for the GPCR (see Fig. S10) family. The plot reveals 8 well conserved average TMSs. However, as many as 6 poorly conserved peaks of hydrophobicity can be seen representing additional potential TMSs in the non-homologous regions of some proteins.
The plot reveals 7 well-conserved peaks of hydrophobicity with moderate amphipathic nature (peaks 2–8 in Fig. 2), as well as a poorly conserved peak (peak 1). This result is expected, given that the majority of the families consist predominantly of 7 TMS proteins. TMS 1 in the 8 TMS homologues is conserved in only a few of the family members. Other less conserved peaks of hydrophobicity are found N- and C-terminal to the 7 well-conserved peaks. A closer look revealed that these peaks are primarily due to the larger PNaS homologues. The 400 residue extension at the N-terminal end of the alignment is attributable in part to the Sko2 protein of the PNaS family. A CDD search identified a member of the Death Domain (DD) superfamily constituting approximately the first 100 residues of the Sko2 N-terminus; DDs participate in protein-protein interactions in signaling pathways by recruiting proteins to complexes that sometimes comprise apoptosis pathways [53]. This accessory signaling domain in some PNaS proteins is not unexpected given their roles in phosphate reabsorption in mammalian tissues [54].
Phylogenetic Analyses of the TOG Superfamily
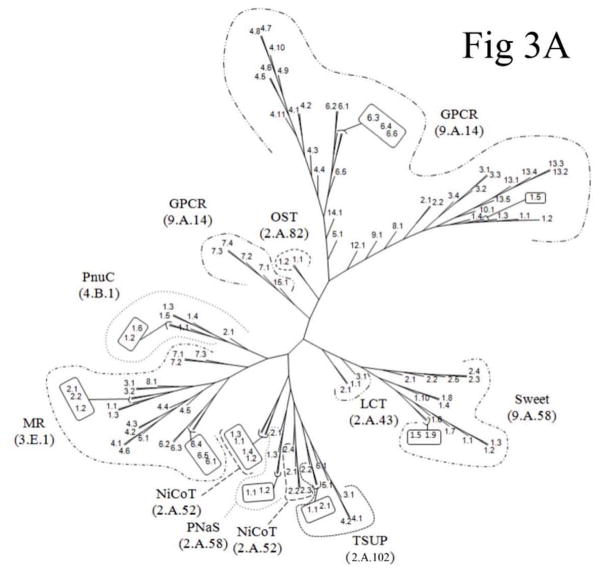
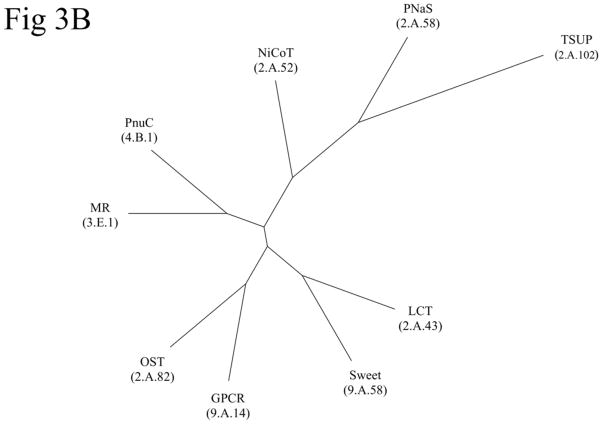
Proteins found in TCDB, representing the various subfamilies within each family (except the GPCR family) of the TOG superfamily, were used to generate a tree using the ClustalX neighbor-joining method (Fig. S11). The same proteins were then used to generate a tree using the BLAST-bit score-based SFT1 method (Fig. 3A) [15–17]. In Figure S11, the ClustalX/TreeView program drew the GPCR family (TC# 9.A.14) in five distinct clusters, the TSUP (TC# 2.A.102) family in three clusters, and the PnuC (TC# 4.B.1), LCT (TC# 2.A.43), NiCoT (TC# 2.A.52) and PNaS (TC# 2.A.58) families each in two clusters. Only members of the MR (TC# 3.E.1), Sweet (TC# 9.A.58) and OST (TC# 2.A.82) families clustered coherently within a single cluster according to their respective TC family assignments. This situation contrasts with the SFT1 tree (Figure 3A), which shows clustering of nearly all protein members coherently according to their respective families with the exceptions of the GPCR and NiCoT families, which are found in two closely related clusters. All members of NiCoT subfamily 1 (TC# 2.A.52.1) comprise one cluster, and all members within NiCoT subfamily 2 (TC# 2.A.52.2) comprise the other. These results reveal the superiority of the SFT1 program over the ClustalX program, an observation now noted for many sequence-divergent superfamilies where multiple alignments are not reliable [15–17]. The SFT2 tree (Fig. 3B) shows the phylogenetic relationships between all nine families within the TOG superfamily. Interestingly, the families that have lost TMS 1 cluster together at the bottom of the tree, suggesting that this event could have occurred before these families diverged from each other.
Figure 3. Phylogenetic (Fitch) trees for the TOG superfamily.
Phylogenetic (Fitch) trees for the TOG superfamily in TCDB as of May, 2012. Three different methods of tree construction were used: (A) The BLAST-derived SFT1 program shows the proteins of families within the TOG superfamily; (B) the SFT2-based tree shows the relationships of the TOG superfamily families to each other. In (A), numbers adjacent to the branches indicate the protein TC#’s (last two digits of the complete protein TC#) while the family designations and family TC numbers are provided in large print in parentheses. In (B), family abbreviations are presented with TC family numbers in parentheses. See TCDB for protein identification. The ClustalX-Tree View tree is presented in supplementary figure S11.
Analyses of Internal Repeats
Shlykov et al. [18] previously reported internal repeats within TSUP family members that corresponded to a 4 transmembrane α-helical (TMS) structural precursor [18], and Zhai et al. [14] demonstrated that TMSs 1–3 are homologous to TMSs 5–7 in the 7 TMS Microbial Rhodopsins (MR). More recently Sun and Saier (unpublished results) demonstrated that TMSs 1–3 are homologous to TMSs 5–7 in members of the PNaS family (14.6 S.D.). Using the AR and GSAT programs [13], comparing TMSs 1–4 with TMSs 5–8 of the 8 TMS TSUP Pas1 protein yielded a comparison score of 15.2 S.D. (Fig. 4), demonstrating that an intragenic 4 TMS duplication event occurred in TSUP family members. The 4 TMS unit duplicated to yield an 8 TMS protein. By the Superfamily Principle, the internal repeats in the TSUP family are applicable to all families within the TOG superfamily. The evolutionary pathway elucidated for the TSUP and MR families explains the alignment of specific transmembrane domains in the two halves of various families within the TOG superfamily (data not shown).
Figure 4. Demonstration of a 4 TMS repeat unit in the Pas1 protein of the TSUP family.

GSAT alignment of TMSs 1–4 of Pas1 (Photorhabdus asymbiotica; gi211638062; 8 TMSs) with TMSs 5–8 of the same protein. A comparison score of 15.2 S.D. was obtained with 50.5% similarity and 30.3% identity. Identical residues are indicated by vertical lines, close similarities are indicated by colons, and more distant similarities are indicated by periods. GSAT was set at default settings with a gap creation penalty of 8 and a gap extension penalty of 2 with 500 random shuffles.
Non-TOG Superfamily proteins previously reported to be related
A recent study [55] described two new Pfam families, one of which (7TMR_DISM) was claimed to be a bacterial family with a domain organization related to mammalian glutamate GPCRs. The other family (7TMR_HD) was reported to be peripherally related to 2.A.102.1.1 of the TSUP family (using Pfam’s HMMs). However, it appears that the comparisons were not performed at the sequence level, and that other 7 TM transporter families were not included in the screen. Using TC- and NCBI PSI-BLAST as well as Protocols 1 and 2 searches, we could not obtain convincing evidence that membrane domains of these sequences are related to the proteins in the TOG superfamily.
