Abstract
A critical observation in sporadic cancers is that not all individuals are equally prone to developing cancer following exposure to a given environmental carcinogen. Epidemiological studies have suggested that the difference in the timing of cancer onset in response to exogenous DNA damage is likely attributable to genetic variations, such as those associated with base excision repair genes. To test this long-standing hypothesis and elucidate how a genetic variation in the base excision repair gene flap endonuclease 1 (FEN1) results in susceptibility to environment insults and causes cancer, we established a mutant mouse model carrying a point mutation (E160D) in Fen1. We demonstrate that the E160D mutation impairs the ability of FEN1 to process DNA intermediate structures in long-patch base excision repair using nuclear extracts or reconstituted purified base excision repair proteins. E160D cells were more sensitive to the base damaging agents methylnitrosourea and hydrogen peroxide, leading to DNA strand breaks, chromosomal breakage, and chromosome instabilities in response these DNA insults. We further show that E160D mice are significantly more susceptible to exposure to methylnitrosourea and develop lung adenocarcinoma. Thus, our current study demonstrates that a subtle genetic variation (E160D) in base excision repair genes (FEN1) may cause a functional deficiency in repairing base damage, such that individuals carrying the mutation or similar mutations are predisposed to chemical-induced cancer development.
Keywords: FEN1, Long-patch base excision repair, methylnitrosourea, tetraploidy, aneuploidy, cancer
Introduction
Cancer onset is associated with the accumulation of genome instabilities (Lengauer et al., 1998). It is generally thought that interactions between genetic variations and environmental insults drive cancer progression. For instance, exposure to tobacco smoke, which contains hundreds of DNA damaging agents and mutagens, is the major risk factor for lung cancer (Vainio and Boffetta, 1994). However, only 15% of smokers develop lung cancer, and non-smokers may also develop lung cancer. These findings suggest genetic backgrounds may mediate susceptibility to the development of smoking-induced lung cancer (Jemal et al., 2002; Landi et al., 2005). It is widely accepted that biological survival requires a delicate balance between genomic stability and genetic variation (Cistulli et al., 2004). DNA repair systems are essential for maintaining genetic stability by detecting, removing and repairing lesions since DNA is under constant attack by numerous endogenous (e.g., free radicals) and exogenous (e.g., radiation and chemicals) DNA damaging agents (van Gent et al., 2001). The main cell DNA repair pathway for removing a damaged base resulting from oxidation, alkylation or abasic (AP) sites, is base excision repair (BER). Although epidemiological studies have suggested linkages between some genetic variations in BER genes and increased cancer risk in humans (Chang et al., 2009; Figueroa et al., 2007; Ming et al., 2009), it is still unclear whether and how these variations impact cancer initiation and progression.
In mammalian BER, at least two major sub-pathways (short-patch and long-patch) have been defined (Asagoshi et al., 2010; Balakrishnan et al., 2009; Ranalli et al., 2002). The simplest and most common form, short patch BER, is initiated by DNA glycosylases (Demple and Harrison, 1994; Krokan et al., 1997; Lindahl, 1993; Lindahl and Wood, 1999; Nakamura et al., 1998; Sun et al., 1995), followed by cleavage of abasic sites by AP endonuclease, DNA polymerase β (Polβ)-mediated gap-filling DNA synthesis (Singhal and Wilson, 1993) and removal of the 5′-deoxyribose phosphate (dRP) of the abasic site (Prasad et al., 2005). However, when the dRP moiety is reduced or oxidized, the dRP lyase cannot remove the modified sugar residue (Klungland and Lindahl, 1997; Liu et al., 2008). In such a case, a few nucleotides are added by polymerase β or δ to produce a short flap, which is subsequently removed by flap endonuclease 1 (FEN1) in complex with the processivity factor proliferating cell nuclear antigen (PCNA) (Gary et al., 1999). This alternative FEN1/PCNA-dependant BER pathway is termed long-patch BER (LP-BER) (Gary et al., 1999). Polβ activity occurs in DNA duplexes that contain a gap, but not a nick, and the exonuclease activity (EXO) of FEN1 is important to cleave the nick substrate and generate the optimal gap intermediate for Polβ during LP-BER (Liu et al., 2005). Liu et al. further suggest a “Hit and Run” model of Polβ and FEN1 action in LP-BER. In this model, FEN1 EXO activity is used to remove one nucleotide, creating a 1-nt gap that allows Polβ to rebind and extend. Subsequent removal of one or several nucleotides by the nuclease from the 5′-end of the nick or short flap leads to another gap for Polβ to fill (Liu et al., 2005). This alternative mechanism for processing of BER intermediates implies that both the FEN and the EXO activities of FEN1 are critical for efficient LP-BER.
The implication that FEN1 EXO activity is important in LP-BER is consistent with our recent study of a Fen1 mutant mouse model carrying the E160D mutation, which broadly represents a group of human FEN1 mutations identified in cancer patients (Zheng et al., 2007). These human FEN1 mutations abolish the EXO activity, but retain the FEN activity (Zheng et al., 2007). E160D mutant mice are viable, but compared to wild type (WT), cells derived from mutants are more sensitive to base damaging agents such as methyl methanesulfonate (MMS) and low dosages of γ-irradiation (Zheng et al., 2007). This suggests that the FEN1 mutation does not affect its activity in RNA primer removal, which is essential for DNA replication and cell survival, but results in a functional deficiency in BER (Zheng et al., 2007). While E160D mutant mice display a high incidence of development of spontaneous cancer, it is difficult to assess the contribution of the BER defect to the spontaneous cancer phenotype because the E160D mutation also caused other defects in DNA metabolism (Zheng et al., 2007). Here, E160D mice were used as a model to examine the effect of a deficiency in FEN1 activity on BER in vivo and to test whether a SNP in BER genes causes individuals to be more sensitive to DNA damaging insults, contributing to the development of DNA damage-induced cancer. BER biochemical assays were used to demonstrate that the E160D FEN1 mutation impairs BER capacity in vitro. Treatment of mutant cells and mice with the alkylating agent methylnitrosourea (MNU), which is similar to the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), was used to elucidate whether and how BER deficiency, due to the FEN1 E160D mutation, causes mice to be susceptible to MNU-treatment and cancer.