Mapping of TC# 9.A.14 and the GRAFS system
HMMER 3.0 was used to map the GPCR family (TC# 9.A.14) and GRAFS in Cytoscape 2.8.3, and a spring embedded layout was applied. A clear clustering pattern was evident, with the Secretin and Adhesion receptor families forming a cluster on one side of the Rhodopsin cluster. Frizzled/Taste2 had two connectors with two sequences also in the Secretin/Adhesion cluster.
The glutamate receptors formed a small cluster. Two sequences within the Secretin/Adhesion receptor family linked these receptors to the Rhodopsin cluster (which includes somatostatin receptors, opioid receptors, galanin and the RF-amide binding receptors (SOG) and Opsin). Five sequences among the Glutamate receptors connected this cluster with the Rhodopsin cluster (all to SOG). All 12 types of Rhodopsins showed good representation with edges to sequences in TCDB. This was clearly the largest and most compact cluster. Twenty six sequences included in TC# 9.A.14 comprise the Rhodopsin GPCRs while 28 sequences in TC# 9.A.14 are non-Rhodopsin members.
TC# and Pfam family correspondance
Specific Pfam families corresponded to our TCDB families: TSUP (TC# 2.A.102) => PF01925 (TauE); LCT (2.A.43) =>PF04193 (PQ-loop); NiCoT (2.A.52) => PF03824 (NicO); PNaS (2.A.58) => PF02690 (Na_Pi_cotrans); OST (2.A.82) => PF03619 (Solute_trans_a); MR (3.E.1) => PF01036 (Bac_rhodopsin); PnuC (4.B.1) =>PF13521 (AAA_28) & PF04973 (NMN_transporter), and Sweet (9.A.58) => PF03083 (MtN3_slv). We also checked the following clans (Pfam’s “clans” are superfamilies of similar Pfam families), to see if obvious relationships could be detected between them: TauE and NicO (same clan), PQ_LOOP and MtN3_slv (same clan), Na_Pi_cotrans (not a member of a clan), Solute_trans_a (not a member of a clan), Bac_rhodopsin (in a large clan called GPCR_A – containing 7TM-7TMR_HD and many GPCRs including the 7tm_1 family, a central node of rhodopsin GPCRs), AAA_28 (in a large clan called P-loop_NTPase) and NMN_transporter (not a member of a clan). Thus, Pfam analyses yielded confirmatory evidence for relatedness among several of the TOG superfamily families.
Statistics of the 9.A.14 network compared with the entire TOG-Pfam network
We used Network Analyzer, treating the network as undirected, comparing TC# 9.A.14 with the entire network. The number of connected components was 4; the network diameter was 16; the network radius was 1; the network centralization was 0.060 (0.175 for TC# 9.A.14); the shortest path is 168890 (87%); the characteristic path length was 6.229; the average number of neighbors was 2.832 (5.074 for TC# 9.A.14); the network density was 0.006 (0.096 for TC# 9.A.14), and the network heterogeneity was 1.210 (0.553 for TC# 9.A.14). This shows that the network of TC# 9.A.14 has higher density and lower heterogeneity than the entire TOG-Pfam network.
Location of the GPCRs within the TOG-Pfam network
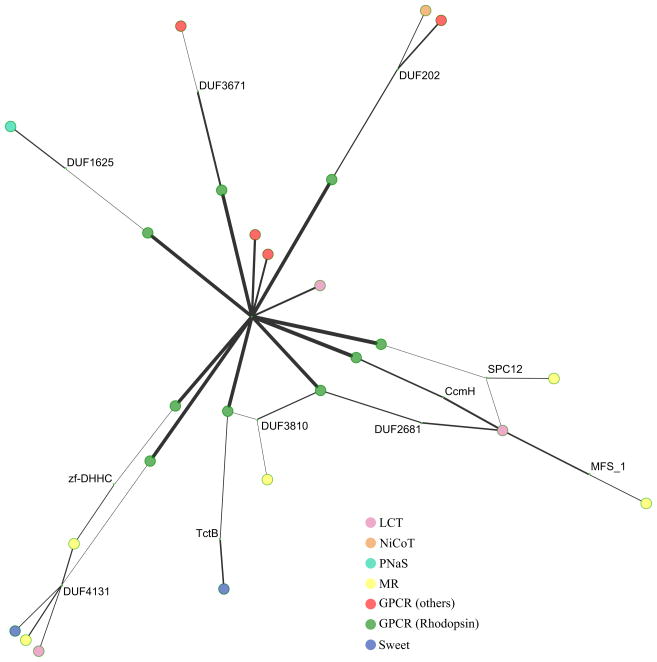
For the small non-Rhodopsin graph component of TC# 9.A.14, the only edges connecting it with the others were from PgaD, DUF4131. From PgaD, there were onward links to the LCT family (TC# 2.A.43.4.1). These were the Glutamate GPCRs, some of which display limited similarity to the SOG group of Rhodopsin GPCRs, as shown by our experimentation. For the large non-Rhodopsin component of GPCRs (TC# 9.A.14), the number of connections is greater to other families, including the Rhodopsin component of the GPCRs (TC# 9.A.14). For example, a direct link between TC# 9.A.14.14.1 (non-Rhodopsin GPCR) was demonstrated to 7tm_1, which is the Pfam family linking many Rhodopsin GPCRs. The same was true for TC# 9.A.14.6.6 and TC# 2.A.43.2.5 (a PQ-loop repeat-containing protein from A. thaliana of the LCT family) which had direct links to Pfam family 7tm_1 (the central node connecting Rhodopsin GPCRs in Fig. 5). The similarity was embedded in motifs found in the MATCH array including LxLxV and KxLLxxVxVF. Even the large non-Rhodopsin component of the GPCRs (TC# 9.A.14) did not show strong direct links to other TCDB families that are members of the TOG Superfamily, showing that the Pfam approach to homology searching is less sensitive than the Superfamily Principle approach described here and in many other publications from our laboratory. The link from Pfam family 7tm_1 to TC# 2.A.43.2.5 (a plant member of the LCT family) is weaker (0.00021) than the link between the GPCR protein TC# 9.A.14.14.1 and 7tm_1 (3.2e−07) as expected, but the former value is still highly significant. The link to the LCT protein TC# 2.A.43.2.5 was much stronger than any link from other GPCRs to other TC families that are members of the TOG Superfamily. This means that the closest neighbor to GPCRs in TC# 9.A.14 was connected via the Rhodopsin component Pfam family (7tm_1), and that the GPCR family (TC# 9.A.14) itself has a non-Rhodospin component (Glutamate GPCRs) that is disconnected from the rest of the GPCRs.
Figure 5. TOG superfamily constituent interrelationships revealed using Pfam.
Cytoscape (using Spring Embedded logic) visualization of our mapping of the proposed TOG superfamily to Pfam using HMMER3 and the default similarity cutoff (10). The Pfam nodes are shown in smaller size, and the edge width represents levels of similarity. The most significant link between any of the GPCR sequences and any of the non-GPCR sequences connects the central node of the Rhodopsin GPCR cluster (“7tm_1”) to an LCT sequence from A. thaliana (colored pink). The LCT sequences are similar to the Sweet sequences (colored violet) and overlap that cluster. The microbial rhodopsins (MR, colored yellow) show connectivity to the rhodopsin GPCR cluster.