Results
E160D causes defects in processing of LP-BER intermediates
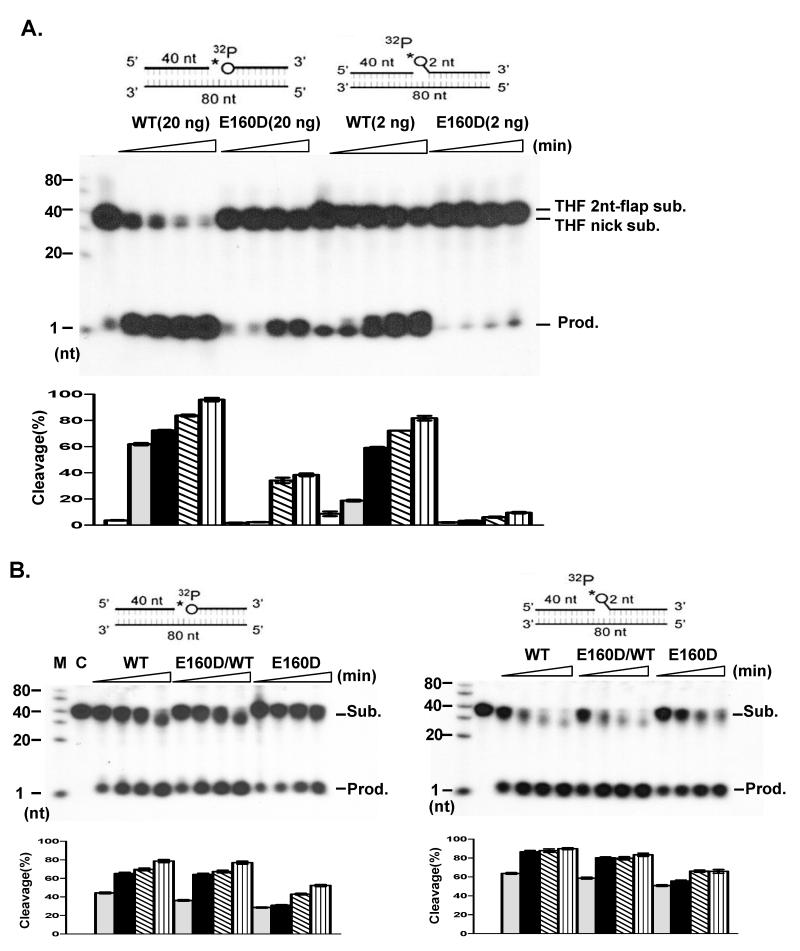
Our previous work showed that E160D mutant cells are more sensitive to DNA damaging agents, including MMS, than WT cells, suggesting a defect in the BER pathway (Zheng et al., 2007). Abolishing 5′ exonuclease activity, which removes one or two nucleotides from the 5′ end of a nicked or gapped DNA duplex, may impair the capability of FEN1 to process BER intermediates that result from AP endonuclease cleavage of AP sites. To test this hypothesis, we prepared a nicked substrate and a short flap (2-nt) substrate, both of which had a tetrahydrofuran (THF) residue at the 5′ end of the downstream oligo, mimicking LP-BER intermediates. Comparison of specific nuclease activity on these substrates showed there was dramatically less cleavage of the nicked-THF or the flap-THF substrate by E160D mutant proteins (Fig. 1A). Although WT FEN1 effectively cleaved over 60-70% of substrate within 10 min, less than 10% of substrate was cleaved by E160D within the same period. To further test if the defect in processing BER intermediates was due to the E160D mutation, nuclear extracts (NE) from WT and E160D cells were isolated and their activity to remove the THF residue was assayed. While incubation of substrates with NE from E160D/WT and E160D cells showed less activity as compared to the WT (Fig. 1B), the loss of activity caused by the E160D NE was considerably less than that caused by purified E160D mutant proteins. This was probably due to partial redundancy with functional homologues of FEN1, such as the 5′ exonuclease 1, in the NEs.
Figure 1.
E160D displays defects in cleavage of LP-BER intermediate substrates (A) Cleavage of nicked or flap substrates with a 5′ THF abasic site (circle). Indicated amounts of purified FEN1 proteins were incubated with 5′ end 32P-labeled DNA substrates for 0, 5, 10, 30 and 60 min. (B) Nuclear extracts (5 μg for nicked substrate or 2 μg for short flap substrate; WT, E160D/WTor E160D) were incubated with 1 pmol 5′ end 32P labeled DNA substrates in a total volume of 10 μl at 37°C for 0, 5, 10, 30 and 60 min.