For the mapping of GPCRs in TCDB, we found that the mapping did not contain disconnected components, and this appeared not to be the result of the addition of missing sequences; it instead depended on a change in the database. Nevertheless, members of the Glutamate GPCR cluster (e.g. TC# 9.A.14.7.3) proved to be poorly connected to other GPCRs. A significant link between the Glutamate cluster and the other GPCRs passed through one Pfam family, PF07077, showing e-values of 0.016 and 0.048. Using the Pfam website, we could not see that PF07077 (DUF1345) and TC# 9.A.14.11.2 (the closest node to the family) were significantly related, even when the threshold was set to 10. The Pfam web service listed the relations as being to 7tm_3 (PF00003) at 3.3e−40, one of the core nodes of the Glutamate GPCRs. In our own mapping, we had the same at 1.2e−42. In summary, these results confirm the conclusion that the GPCR cluster is not held together by strong edges, especially Glutamate GPCRs. In fact, these edges were weaker than the edges between Rhodopsin GPCRs and the LCT and Sweet families.
HMM:HMM comparisons
For the most significant result of our HMM:HMM comparison, we scored 30.6% between the Opsin cluster of the human Rhodopsin GPCR (cluster α) in the GRAFS classification system and the MR family (TC# 3.E.1). This is a significant result, indicating homology [56]. In fact, we had 753 match columns, and the e-value was 0.00044. The hit was broken into two halves, the first half showing a 30.6% probability over 104 columns, and the second showing 14.2% probability over 85 columns. The consensus alignment is presented at the bottom of Table S10.
Using hmmsearch (searching profile(s) against a sequence database) in HMMER 3.0, we confirmed that bovine rhodopsin was a member of the Opsin cluster; the score was 1.5e−136 against the Opsin HMM, but only 4.8e−37 against the Amine HMM. The term “opsin” means Rhodopsin without retinal, and both microbial and bovine rhodopsins link retinal to the corresponding lysine amino acyl residue in TMS7. This can be compared with other significant results between TC families. For example, OST (TC# 2.A.82) and PNaS (TC# 2.A.58) scored a 31% probability of homology in HHsearch (hhsuite-2.0.16); MR (TC# 3.E.1) and NiCoT (TC# 2.A.52) scored 12%; MR (TC# 3.E.1) and LCT (TC# 2.A.43) scored 26.7%, and Sweet (TC# 9.A.58) and LCT (TC# 2.A.43) scored 97%.
Structural superposition of visual rhodopsin and microbial rhodopsin
Differences in signaling systems of visual (e.g. squid [57]) and microbial rhodopsins, and the conformational changes of retinal isomerization and helix movements of spectroscopically distinct intermediates have been presented elsewhere [58–59]. A superimposition of the bovine visual rhodopsin (VR) structure (PDB:1F88, chain A [60]), and a microbial rhodopsin (MR) structure (PDB:3NS0), was performed in Chimera 1.7 (Figure 6; Video S1). The individual TM helices matched up in the superimposed configuration, although they were not perfectly aligned, often being tilted at somewhat different angles. Nevertheless, when we oriented the structures to view them through the pore and place TMS 1 at the top, we could count the 7 TMSs clockwise from one side in a corresponding manner between the structures. It was clear that the N-termini, the C-termini, and all the loops between the TMSs corresponded.
When the 7 TMSs of the human adenosine receptor (PDB:2YDO) (3 Ångström resolution structure, without the non-membrane helix after position 298), were compared with the 7 TMSs in the α-Rhodopsin GPCR/MECA cluster, and these were compared with VR (PDB:1F88, chain A), we found that the adenosine receptor could be oriented so that TMS1 is on top, and the consecutive helices could then be counted clockwise. This superposition was better than the one noted above with MR. Here, the RMSD was 4.925 for all α-carbons. A similar result was obtained with the opioid (Rhodopsin GPCR) receptor (PDB:4EA3, chain A), as the same clockwise arrangement of TMSs was observed, and the RMSD was 4.467 for all α-carbons. These results showed that while the clockwise arrangement of the 7 TMSs is shared between VR and MR, the structural superimposition is better between adenosine, opioid and VR, as expected considering their relative phylogenetic distances.
To evaluate how the superimposition is influenced by the various states in which both MR and VR can exist, we compared them in their different states. 1F88-A is the dark-adapted conformation of VR, similar to 4A4M (constitutively active light-adapted VR). Previoulsy, when we compared 1F88-A (VR) with 3NS0 (ground state MR), the RMSD was 7.176 for all α-carbons. However, if we took 1KGB (another ground state of MR), there was an improvement; the RMSD was 7.019 for all α-carbons. With the K intermediate of MR (1IXF), the RMSD was 7.639 for all α-carbons, and with the L intermediate of MR (1UCQ), the RMSD was 8.401 for all α-carbons. For the early M intermediate (1KG8), we observed an RMSD of 7.118 for all α-carbons, and with the actual M intermediate (1IW9), the RMSD was 7.638 for all α-carbons. Hence, the early M intermediate of MR (1KG8) is notably more similar to the dark-inactivated conformation of VR (1F88-A) than the K, L or actual M intermediates. However, when we considered all of the atoms, the best RMSD between VR (1F88-A) and MR was with the ground state of MR, especially 1KGB. Such RMSD values only demonstrate a similar fold, not homology. However, taken together with the other statistical results reported in this study, these results provide confirmation of homology.
Discussion
The analyses reported here allowed us to interlink nine integral membrane protein families of diverse mechanistic types to form the novel TOG superfamily (see Table 1 and Figure 1). No other transport protein superfamily in TCDB [3–4] exhibits functional and mechanistic diversity as great as that of the TOG superfamily. This unexpected quality is highlighted by the presence of known and putative secondary carriers, group translocators, light-driven pumps, channels, transmembrane chaperone proteins, photoreceptors and G-protein-coupled receptors. Clearly, this is a case where superfamily assignment is not a guide to energy coupling mechanism or mode of transport. Indeed, in contrast to most superfamilies of integral membrane transport proteins [6], several TOG superfamily members are not transporters at all, and there is no correlation between mode of transport or substrate specificity and position in the phylogenetic (SFT) tree (see Fig. 3). The results illustrate the potential of some superfamilies, but not others, to diverge into proteins with different modes of action. For example, no member of the Major Facilitator Superfamily (MFS) functions in transport by a mechanism other than by secondary transport, and the only alternative function is that of transmembrane receptor [13, 20, 61]. Even this alternative functional type is exceedingly rare in nature.
A 2 TMS precursor could have duplicated to yield the 4 TMS unit that gave rise to all nine families in the TOG superfamily, but this has not been demonstrated. More importantly, the comparisons presented resolve some of the uncertainties in the evolutionary pathways of families exhibiting odd numbered topologies, such as the PnuC family. Topological analyses of the entire superfamily reveal 7 well conserved average TMSs, as is expected given the dominant 7 TMS topology in most families within the superfamily. The N-terminal TMS of the 8 TMS topology was usually lost in the families under study, while loss of the C-terminal TMS occurred with a much lower frequency. Nevertheless, the N-terminal TMS, lost in many homologues, is present in enough members of the TOG superfamily to be visualized in the AveHAS plot (Figure 2).
The greatest topological variation is observed for the Sweet, NiCoT and PNaS families. In the Sweet family, 3 TMS homologues are found in prokaryotes in addition to the 7 TMS proteins found ubiquitously. In the NiCoT family, some members appear to have only 6 TMSs, due to loss of both TMSs 1 and 8 of the 8 TMS precursor. In the PNaS family, additional TMSs present in several family members proved to be due to fusions or late duplication events. Thus, 4 extra TMSs are homologous to the last 4 TMSs in some family members. This fact suggests that at least some members of the PNaS family arose by triplication of the 4 TMS repeat unit.