E160D results in defects in LP-BER in vitro
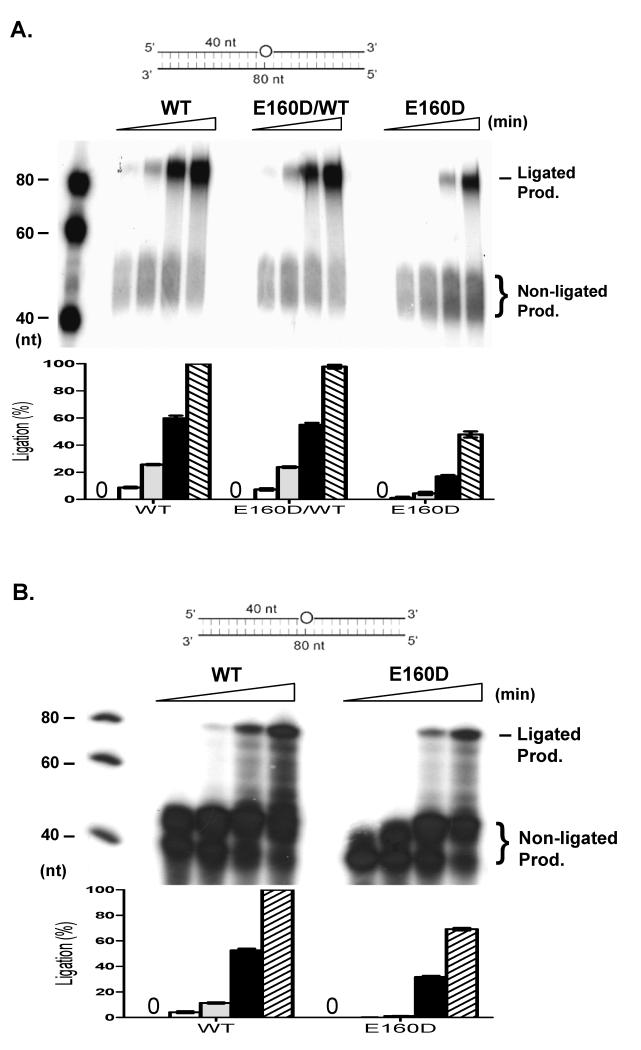
To further determine if the E160D FEN1 mutation affected overall BER efficiency, we prepared a BER model substrate with a THF in the middle of a DNA duplex (80-nt) and conducted in vitro BER assays with reconstituted purified BER proteins or NEs isolated from WT, E160D/WT, and E160D mouse embryonic fibroblasts (MEF). Following removal of the THF and nucleotide incorporation, FEN1 processing of the intermediate and Lig1-mediated DNA ligation generated a repaired product of 80-nt (Fig. 2A). We found that WT NE efficiently repaired the THF damage, resulting in an intense 80-nt band, but the E160D NE produced much less repaired product (Fig. 2A). We further reconstituted the key chemical reactions in LP-BER by using purified recombinant BER proteins. Similar to what was observed with NE, reconstitution with the E160D mutant protein was considerably less efficient at repairing THF damage than that with WT FEN1 (Fig. 2B). In addition, overall 32P incorporation was considerably less in reactions with E160D versus WT FEN1. This was consistent with data from Wilson’s group demonstrating that FEN1 exonuclease activity is important for the generation of ideal substrates for Polβ in LP-BER (Liu et al., 2005).
Figure 2.
E160D impairs overall LP-BER efficiency. (A) LP-BER in nuclear extract (NE) isolated from WT, E160D/WT or E160D MEF cells. NE (2 μg) was incubated with LP-BER model substrates. Bottom panel shows the relative ratio of ligation product. (B) Reconstitution of LP-BER by purified human recombinant proteins. APE1 (20 ng), Polβ (10 ng), DNA LigI (50 ng) and WT FEN1 or E160D (1 ng) were mixed. The mixture was then incubated with THF model LP-BER substrate in reaction buffer containing 5 μCi [α-32P] dCTP and 50 μM each of dATP, dGTP, and dTTP in a total volume of 10 μl at 37°C for 0, 5, 10, 30, 60 min. Bottom histogram presents the ligated product means ± s.d. of three independent experiments .
E160D increases collapse of DNA replication forks in response to chemical treatment
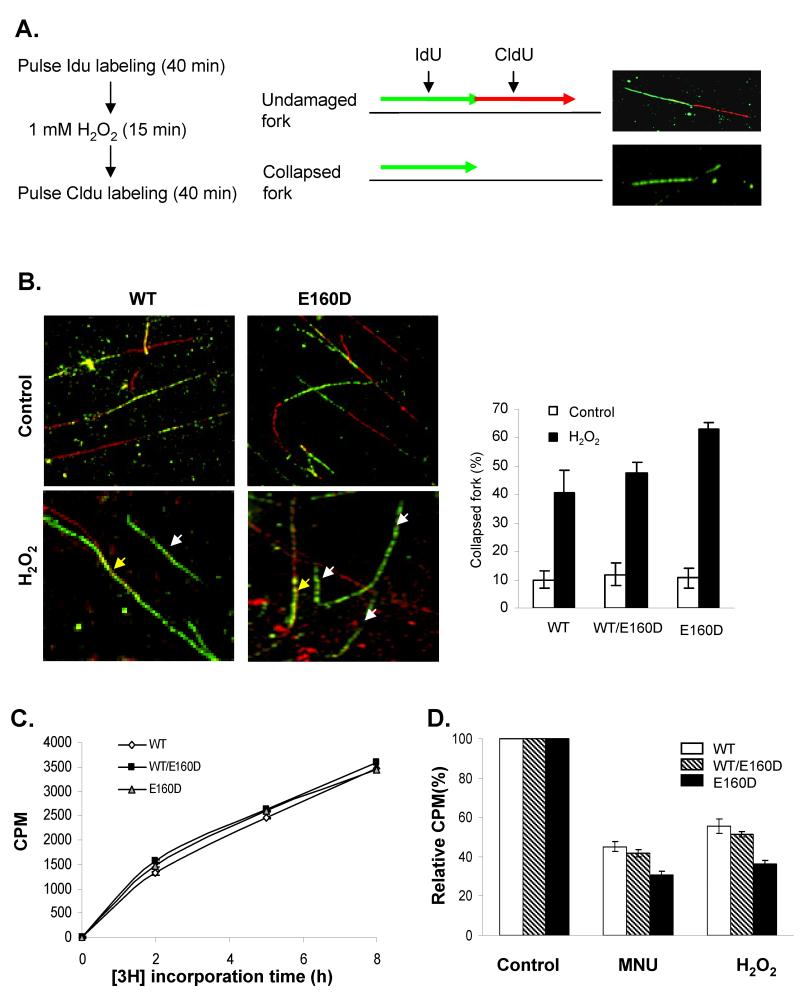
Unligated nicks in the genome that result from E160D if encounter the DNA replication fork, may collapse the ongoing fork, leading to DNA double strand breaks. To test this hypothesis, we first conducted DNA fiber assays in the presence or absence of the DNA damaging agent H2O2 (Fig. 3A). The DNA fiber assay allows us to analyze the progression of DNA replication fork including determination of the number of the stalled or collapsed forks with or without treatments with DNA damaging agents (Merrick et al., 2004). Cells were pulse labeled with IdU (green), treated or untreated with H2O2, and labeled with CldU (red) (Figure 3A). We observed three typical patterns: green/red, red only, and green only (Figure 3A and 3B), which can be interpreted as undamaged forks, newly fired forks, and collapsed forks (Merrick et al., 2004). In the absence of H2O2, approximately 90% of tracks in WT, WT/E160D and E160D DNA samples, were undamaged (green/red or red-only tracks, Fig 3B), indicating that the E160D mutation did not induce stalled replication forks. However, treatment of the WT WT/E160D and E160D/E160D MEF cells with 1 mM H2O2 after pulse-labeling the cells with IdU, dramatically increased the levels of green-only tracks in DNA samples from these cells, indicating that H2O2-induced ssDNA breaks collapsed DNA replication forks (Fig 3B). Furthermore, the number of green-only tracks in the E160D/E160D sample was considerably more than that in the WT sample, suggesting that the E160D cells accumulated more DNA breaks in response to H2O2 treatments (Fig. 3B).