For the most part, the nine families within the TOG superfamily do not cluster according to mechanistic type or substrate specificity. Instead families are interspersed (Figs. 3A and 3B). The diversity within the TOG superfamily is reminiscent of the demonstrated or hypothesized alternative energy coupling mechanisms used by members of certain families found in TCDB. The ArsB transporters (TC# 3.A.4), members of the IT superfamily [62], can function either as secondary carriers or as ATP-driven primary active transporters, depending on the availability of the ArsA ATPase, and the same may be true of Acr3 porters (TC# 2.A.59) (see Castillo and Saier [63] and references cited therein). Members of the PTS Galactitol (Gat) family (TC# 4.A.5), but not members of the related PTS L-Ascorbate (L-Asc) family (TC# 4.A.7), appear to be capable of functioning either by group translocation involving the PTS energy coupling proteins or by secondary active transport when these proteins are lacking [64–65]. Evidence supports the suggestion that members of the PnuC family TC# 4.B.1 within the TOG superfamily can function by group translocation using ATP-dependent nicotinamide ribonucleoside kinase as the energy-coupling enzyme. However, many members of this family are encoded in genomes that lack this enzyme, supporting the conclusion that these porters function as secondary carriers [66]. Comparable studies have shown that members of the NaT-DC family TC# 3.B.1 catalyze sodium efflux in a process driven by decarboxylation of various carboxylic acids. However, all other members of the CPA superfamily function as secondary carriers (see the CPA superfamily in TCDB). A similar situation has been demonstrated for members of the ECF sub-superfamily of the ABC superfamily [67]. Some of these porters can transport vitamins such as biotin and thiamin either by ATP-dependent primary active transport or by pmf-driven secondary active transport [67] (Sun and Saier, unpublished results).
It is a common assumption that sequence similarity between visual rhodopsins and microbial rhodopsins is undetectable [68]. The GPCR family (TC# 9.A.14) sequence set is spread out in three separate network components, one of which, the Glutamate GPCR set, is distantly connected to the others, having only weak links to other members of the TOG superfamily. One sequence, TC# 2.A.43.2.5 (PQ-loop repeat-containing protein from A. thaliana), which is a member of Lysosomal Cystine Transporter (LCT) family and closely related to Sweet, has a similarity of 0.00021 to Pfam family 7tm_1 which is the central node of the Rhodopsin GPCRs. Despite LCTs being larger than MRs, and while the LCT family is found exclusively in the eukaryotic domain, this is the most significant Pfam connection between any member of the GPCRs (TC# 9.A.14) and the rest of the TOG superfamily network. The similarity between LCT and Rhodopsin GPCRs has been noted elsewhere [14]. In summary, the connection between Rhodopsin GPCRs and the Lysosomal Cystine Transporter (LCT) and Sweet families is stronger than the intra-GPCR connections of many GPCRs to the most divergent Glutamate GPCRs. Thus, the concept of 7 TMS GPCRs being a closely related group of sequences, often taken for granted, is not valid, while the conclusion that sequence similarity between visual rhodopsin and bacteriorhodopsin is non-detectable is equally invalid.
Using HMM:HMM comparisons (hhsuite-2.0.16) and the GRAFS system for GPCR classification, we detected a 30.6% probability of significant homology between the MR family (3.E.1) and the Opsin cluster of α-Rhodopsin GPCRs (Table S10). As a comparison, the Glutamate cluster of the GPCRs scores only a 1.2% chance of a distant homologous relationship with the Opsin cluster. This observation confirms the conclusion that different GPCRs are more divergent from each other than microbial and visual rhodopsins are from each other.
We are aware that sequence convergence is potentially capable of explaining some degree of sequence similarity when the regions compared are short, but skepticism is appropriate [69]. The need for stable transmembrane segments, along with functional requirements, may dictate sequence convergence in somewhat longer sequences [70–72]. However, we believe that convergence cannot explain a degree of similarity sufficient to give a comparison score of 12–14 S.D. for a stretch of over 60 aas, particularly where two or more α-helical domains are aligned in a manner that makes evolutionary sense and fits a proposed pathway. The results of our control experiments using family members that evolved independently of the TOG superfamily support a current threshold of 12.0 S.D.s for establishing homology.
The elucidation of superfamily relationships is likely to open up new fields of study by allowing extrapolation of structural data from a well-characterized superfamily homologue to all or most members of the same superfamily. However, when the evolutionary process gives rise to homologues of differing topologies, extrapolation of structural data from one superfamily member to another may not be justified [13]. Future studies will be required to reveal the degrees of structural dissimilarity that result from sequence divergence and topological variation within a superfamily.
Methods
Obtaining Homologues and Removing Redundancies
Query sequences used to identify members of each family were (1) bacteriorhodopsin of Halobacterium salinarum (GenBank:gi# 114811, TC# 3.E.1.1.1), (2) MtN3 of Medicago truncatula (gi# 75220431, TC# 9.A.58.1.1), (3) PnuC of Haemophilus influenzae (gi# 81335937, TC# 4.B.1.1.2), (4) Orf of Pyrococcus abyssi (gi# 74545625, TC# 2.A.102.4.1), (5) YfcA of Escherichia coli (gi# 82592533, TC# 2.A.102.3.1), (6) Orf of Oryza sativa (gi# 75252893, TC# 2.A.102.5.1), (7) RcnA of E. coli (gi# 3025266, TC# 2.A.52.2.1), (8) Ost α of Raja erinacea (gi# 82108802, TC# 2.A.82.1.1), (9) NptA of Vibrio cholera (gi# 81345622, TC# 2.A.58.1.2), (10) CTNS of Homo sapiens (gi# 269849555, TC# 2.A.43.1.1), and (11) ROP of Homo sapiens (gi# 129219, TC# 9.A.14.1.1). Analyses dealing with the HORC family TC# 1.A.69 were performed utilizing OR83b of Drosophila melanogaster (gi# 14285640, TC# 1.A.69.1.1) as the query. NCBI PSI-BLAST searches with two iterations (e−4; e−6 cutoffs, respectively) were performed using Protocol1 [13, 73] to identify members of each family. The Protocol1 program compiles homologous sequences from the BLAST searches into a single file in FASTA format, eliminates redundancies and fragmentary sequences, and generates a table of the obtained sequences containing protein abbreviations, sequence descriptions, organismal sources, protein sizes, gi numbers, organismal groups or phyla, and organismal domains (see supplementary tables). Protocol1’s CD-HIT option was used to remove redundancies and highly similar sequences [13, 17]. An 85% identity cut-off was used in establishing homology between family members, while a 70% identity cut-off was used to create more easily viewed average hydropathy plots and phylogenetic trees. These percent identity values thus refer to the values above which redundant sequences were removed. Thus, an 85% cutoff means that no two protein sequences retained for analysis were more than 85% identical. FASTA files from Protocol1 were considered representative of each respective protein family, although selected proteins that demonstrated homology between families were confirmed with NCBI’s Conserved Domain Database [74] and PSI-BLAST results.
Multiple Alignments and Topological Analyses
The ClustalX program was used to create a multiple alignments of homologous proteins, and the few sequences that introduced large gaps into the alignment (usually a reflection of fragmentation, inclusion of introns, or incorrect sequences) were removed. This allowed the generation of a coherent multiple alignment where all or most sequences are homologous throughout most of their lengths. Results obtained with this program have been compared with 5 other programs, and when sequence similarity was sufficient to give reliable multiple alignments, phylogenetic trees obtained with the six programs (Neighbor Joining or Parsimony) were very similar [75]. For topological analyses of single protein sequences, the WHAT, TMHMM 2.0, and HMMTOP programs were used [76–77]. Inputting the multiple alignment files generated by ClustalX into the Average Hydropathy, Amphipathicity and Similarity (AveHAS) program facilitated more accurate topological assessments of multiple proteins or entire families. CDD was also used to analyze protein sequence extensions identified using the AveHAS plot. Motif analyses were performed using the MEME/MAST programs [78–79].