Figure 3.
E160D MEF cells had more collapsed replication forks and lower DNA replication rates in response to DNA damage. (A) and (B) Replication tracks of WT, WT/E160D and E160D/E160D cells in the absence or presence of H2O2. (A) Outline for labeling newly synthesized DNA with IdU (green) and CldU (red) and treatment with H2O2. Images indicate the typical undamaged replication fork (green/red track) and collapsed replication fork (green-only track). Red-only track that would represent the newly fired replication fork is not shown. (B) Representative images of DNA fibers of WT and E160D cells untreated or treated with H2O2. In left bottom panels, yellow arrows specified the normal replication forks (green/red tracks) without DNA damage, and white arrows indicated the stalled replication forks due to DNA damage (green only tracks). Replication tracks were assessed and quantified and the relative level of collapsed replication forks in each sample was expressed as the percentage of the total number of incorporated DNA forks (all tracks). Values are means ± s.d. of three independent experiments (Right panel). (C) DNA replication rates of WT, WT/E160D and E160D/E160D cells under normal conditions. MEF cells were cultured in DMEM medium with 1 μCi/ml [3H] thymidine, and were collected after a specific time (0, 2, 5, or 8 h). Cells were extensively washed and lysed. DNA was precipitated and the radioactivities of all DNA samples were counted by a liquid scintillation counter. Values are means ± s.d of three independent assays. (D) MEF cells were treated (or not) with MNU (200 μg/ml, 1h) or H2O2 (1 mM, 15 min). Cells were then incubated in DMEM medium containing 1 μCi/ml [3H] thymidine (5 h). The incorporation of [3H] thymidine was quantified and the relative [3H] thymidine incorporation rate of each DNA sample was calculated. The incorporation rate of corresponding untreated cells was arbitrarily set as 100.
Next, we used the [3H]-thymidine incorporation assay to analyze the DNA replication rate in the presence or absence of DNA damaging agents. In normal growth conditions, the WT/E160D and E160D/E160D cells incorporated [3H]-thymidine at rates similar to WT cells (Fig. 3C), which is consistent with the observation that the E160D mutation on its own did not result in replication fork collapse. However, treating cells with H2O2 or MNU greatly decreased the incorporation rate (Fig. 3D). Moreover, the presence of MNU or H2O2 reduced the incorporation rate by 70% or 65%, respectively, in the E160D/E160D cells as compared to 45% (MNU) or 41% (H2O2) in the WT cells (Fig. 3D). This is consistent with the DNA fiber assay showing that DNA damage induced more stalled replication forks in the E160D cells than in the WT cells.