Establishing Homology Between Families
Initially, a large screen was performed comparing distantly related TSUP family members [18] against all families of the TC# 2.A, 3.E and 9.A subclasses. The Targeted Smith-Waterman Search (TSSearch) feature of Protocol2 [13] was then run in order to compare each family to all other TOG superfamily members [12, 18]. TSSearch uses a rapid search algorithm to find distant homologues between two different FASTA files that may not readily be apparent in BLAST or PSI-BLAST searches [13]. The most promising comparisons between proteins were automatically analyzed using the Global Sequence Alignment Tool (GSAT) [80] feature of Protocol2 [13]. Comparisons using the GSAT feature of Protocol2 are reported in standard deviations (S.D.), which refers to the number of S.D.s a given score is from the mean, raw local bit score of pairwise scores of 200 shuffled residues. Scores are calculated with the Needleman-Wunsch algorithm. Promising results with a comparison score of 12.0 S.D. or greater were confirmed and analyzed further using the GSAT and GAP programs set at default settings with a gap creation penalty of 8 and a gap extension penalty of 2 with 2,000 random shuffles; assuming a Gaussian distribution, a comparison score of 12.0 S.D. corresponds to a probability of 1.77×10−33 that the degree of similarity between two proteins arose by chance (See supplementary Fig. S9) [81]. In spite of this conclusion by Dayhoff et al. [81], Gaussian skew can increase the probability of chance similarity for any given standard deviation value [82].
Probabilities for comparison scores were calculated using Mathematica (Wolfram Research, Inc., Champaign, IL, USA). Comparisons involving at least 60 amino acyl residues, the average size of a prototypical protein domain, alignment of 2 or more α-helical domains between compared proteins, and a comparison score of at least 12.0 S.D. were considered sufficient to provide strong evidence for homology between two proteins or internal repeat units in the studies reported [1, 4, 17, 81]. Convergent sequence evolution is possible and has been demonstrated for short motifs but never for large segments of proteins such as entire domains. One reason we use a minimum of 60 amino acyl residues in defining homology is that for such a long sequence, convergence to give 12 S.D. is exceedingly unlikely.
Optimization of the GSAT/GAP alignments was performed on sequences by maximizing the number of identities, minimizing gaps, and removing non-aligned sequences at the ends of the alignment, but never in central regions of the alignment. Optimization usually yields a higher comparison score that better represents the level of similarity between two internal sequences.
The Ancient Rep (AR) program [13] was used to look for internal repeats, and results were confirmed using the GSAT/GAP and HHRep programs [83]. The AR program compares potential transmembrane repeat sequences (hydrophobic TMS regions predicted by HMMTOP) within a single protein and between proteins in a FASTA file, giving a comparison score in S.D.s in the same format as Protocol2.
Controls
A large screen was performed with all members of the TOG superfamily against the Major Intrinsic Protein (MIP; TC# 1.A.8) and the Mitochondrial Carrier (MC; TC# 2.A.29) families, two large families whose known evolutionary pathways and topologies differ from each other and those of the proposed TOG superfamily [49–50, 84]. Comparisons between each family were conducted using the same techniques and programs in establishing homology between TOG superfamily members (Protocol1, Protocol2, GSAT). The best comparison scores were selected using the same criteria as outlined previously; selected comparisons contained at least 2 or more aligned α-helical domains and involved at least 60 residues. The evolutionary pathway was not considered in selections. Precise scores of the best alignments fitting these criteria were obtained with GSAT and GAP programs set at default settings with a gap creation penalty of 8 and a gap extension penalty of 2 with 2,000 random shuffles. These scores were then compared against alignments demonstrating homology between members of the TOG superfamily.
As controls, we looked for similarities between members of the MIP family (TC# 1.A.8) and the MC family (TC# 2.A.29) using Pfam-A, a database of HMMs of protein domains. We used HMMER3 to search the current version of TCDB, using the default cutoff (10). We loaded all edges connecting either MC or MIP proteins in Cytoscape 2.8.3 to view the results. Significant similarities were not found.
Phylogenetic and Sequence Analyses
The ClustalX program [51] was used to create multiple alignment for homologous sequences using default settings, and a neighbor-joining phylogenetic tree for the TOG superfamily was created using the TreeView program [52]. Phylogenetic trees for individual families were also drawn using the FigTree program (http://tree.bio.ed.ac.uk/software/figtree/). To depict phylogenetic relationships more accurately than possible using the multiple alignments provided by the ClustalX program, the SFT programs [15–17] were used to generate SFT1 and SFT2 phylogenetic trees using tens of thousands of BLAST bit scores instead of multiple alignments [17]. The SFT1 phylogenetic tree was generated to visualize relationships between all subfamilies within families of the TOG superfamily. The SFT2 tree, drawn with the TreeView program [52], consolidated individual members into their respective families for visualizing phylogenetic relationships between families within the TOG superfamily.
Obtaining sequences from the GRAFS system
Our starting point for mapping to the Rhodopsin GPCRs was the well-known classification system for human GPCRs, published shortly after completion of the human genome sequence, the so called GRAFS system [35]. Not all reported sequences are available, but as many as possible were extracted. We obtained the list of IDs from the GRAFS system: Secretin (15), Adhesion (24), Glutamate (15) and Frizzled/Taste2 (24). Of these, all 15 Secretins, 24 Adhesions, 15 Glutamates and 23 Frizzled/Taste2 entries were used; one ID was a duplicate in the original publication. For the Adhesion family, 8 sequences were nucleotide entries.
The α-group of Rhodopsin Receptors (89) contains the prostaglandin receptor cluster (15). Of these, we eliminated 2 NT entries because they did not refer to a single translated sequence and because the actual gene names were not present in the current entry. This left 13 sequences in the prostaglandin receptor cluster. In the amine receptor cluster (40), one entry had been removed at the submitter’s request, leaving 39. For the opsin receptor cluster (9), the melatonin receptor cluster (3) and the MECA receptor cluster (22), all sequences were retrievable.
In the β-group of Rhodopsin Receptors (35), one sequence (NP_004113.2) did not exist, leaving 34. In the γ-group of Rhodopsin Receptors (59), all of the sequences in the SOG receptor cluster (15), the MCH receptor cluster (2), and the chemokine receptor cluster (42) were retrievable except (NP_002021.1) in the chemokine receptor cluster, leaving 41 sequences in that cluster.
In the δ-group of Rhodopsin Receptors (58), one sequence in the MAS-related receptor cluster (8), (NP_089843.1), did not exist, leaving 7. All sequences in the glycoprotein receptor cluster (8) were retained. We eliminated one NT entry (NT_006337.5) in the purine receptor cluster (42), leaving 41. These sequences were sent to FASTA files and parsed so that the header only contained the sequence ID, and the sequences were each on single lines in lower case letters.
Training HMMs on the GRAFS sequences
MAFFT v7.023b (2013/02/03) [85] was used to make alignments with “E-INS-I” program in the “accurate” mode. The alignments were converted to Stockholm format. Using HMMER 3.0, we built an HMM for each alignment representing the major GRAFS groupings, creating 4 files for the non-rhodopsin groupings (Adhesion.hmm, Frizzled.hmm, Glutamate.hmm and Secretin.hmm) as well as 5 files for the first major cluster of rhodopsin sequences (Amine.hmm, MECA.hmm, Melatonin.hmm, Opsin.hmm and Prostaglandin.hmm) and 1 file for the second major cluster of rhodopsin sequences (β.hmm) and 3 files for the third major cluster of rhodopsin sequences (chemokine.hmm, MCH.hmm and SOG.hmm) and 3 files for the fourth major cluster of rhodopsin sequences (glycoprotein.hmm, MAS.hmm and purin.hmm). This left 16 HMMs.
HMMSEARCH was used, using the default similarity threshold (10.0) to search these 16 HMMs against 54 sequences in the GPCR family (TC# 9.A.14). Since they are not listed in the GRAFS paper, we ignored the olfactory receptor cluster (estimated at 460), and when listed, the other 7 TMS Receptors (23).