Increased accumulation of MNU- and hydrogen peroxide-induced double strand breaks in E160D cells
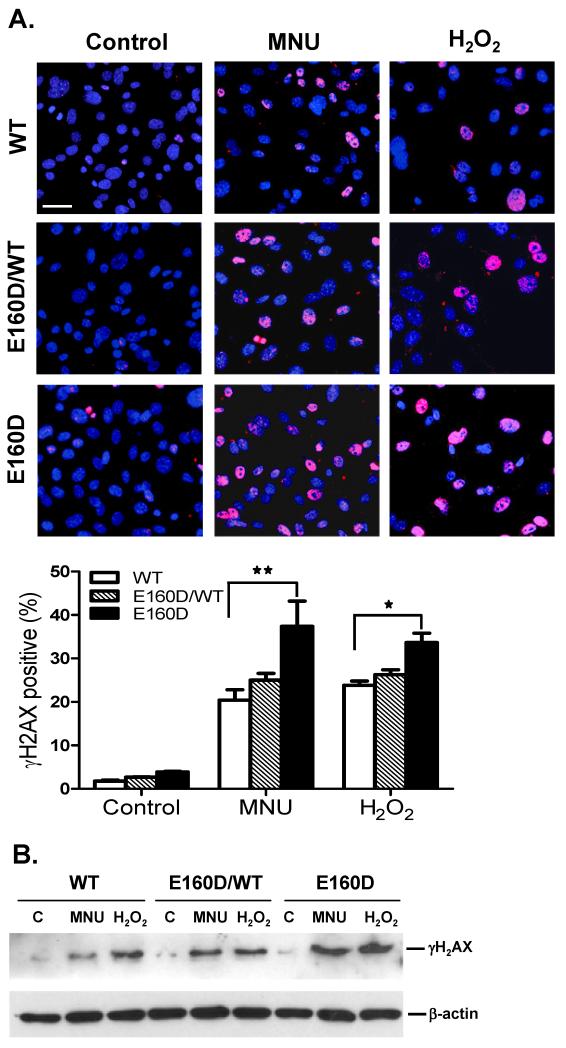
Our data suggested the E160D mutation affected the cleavage of BER intermediates and overall BER efficiency. The un-sealed DNA nicks or gaps might collapse DNA replication forks and subsequently lead to double strand breaks (DSBs) in response to DNA base damage (Pascucci et al., 2005). To test whether the E160D mutation increased the induction of DSBs, we treated WT and E160D cells with a DNA alkylation (MNU) or oxidative agent (H2O2), and stained the treated cells with an antibody against phosphorylated histone H2AX (γH2AX). Histone H2AX is rapidly phosphorylated at the site of each nascent DSB and γH2AX focus formation is considered a sensitive and selective signal for the presence of DSBs (Bonner et al., 2008). Our findings suggested that, in the absence of MNU or H2O2, WT and E160D cells had low levels of γH2AX-positive nuclei; however, chemical treatment resulted in a significant increase in γH2AX-positive cells (Fig 4A). Furthermore, a greater number of γH2AX-positive nuclei were observed in E160D than in WT cells after MNU or H2O2 treatment (*P=0.025; **P=0.0003; Two-tailed Fisher Exact) (Fig. 4A). γH2AX levels in NE were further analyzed by Western blotting and were consistent with immunostaining data. γH2AX band intensity in MNU- or H2O2-treated WT, E160D/WT, or E160D NE was considerably greater than in untreated samples. Moreover, the γH2AX band intensity of MNU- or H2O2-treated E160D NE was significantly greater than that of MNU- or H2O2-treated WT NE (Fig. 4B).
Figure 4.
E160D MEF cells accumulate more MNU- and H2O2-induced DNA strand breaks than WT cells. MEF cells were treated either with MNU (200 μg/ml, 1h), H2O2 (1 mM, 15 min) or left untreated. (A) Immunocytochemistry staining for H2AX phosphorylation (Ser 139, red) in MEF cells with DNA counterstaining by DAPI (blue). Bottom panel shows the ratio of γH2AX-positive nuclei to DAPI-positive nuclei. Scale bar, 60 μm. *P=0.025; **P=0.0003, Two-tailed Fisher Exact (B) Western blot of γH2AX levels using nuclear extracts from treated and untreated MEF cells.
E160D cells have more chemical-induced chromosomal aberrations
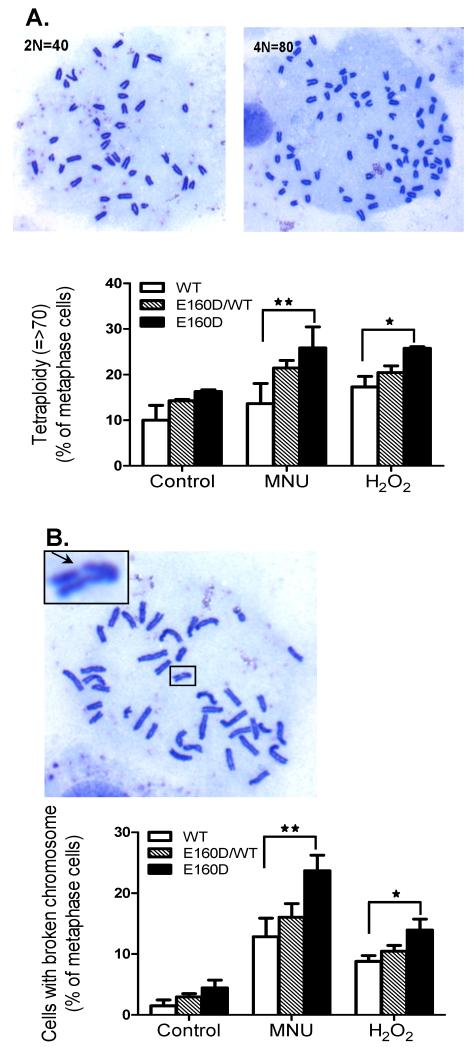
Accumulation of DNA strand breaks in the genome may cause aneuploidy and chromosome breaks (Ganem et al., 2007; van Gent et al., 2001). Therefore, we next determined if DNA damaging agents induced such chromosomal aberrations. WT, E160D/WT and E160D cells were treated with MNU or H2O2, and the number of chromosome was counted. We found that MNU or H2O2 greatly induced aneuploidy in the E160D cells but not the WT cells. In normal growth conditions, the level of near-tetraploid aneuploidy was 10%, 14%, and 17% in the WT, WT/E160D, and E160D/E160D cells, respectively (Fig 5A). However, treatment of the cells with MNU for 1 h increased the level of near tetraploid aneuploidy to 25% in the E160D/E160D cells, compared to only 13% for the WT (**P= 0.002, Two-tailed Fisher Exact) (Fig 5A). A similar phenomenon was observed with the H2O2-treated E160D and WT cells (*P= 0.026, Two-tailed Fisher Exact) (Fig 5A). Furthermore, we found that the E160D cells were more susceptible to generation of chromosomal breaks in response to these DNA damaging agents. In normal growth conditions, less than 5% of WT, WT/E160D, or E160D/E160D cells had chromosomal breaks; however, treatment of the cells with MNU resulted in chromosomal breaks in 13%, 17% and 23% of WT, E160D/WT, and E160D/E160D cells, respectively (**P= 0.0004, Two-tailed Fisher Exact) (Fig. 5B). Similar to MNU, H2O2 induced chromosomal breaks in 8%, 10%, and 14% of WT, E160D/WT, and E160D/E160D cells (*P=0.011, Two-tailed Fisher Exact) (Fig. 5B).