Training HMMs on TOG sequences
On March 4, 2013, we downloaded 8,790 proteins from TCDB (http://www.tcdb.org/public/tcdb). Because some multicomponent systems have multiple sequences under a single TC number (e.g. MexA and MexB), there were only 6,316 unique IDs. We added a letter (A,B,C…) after the TC# when this was the case to distinguish the sequences. We found 167 sequences in the 10 families (including the odorant receptors, not established members of this superfamily) comprising the TOG Superfamily: TC#s 1.A.69, 2.A.43, 2.A.52, 2.A.58, 2.A.82, 2.A.102, 3.E.1, 4.B.1, 9.A.14 and 9.A.58. We used MAFFT v7.023b (2013/02/03) “E-INS-I” (accurate). Each alignment was converted to Stockholm format, and HMMER 3.0 was used to build HMMs for each. Pfam-A was downloaded from ftp://ftp.sanger.ac.uk/pub/databases/Pfam/current_release/Pfam-A.hmm.gz. Pfam-A was searched against the 10 families comprising the TOG Superfamily using a relatively stringent value of 1e−20, as well as the default (10.0) similarity threshold.
Mapping TOG to Pfam
The mapping from the default cutoff was used as our main mapping (parts of which can be seen in Fig. 5). This mapping contained 623 edges, mostly weak links to distantly related Pfam families. A scale of 10 different edge widths was used to represent each tenth of the distribution of e-values, thick lines representing higher similarity. Using a spring embedded layout, the nodes representing Pfam families were decreased in size.
We imported the Pfam mapping of the 10 TCDB families to Pfam-A using an evalue threshold of 10 in Cytoscape 2.8.3. In total, 440 nodes and 623 edges were imported. We set the visual style to Nested Network Style and applied the Spring-embedded logic. We could highlight sets of nodes, such as TC# 9.A.14 using the Node Attribute Batch Editor. Visual Mapping Bypass was first used to establish a Node Attribute with Node Size 5 as default and 30 for TCDB nodes. In VizMapper™, we used a Discrete Mapper for Edge Width based on interaction.
Supplementing the TCDB sequence set with GRAFS sequences
We added three new GPCRs to TCDB in order to have representatives from all classes in the GRAFS system. The most recent additions were FZD1 (TC# 9.A.14.16.1), TAS2 (TC# 9.A.14.17.1) and KiSS (TC# 9.A.14.18.1). To ensure that the poor connectedness between TC# 9.A.14 sequences in the mapping of the TOG superfamily to Pfam (Figure 5) was not due to the lack of representation of these families, we took the 8,843 sequences in TCDB, on March 14, 2013. Of these, 62 were GPCRs, containing representatives from all branches of the GPCR system. We used default settings in HMMER3, using a threshold e-value of 10 to map these against Pfam-A. Since the evalues depend on the database size, the exact e-values are not directly comparable with the other mapping. We executed a spring-embedded logic on 314 edges in Cytoscape 2.8.3, using a passthrough mapper on a 1–10 scale representing edge width bands, subdividing our edges.
HMM:HMM comparisons
We downloaded and installed the HHSuite (hhsuite-2.0.16) for HMM-HMM comparisons and used HHMAKE (HHmake version 2.0.15) to retrain HHMs in our proposed superfamily. HMMs were not used, as use of HMMER format as input results in severe loss of sensitivity for the nine families (not including TC# 9.A.14, the GPCRs). We used the -M 50 flag for FASTA columns and HHsearch (2.0.15) to compare each of the α, β, γ and δ clusters of Rhodopsin GPCRs [35]. In total, 12 HMMs were used from these groups: Amine, MECA, Melatonin, Opsin, Prostaglandin, β, Chemokine, MCH, SOG, Glycoprotein, MAS, and Purin [35]. These were compared with HMMs representing the nine TC families MR (TC# 3.E.1), Sweet (TC# 9.A.58), PnuC (TC# 4.B.1), TSUP (TC# 2.A.102), NiCoT (TC# 2.A.52), OST (TC# 2.A.82), PNaS (TC# 2.A.58), LCT (TC# 2.A.43), and HORC (TC# 1.A.69). For each comparison, we recorded the HHsearch (2.0.15) percentage probability, representing the probability of homology. The relevant TCDB families were also compared internally. The results are presented in Table S10.
Supplementary Material
Figure S1. AveHAS plot, phylogenetic tree for the LCT family, homology between members of the Sweet and LCT families, and homology between members of the PNaS and LCT families.
Figure S2. AveHAS plot, phylogenetic tree for the MR family.
Figure S3. AveHAS plot, phylogenetic tree for the NiCoT family, and homology between members of the TSUP and NiCoT families.
Figure S4. AveHAS plot, phylogenetic tree for the OST family, homology between members of the TSUP and OST families, binary alignment of members of the TSUP and OST families, GSAT alignment of TSUP and OST.
Figure S5. AveHAS plot, phylogenetic tree for the Sweet family, homology between members of the Sweet and OST families, binary alignment of members of the Sweet and OST families, GSAT alignment of Sweet and OST.
Figure S6. AveHAS plot, phylogenetic tree for the PNaS family, homology between members of the Sweet and PNaS families.
Figure S7. AveHAS plot, phylogenetic tree for the PnuC family, homology between members of the PnuC and TSUP families, binary alignment of members of the PnuC and TSUP families, GSAT alignment of PnuC and TSUP.
Figure S8. AveHAS plot, phylogenetic for the GPCR family, homology between members of the GPCR and the microbial rhodopsin (MR) families, binary alignment of members of the GPCR and MR families, GSAT alignment of GPCR and MR.
Figure S9. Schematic representation of the mean bit score of a given alignment, algorithm input into Mathematica 7.0, probability of notable Z scores.
Figure S10. AveHAS plots representing all families within the TOG superfamily.
Figure S11. Results from the ClustalX-based neighbor-joining program.
Video file of rotation of the superimposition of B-rho and M-rho.
Acknowledgments
The work reported was supported by NIH Grant 2 RO1 GM077402-05A1. We thank Carl Welliver for assisting in the preparation of this manuscript, and Benjamin Hass for assisting in statistical interpretations.
Abbreviations
- TOG
Transporter-Opsin-G protein-coupled
- MR
Microbial rhodopsin
- VR
Visual rhodopsin
- TSUP
4-Toulene Sulfonate Uptake Permease
- PnuC
Nicotinamide Ribonucleoside Uptake Permeases
- NiCoT
Ni2+-Co2+ Transporters
- OST
Organic Solute Transporters
- PNaS
Phosphate:Na+ Symporters
- LCT
Lysosomal Cystine Transporters
- HORC
Heteromeric Odorant Receptor Channel
- HMM
Hidden Markov model
- RMSD
Root mean square deviation
Footnotes
Competing financial interests
The authors declare no competing interests.
Author contributions
Conceived and designed the experiments: DCY, MAS, SA, EIS, MHS. Performed the experiments: DCY, MAS, SA, AJV. Analyzed the data: DCY, MAS, AJV, MHS. Contributed reagents/materials/analysis tools: VSR. Wrote the paper: DCY, MAS, EIS, VSR, MHS.
References
- 1.Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saier MH., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–6. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–8. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busch W, Saier MH., Jr The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 6.Lam VH, Lee JH, Silverio A, Chan H, Gomolplitinant KM, Povolotsky TL, Orlova E, Sun EI, Welliver CH, Saier MH., Jr Pathways of transport protein evolution: recent advances. Biol Chem. 2011;392:5–12. doi: 10.1515/BC.2011.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang AB, Lin R, Keith Studley W, Tran CV, Saier MH., Jr Phylogeny as a guide to structure and function of membrane transport proteins. Mol Membr Biol. 2004;21:171–81. doi: 10.1080/09687680410001720830. [DOI] [PubMed] [Google Scholar]
- 8.Mansour NM, Sawhney M, Tamang DG, Vogl C, Saier MH., Jr The bile/arsenite/riboflavin transporter (BART) superfamily. Febs J. 2007;274:612–29. doi: 10.1111/j.1742-4658.2006.05627.x. [DOI] [PubMed] [Google Scholar]
- 9.Furutani Y, Kandori H. Internal water molecules of archaeal rhodopsins (Review) Mol Membr Biol. 2002;19:257–65. doi: 10.1080/0968768021000035069. [DOI] [PubMed] [Google Scholar]
- 10.Hirai T, Subramaniam S, Lanyi JK. Structural snapshots of conformational changes in a seven-helix membrane protein: lessons from bacteriorhodopsin. Curr Opin Struct Biol. 2009;19:433–9. doi: 10.1016/j.sbi.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou XE, Melcher K, Xu HE. Structure and activation of rhodopsin. Acta Pharmacol Sin. 2012;33:291–9. doi: 10.1038/aps.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearson WR. Empirical statistical estimates for sequence similarity searches. J Mol Biol. 1998;276:71–84. doi: 10.1006/jmbi.1997.1525. [DOI] [PubMed] [Google Scholar]
- 13.Reddy VS, Saier MH., Jr BioV Suite--a collection of programs for the study of transport protein evolution. Febs J. 2012;279:2036–46. doi: 10.1111/j.1742-4658.2012.08590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai Y, Heijne WH, Smith DW, Saier MH., Jr Homologues of archaeal rhodopsins in plants, animals and fungi: structural and functional predications for a putative fungal chaperone protein. Biochim Biophys Acta. 2001;1511:206–23. doi: 10.1016/s0005-2736(00)00389-8. [DOI] [PubMed] [Google Scholar]
- 15.Chen JS, Reddy V, Chen JH, Shlykov MA, Zheng WH, Cho J, Yen MR, Saier MH., Jr Phylogenetic characterization of transport protein superfamilies: superiority of SuperfamilyTree programs over those based on multiple alignments. J Mol Microbiol Biotechnol. 2011;21:83–96. doi: 10.1159/000334611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yen MR, Chen JS, Marquez JL, Sun EI, Saier MH. Multidrug resistance: phylogenetic characterization of superfamilies of secondary carriers that include drug exporters. Methods Mol Biol. 2010;637:47–64. doi: 10.1007/978-1-60761-700-6_3. [DOI] [PubMed] [Google Scholar]
- 17.Yen MR, Choi J, Saier MH., Jr Bioinformatic analyses of transmembrane transport: novel software for deducing protein phylogeny, topology, and evolution. J Mol Microbiol Biotechnol. 2009;17:163–76. doi: 10.1159/000239667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shlykov MA, Zheng WH, Chen JS, Saier MH., Jr Bioinformatic characterization of the 4-Toluene Sulfonate Uptake Permease (TSUP) family of transmembrane proteins. Biochim Biophys Acta. 2012;1818:703–17. doi: 10.1016/j.bbamem.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung YJ, Krueger C, Metzgar D, Saier MH., Jr Size comparisons among integral membrane transport protein homologues in bacteria, Archaea, and Eucarya. J Bacteriol. 2001;183:1012–21. doi: 10.1128/JB.183.3.1012-1021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saier MH, Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jahn PS, Lew K, Liu J, Pao SS, Paulsen IT, Tseng TT, Virk PS. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–79. [PubMed] [Google Scholar]
- 21.Iwig JS, Rowe JL, Chivers PT. Nickel homeostasis in Escherichia coli - the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol Microbiol. 2006;62:252–62. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 22.Rodrigue A, Effantin G, Mandrand-Berthelot MA. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J Bacteriol. 2005;187:2912–6. doi: 10.1128/JB.187.8.2912-2916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawson PA, Hubbert M, Haywood J, Craddock AL, Zerangue N, Christian WV, Ballatori N. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J Biol Chem. 2005;280:6960–8. doi: 10.1074/jbc.M412752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seward DJ, Koh AS, Boyer JL, Ballatori N. Functional complementation between a novel mammalian polygenic transport complex and an evolutionarily ancient organic solute transporter, OSTalpha-OSTbeta. J Biol Chem. 2003;278:27473–82. doi: 10.1074/jbc.M301106200. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Seward DJ, Li L, Boyer JL, Ballatori N. Expression cloning of two genes that together mediate organic solute and steroid transport in the liver of a marine vertebrate. Proc Natl Acad Sci U S A. 2001;98:9431–6. doi: 10.1073/pnas.161099898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takanaga H, Frommer WB. Facilitative plasma membrane transporters function during ER transit. Faseb J. 2010;24:2849–58. doi: 10.1096/fj.09-146472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, Chermak D, Antony G, White FF, Somerville SC, Mudgett MB, Frommer WB. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–32. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saier MH., Jr Tracing pathways of transport protein evolution. Mol Microbiol. 2003;48:1145–56. doi: 10.1046/j.1365-2958.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- 29.Ghezzi C, Murer H, Forster IC. Substrate interactions of the electroneutral Na+-coupled inorganic phosphate cotransporter (NaPi-IIc) J Physiol. 2009;587:4293–307. doi: 10.1113/jphysiol.2009.175596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foster JW, Park YK, Penfound T, Fenger T, Spector MP. Regulation of NAD metabolism in Salmonella typhimurium: molecular sequence analysis of the bifunctional nadR regulator and the nadA-pnuC operon. J Bacteriol. 1990;172:4187–96. doi: 10.1128/jb.172.8.4187-4196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merdanovic M, Sauer E, Reidl J. Coupling of NAD+ biosynthesis and nicotinamide ribosyl transport: characterization of NadR ribonucleotide kinase mutants of Haemophilus influenzae. J Bacteriol. 2005;187:4410–20. doi: 10.1128/JB.187.13.4410-4420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penfound T, Foster JW. NAD-dependent DNA-binding activity of the bifunctional NadR regulator of Salmonella typhimurium. J Bacteriol. 1999;181:648–55. doi: 10.1128/jb.181.2.648-655.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurnasov OV, Polanuyer BM, Ananta S, Sloutsky R, Tam A, Gerdes SY, Osterman AL. Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis. J Bacteriol. 2002;184:6906–17. doi: 10.1128/JB.184.24.6906-6917.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh SK, Kurnasov OV, Chen B, Robinson H, Grishin NV, Osterman AL, Zhang H. Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities. J Biol Chem. 2002;277:33291–9. doi: 10.1074/jbc.M204368200. [DOI] [PubMed] [Google Scholar]
- 35.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–72. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 36.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–57. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 37.Civelli O, Reinscheid RK, Zhang Y, Wang Z, Fredriksson R, Schioth HB. G protein-coupled receptor deorphanizations. Annu Rev Pharmacol Toxicol. 2013;53:127–46. doi: 10.1146/annurev-pharmtox-010611-134548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnan A, Almen MS, Fredriksson R, Schioth HB. The origin of GPCRs: identification of mammalian like Rhodopsin, Adhesion, Glutamate and Frizzled GPCRs in fungi. PLoS One. 2012;7:e29817. doi: 10.1371/journal.pone.0029817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordstrom KJ, Sallman Almen M, Edstam MM, Fredriksson R, Schioth HB. Independent HHsearch, Needleman--Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families. Mol Biol Evol. 2011;28:2471–80. doi: 10.1093/molbev/msr061. [DOI] [PubMed] [Google Scholar]