Figure 5.
MNU- or H2O2-treatment induced more chromosomal abnormalities in E160D MEF cells than in WT (A) The percentage of tetraploid and near tetraploid (n>70) chromosomes in MEF cells in WT, E160D/WT and E160D. This includes spontaneous abnormalities and those arising after chemical treatments with MNU (200 μg/ml, 1 h) or H2O2 (1 mM, 15 min). The chromosome number of each mitotic cell was counted; approximately 200 mitotic cells per cell type, per treatment were analyzed. Shown are the results of three independent experiments. *P= 0.026; **P= 0.002, Two-tailed Fisher Exact. (B) Quantification of mitotic cells (WT, E160D/WT and E160D) with broken chromosomes. The ratio was calculated by dividing the number of cells with broken chromosome by the total number of mitotic cells. *P=0.011; **P= 0.0004, Two-tailed Fisher Exact.
E160D mice have greater lung tumor incidence than WT mice after chemical induction
That the E160D FEN1 mutation caused defects in the BER pathway and subsequently resulted in DNA damage-induced chromosomal aberrations suggested that E160D mice would be more susceptible to development of cancer following exposure to environmental insults. To test this hypothesis, WT (n = 26), WT/E160D (n = 25), and E160D/E160D (n = 26) mice were treated with a single dose of the base damaging agent MNU or PBS buffer as a control. After 8 months, 90% and 100% of treated E160D/WT and E160D mice developed lung adenocarcinoma, compared to 70% of treated WT mice; whereas no mice in the control group developed cancer (Fig. 6A). Cancer in other organs was rare in MNU-treated WT, E160D/WT, and E160D mice. On average, E160D/WT and E160D mice had 2.5 and 7.0 MNU-induced tumors, respectively, compared to 1.0 in WT (P<0.0001, Student’s t-test) (Fig 6B).
Figure 6.
E160D mice are more susceptible to the development of MNU-induced lung cancer. For each group of transgenic mice (WT, E160D/WT and E160D) 24 randomly selected 129S1 mice (6-8 weeks old) were injected with either a single dose of MNU solution (50 mg/kg) in PBS buffer or PBS butter as a control. Mice were fed normally then sacrificed after 8 months. The number of tumors was counted (A) Lung cancer frequency in WT, E160D/WT and E160D mice after MNU treatment. Transgenic mice were randomly chosen for MNU (50 mg/Kg) injection. No mouse had lung tumors at that time of treatment. (B) Tumor numbers of each MNU-treated mouse. P<0.0001, Student’s t-test.
Discussion
Genetic variations, such as SNPs in BER genes, have long been identified in humans (Chang et al., 2009; Figueroa et al., 2007). Although epidemiological studies have demonstrated strong linkage between BER SNPs and cancer incidence (Chang et al., 2009; Figueroa et al., 2007), whether and how BER SNPs contribute to cancer development is still not completely understood. In mammals, efficient completion of BER via the long-patch pathway depends on FEN1, a multi-functional nuclease that has been implicated to play a vital role in various DNA metabolic pathways (Liu et al., 2004; Shen et al., 2005). However, the in vivo requirement of specific FEN1 activity in DNA replication and repair and its role in cancer avoidance has not been fully elucidated. E160D mutant mice, used to model cancer patients harboring FEN1 mutations deficient in EXO and GEN activity, developed spontaneous lung and other cancers at high incidence (Zheng et al., 2007). However, it is difficult to link spontaneous cancer development to BER deficiency because multiple defects are observed in DNA metabolism pathways in E160D mice. The current study demonstrates that E160D mutant mice develop lung adenocarcinoma at their early stage (8 months) following exposure to the base damaging agent MNU, while their control untreated counterparts do not, suggesting that the cancer phenotype in E160D mice is likely the outcome of environmental insults and BER deficiency. This in vivo experimental evidence supports the long-standing hypothesis that a subtle genetic variation (E160D point mutation) in a BER gene (Fen1) causes individuals to be susceptible to environmental insults and cancer development.
Our study further elucidates the molecular mechanism by which the E160D Fen1 mutation impairs BER and promotes genome instability in response to environmental insults. E160D mutant proteins in a purified form, or in NE, display significantly reduced ability to cleave nicked and short flap THF substrate, compared to WT (Fig. 1). Our data suggest that failure or a delay in the removal of the damaged base has two distinct effects on LP-BER. First, failure to remove the damaged base prevents DNA ligase I from sealing the DNA ends and generating an intact DNA duplex. Second, it affects Polβ polymerase activity. In addition, our in vivo data further support studies by the Wilson group suggesting that Polβ and FEN1 exonuclease activity employ a “gap translation” mechanism to replace the damaged base and additional nucleotides during processing of BER intermediates, producing ligatable ends for Lig1 (Liu et al., 2005). In support of this model, we demonstrate that levels of Polβ-mediated nucleotide incorporation in reconstituted BER reactions with E160D were significantly less than those observed in WT. In addition, the E160D mutation did not affect the interaction between FEN1 and Polβ. This leads us to believe that the low nucleotide incorporation activity is likely due to a deficiency in FEN1 exonuclease activity, which is responsible for conversion of the nicked substrate into the gapped substrate for Polβ.