- 40.Schioth HB, Fredriksson R. The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen Comp Endocrinol. 2005;142:94–101. doi: 10.1016/j.ygcen.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 41.Almen MS, Nordstrom KJ, Fredriksson R, Schioth HB. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009;7:50. doi: 10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–56. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang XL, Wang Y, Li DL, Luo J, Liu MY. Orphan G protein-coupled receptors (GPCRs): biological functions and potential drug targets. Acta Pharmacol Sin. 2012;33:363–71. doi: 10.1038/aps.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–6. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 46.Smart R, Kiely A, Beale M, Vargas E, Carraher C, Kralicek AV, Christie DL, Chen C, Newcomb RD, Warr CG. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol. 2008;38:770–80. doi: 10.1016/j.ibmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Touhara K. Insect olfactory receptor complex functions as a ligand-gated ionotropic channel. Ann N Y Acad Sci. 2009;1170:177–80. doi: 10.1111/j.1749-6632.2009.03935.x. [DOI] [PubMed] [Google Scholar]
- 48.Park JH, Saier MH., Jr Phylogenetic, structural and functional characteristics of the Na-K-Cl cotransporter family. J Membr Biol. 1996;149:161–8. doi: 10.1007/s002329900016. [DOI] [PubMed] [Google Scholar]
- 49.Kuan J, Saier MH., Jr The mitochondrial carrier family of transport proteins: structural, functional, and evolutionary relationships. Crit Rev Biochem Mol Biol. 1993;28:209–33. doi: 10.3109/10409239309086795. [DOI] [PubMed] [Google Scholar]
- 50.Kunji ER, Robinson AJ. Coupling of proton and substrate translocation in the transport cycle of mitochondrial carriers. Curr Opin Struct Biol. 2010;20:440–7. doi: 10.1016/j.sbi.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 51.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Zhai Y, Tchieu J, Saier MH., Jr A web-based Tree View (TV) program for the visualization of phylogenetic trees. J Mol Microbiol Biotechnol. 2002;4:69–70. [PubMed] [Google Scholar]
- 53.Park HH. Structural analyses of death domains and their interactions. Apoptosis. 2011;16:209–20. doi: 10.1007/s10495-010-0571-z. [DOI] [PubMed] [Google Scholar]
- 54.de la Horra C, Hernando N, Lambert G, Forster I, Biber J, Murer H. Molecular determinants of pH sensitivity of the type IIa Na/P(i) cotransporter. J Biol Chem. 2000;275:6284–7. doi: 10.1074/jbc.275.9.6284. [DOI] [PubMed] [Google Scholar]
- 55.Anantharaman V, Aravind L. Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics. 2003;4:34. doi: 10.1186/1471-2164-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–60. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 57.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–7. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 58.Vinothkumar KR, Henderson R. Structures of membrane proteins. Q Rev Biophys. 2010;43:65–158. doi: 10.1017/S0033583510000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–94. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 60.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 61.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prakash S, Cooper G, Singhi S, Saier MH., Jr The ion transporter superfamily. Biochim Biophys Acta. 2003;1618:79–92. doi: 10.1016/j.bbamem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 63.Castillo R, Saier MH. Functional Promiscuity of Homologues of the Bacterial ArsA ATPases. Int J Microbiol. 2010;2010:187373. doi: 10.1155/2010/187373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou XF, Saier MH., Jr The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur J Biochem. 2003;270:799–813. doi: 10.1046/j.1432-1033.2003.03418.x. [DOI] [PubMed] [Google Scholar]
- 65.Saier MH, Hvorup RN, Barabote RD. Evolution of the bacterial phosphotransferase system: from carriers and enzymes to group translocators. Biochem Soc Trans. 2005;33:220–4. doi: 10.1042/BST0330220. [DOI] [PubMed] [Google Scholar]
- 66.Rodionov DA, Hebbeln P, Eudes A, ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD, Eitinger T. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol. 2009;191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hebbeln P, Rodionov DA, Alfandega A, Eitinger T. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci U S A. 2007;104:2909–14. doi: 10.1073/pnas.0609905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Josefsson LG. Evidence for kinship between diverse G-protein coupled receptors. Gene. 1999;239:333–40. doi: 10.1016/s0378-1119(99)00392-3. [DOI] [PubMed] [Google Scholar]
- 69.Doolittle RF. Convergent evolution: the need to be explicit. Trends Biochem Sci. 1994;19:15–8. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- 70.Baeza-Delgado C, Marti-Renom MA, Mingarro I. Structure-based statistical analysis of transmembrane helices. Eur Biophys J. 2013;42:199–207. doi: 10.1007/s00249-012-0813-9. [DOI] [PubMed] [Google Scholar]
- 71.Remmert M, Biegert A, Linke D, Lupas AN, Soding J. Evolution of outer membrane beta-barrels from an ancestral beta beta hairpin. Mol Biol Evol. 2010;27:1348–58. doi: 10.1093/molbev/msq017. [DOI] [PubMed] [Google Scholar]
- 72.Ried CL, Kube S, Kirrbach J, Langosch D. Homotypic interaction and amino acid distribution of unilaterally conserved transmembrane helices. J Mol Biol. 2012;420:251–7. doi: 10.1016/j.jmb.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 73.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 74.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–52. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Young GB, Jack DL, Smith DW, Saier MH., Jr The amino acid/auxin:proton symport permease family. Biochim Biophys Acta. 1999;1415:306–22. doi: 10.1016/s0005-2736(98)00196-5. [DOI] [PubMed] [Google Scholar]
- 76.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–50. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 77.Zhai Y, Saier MH., Jr A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J Mol Microbiol Biotechnol. 2001;3:501–2. [PubMed] [Google Scholar]
- 78.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 79.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 80.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–95. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dayhoff MO, Barker WC, Hunt LT. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–45. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 82.O’Hagan A, Leonard T. Bayes estimation subject to uncertainty about parameter constraints. Biometrika. 1976;63:201–202. [Google Scholar]
- 83.Soding J, Remmert M, Biegert A. HHrep: de novo protein repeat detection and the origin of TIM barrels. Nucleic Acids Res. 2006;34:W137–42. doi: 10.1093/nar/gkl130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park JH, Saier MH., Jr Phylogenetic characterization of the MIP family of transmembrane channel proteins. J Membr Biol. 1996;153:171–80. doi: 10.1007/s002329900120. [DOI] [PubMed] [Google Scholar]
- 85.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. AveHAS plot, phylogenetic tree for the LCT family, homology between members of the Sweet and LCT families, and homology between members of the PNaS and LCT families.
Figure S2. AveHAS plot, phylogenetic tree for the MR family.
Figure S3. AveHAS plot, phylogenetic tree for the NiCoT family, and homology between members of the TSUP and NiCoT families.
Figure S4. AveHAS plot, phylogenetic tree for the OST family, homology between members of the TSUP and OST families, binary alignment of members of the TSUP and OST families, GSAT alignment of TSUP and OST.
Figure S5. AveHAS plot, phylogenetic tree for the Sweet family, homology between members of the Sweet and OST families, binary alignment of members of the Sweet and OST families, GSAT alignment of Sweet and OST.
Figure S6. AveHAS plot, phylogenetic tree for the PNaS family, homology between members of the Sweet and PNaS families.
Figure S7. AveHAS plot, phylogenetic tree for the PnuC family, homology between members of the PnuC and TSUP families, binary alignment of members of the PnuC and TSUP families, GSAT alignment of PnuC and TSUP.
Figure S8. AveHAS plot, phylogenetic for the GPCR family, homology between members of the GPCR and the microbial rhodopsin (MR) families, binary alignment of members of the GPCR and MR families, GSAT alignment of GPCR and MR.
Figure S9. Schematic representation of the mean bit score of a given alignment, algorithm input into Mathematica 7.0, probability of notable Z scores.
Figure S10. AveHAS plots representing all families within the TOG superfamily.
Figure S11. Results from the ClustalX-based neighbor-joining program.
Video file of rotation of the superimposition of B-rho and M-rho.