LP-BER deficiency results in the accumulation of nicked or gapped BER intermediates, leading to DSBs, likely due to collapse of DNA replication forks. Supporting this hypothesis, we showed the E160D cells were more susceptible than WT cells to treatments with DNA damaging agents and displayed more collapsed replication forks and DSBs, which may contribute to chromosome breakage or disturb chromosome segregation and cell division, leading to tetraploidy and aneuploidy. Consistent with this notion, treatments with MNU or H2O2 induced chromosome breakage and promoted tetraploidization in both WT and E160D cells. In any case, the impact of environmental insults on E160D cells is significantly greater than on WT cells. These findings further implicate the E160D mutation as a causative factor in BER deficiency and in the increased sensitivity of mutant cells to DNA damaging agents.
The increase in chemically-induced DSBs and chromosome instability and breakage observed in E160D mice treated with MNU may play a key role in the development of MNU-induced cancer. Numerous studies have found an association between DSBs, genome instability and cancer predisposition syndromes (Gorgoulis et al., 2005; Rotman and Shiloh, 1998; van Gent et al., 2001; Wang et al., 2005). It is generally accepted that alterations in chromosomal stability, including chromosome number and structure seen in cancer cells, confer some selective advantage to the evolving tumor (Ganem et al., 2007; Rajagopalan and Lengauer, 2004; van Gent et al., 2001). Tetraploidy and aneuploidy are commonly observed in human cancers (Ganem et al., 2007; Rajagopalan and Lengauer, 2004) and have been implicated as a driving force for cancer initiation and/or progression. Large chromosome deletions may also contribute to the inactivation of tumor suppressor genes or loss-of-heterozygosity (Ganem et al., 2007; Rajagopalan and Lengauer, 2004), both of which are key molecular events in cancer development. A previous study showed a RPA mutation in mutant mice caused spontaneous DSBs, resulting in chromosome breaks, gross chromosomal rearrangements, and aneuploidy (Wang et al., 2005), and the development, in later life, of spontaneous lymphoma in mutant mice. Consistent with these studies, our data demonstrate that DNA-damaging insults induce tetraploidy and aneuploidy in E160D cells, accompanied by chromosome breakage. E160D mice, in turn, developed lung adenocarcinoma at an early age following a single dosage treatment with MNU. Taken together, our data demonstrate that the E160D mutation in the BER gene FEN1 partly abolishes FEN1’s nuclease activities and impairs its capacity to process intermediate structures during LP-BER, leading to non-ligated DNA strand nicks and gaps and subsequently DSBs. As a consequence, the E160D cells are more susceptible to DNA insults and develop chemical-induced chromosome instabilities and structural aberrations, contributing to cancer development.
Materials and methods
Preparation of nuclear extracts
MEF cells, untreated or treated with MNU or H2O2, were harvested, washed and suspended with a cell extract (CE) buffer (10 mM HEPES, 60 mM KCl, 1 mM DTT, 1 mM EDTA, and proteinase inhibitor cocktail, 0.075% NP-40, pH 7.5-8.0) for 10 min. Nuclei were washed with CE buffer (without NP-40) once, and incubated (90 min) with a nuclear extract (NE) buffer (25% glycerol, 20 mM Tris-HCl, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, and proteinase inhibitor cocktail). After centrifugation, the supernatant was stored at −80°C. Phosphatase inhibitor cocktails 1 & 2 (Sigma, St. Louis, MO) were added to both CE and NE buffer prior to Western blot analysis for γH2AX.
Protein expression and purification
Purified human APE1, FEN1 and Polβ for reconstitution LP-BER assay were prepared as previously described (Frank et al., 2001; Guo et al., 2009). Briefly, target genes were cloned into expression vector pET28b, and transformed into Escherichia coli strain BL21 (DE3). Cells were amplified and induced by IPTG. Harvested cells were extracted and purified by PrepEase™ high specificity Histidine-tagged Protein Purification kit (USB, Cleveland, Ohio). Eluted proteins were concentrated in dialysis buffer (50% glycerol, 2 mM DTT, 20 mM Tris-HCl pH 8.0, 1 mM MgCl2, 20 mM NaCl, 0.02% NaN3). The protein concentration was determined using Bio-Rad (Bradford) protein assay and protein purity was evaluated by SDS-PAGE. Human DNA LigI was purchased from ALEXIS (Plymouth Meeting, PA).
FEN1 nuclease activity assay
32P-labeled nick or flap DNA substrates that contained THF analogue were prepared as described previously (Zheng et al., 2008). The binding specificity of THF reflects sugar analogue binding to the natural deoxyribose phosphate-containing BER intermediate (Wong and Demple, 2004). To assay nuclease activity, NE, WT FEN1 or E160D mutant protein was incubated (37°C) with 1 pmol DNA substrate for specified times in a reaction buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, and 0.2 mg/ml BSA. Reactions were stopped by addition of 20 μl gel loading buffer (90% formamide dye, 3 M EDTA, 0.02% bromophenol blue and 0.02% xylene cyanol), resolved on a 15% denaturing PAGE and visualized by an autoradiograph. Target bands were quantified by Image J software.
In vitro LP-BER assay
Nuclear extract LP-BER activity was assayed using a synthetic DNA duplex that harbored a THF at residue 40 (Zheng et al., 2008). Reactions were conducted according to published procedures (Zheng et al., 2008), but with a specified time course. For LP-BER reconstitution, proteins were incubated with the indicated DNA substrates in a reaction buffer containing 50 mM HEPES-KOH (pH 7.5), 45 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 2 mM ATP, 200 units creatine-phosphokinase, 0.5 mM NAD and 5 mM phosphocreatine. Repair synthesis was monitored by the incorporation of [32P] dCTP. Reactions were terminated and radioactivity measured as described above.
Establishment and culture of MEF cells
To establish primary MEF cells of WT, E160D/WT, and E160D genetic background the following crosses were performed: WT (female) with WT (male), WT (female) with E160D (male), and E160D (female) with E160D (male), respectively. All mice were of 129S1 pure genetic background. At E13.5, pregnant mice were sacrificed and embryos were isolated. The heads, liver, intestines and heart were removed from the embryos and the remaining carcass was incubated in trypsin solution. Large debris was removed and single cells collected and cultured (37°C, 5% CO2) in DMEM containing 10% FBS, 1% pen strep, 1% Non-Essential amino acids and β-mercaptoethanol.
Cell treatment
Methylnitrosourea (MNU) (Sigma, St. Louis, MO) was dissolved in PBS buffer (stock concentration 10 mg/ml). Before cell treatment, MNU was diluted to 200 μg/ml and H2O2 (stock concentration 10 M) was diluted to 1 mM in DMEM. Cells were treated with MNU for 1 h or H2O2 for 15 min. Treatments were stopped by washing cells with cell culture medium.
[3H]Thymidine incorporation assay
For cell proliferation assay, 1 × 105 MEF cells were plated in 12-well plates. [3H]thymidine (1 μCi/ml) was added 24 h later and incubated for 0, 2, 5 or 8 h. Cells were then washed twice with ice-cold PBS and incubated with 10% ice-cold trichloroacetic acid containing 10 mM thymidine for 15 min at 4°C to precipitate DNA. Cells then were washed twice with cold PBS and lysed with 0.5 M NaOH. In the meantime, the same number of cells was treated with MNU or H2O2. After chemical induction, cells were washed twice with the fresh medium and incubated in the medium with [3H]Thymidine for 5 h, followed by precipitation and lysis as described. Radioactivities of the samples were counted by a liquid scintillation counter.
DNA fiber assays
DNA was pulse labeled with IdU (Sigma, St. Louis, MO) and CldU (Sigma, St. Louis, MO), and DNA fibers were spread as previously described (Jackson and Pombo, 1998; Merrick et al., 2004). Briefly, MEF cells (50-70% confluence) were incubated in DMEM containing 50 μM IdU for 40 min. The cells were then left untreated or treated with 1 mM H2O2 for 20 min. After washing, the cells were incubated with DMEM containing 100 μM CldU for 40 min. Cells were then harvested and suspended in ice-cold PBS at 3 × 106 cells/ml. Labeled cells (25 μl) were mixed with an equal volume of unlabeled cells (2 × 107 cells/ml) and lysed with 150 μl lysis buffer (0.5% SDS in 200 mM Tris-HCl, pH 7.4, and 50 mM EDTA) for 10 min. Lysates (20 μl) were dropped onto a clean glass slide, which was tilted at ~15° to allow the cell lysate to slowly move down the slides to form the DNA fiber spread. The resulting DNA spreads were air-dried and fixed in 3:1 methanol:acetic acid at 4°C overnight.
For detection of IdU- and CldU-labeled DNA fiber spreads, the slides were treated with 2.5 M HCl for 1 h followed by extensive washing with PBS for 10 min. (twice), 100 μM sodium borate for 15 min, and PBS buffer for 10 min. Slides were blocked with Image-iT™ FX signal enhancer (Invitrogen, Carlsbad, CA) for 35 min. Slides were then incubated at room temperature for 1 h with 1:50 rat anti-BrdU (OBT0030G, Accuratechemical.com, detection of CldU) followed by incubation with 1:50 mouse anti-BrdU (Becton Dickinson, Franklin Lakes, NJ, detection of IdU). After incubation with the first antibodies, the extensively washed slides were then incubated with 1:200 Alexa Fluor 633-conjugated anti-rat (Invitrogen, Carlsbad, CA) and 1:200 Alexa Fluor 488 anti-mouse (Invitrogen, Carlsbad, CA) for 1 h.
Immunofluorescence microscopy
For DSB staining, MEF cells were grown on coverslips, treated (controls received equal volume of vehicle), fixed with 4% paraformaldehyde, washed extensively with PBS buffer, permeabilized (4 min, room temperature) with 0.2% Triton X-100, washed with PBS buffer, and then blocked (30 min) with Image-iT™ FX signal enhancer (Invitrogen, Carlsbad, CA). The cells were then incubated (90 min) with the monoclonal γH2AX antibody (Abcam, Cambridge, MA), washed with PBS, and incubated with a fluorescence-labeled secondary antibody. Nuclei were stained with 200 ng/ml DAPI. All slides were examined by fluorescence microscopy (AX70, Olympus, Center Valley, PA) or confocal fluorescence microscopy (Zeiss LSM510 Carl Zeiss, Thornwood, NY).
Metaphase spread preparation and chromosome counting
MEF cells, treated (or not) for 4 h, were harvested and treated with colcemide to arrest cells at metaphase. Cells were incubated (20 min, room temperature) with hypotonic solution (75 mM KCl) then placed in a 37°C water bath (5 min) and fixed with Carnoy’s solution. The fixation process was repeated three times and a dropped was used to place cells onto a clean slide. The cell spread was incubated (55°C overnight), stained with Giemsa solution, and scanned under a microscope for mitotic cells. Images were recorded and analyzed with ImagePro 7.0, and the chromosomes in each metaphase cell were counted.
Statistical analysis
Phenotypic and molecular differences, due to genetic differences or environmental treatments, in cultured cells or mice were assessed by statistical analysis, including Student’s t-test and Fisher exact test, as indicated in figure legends, using the n-Query program (Statistical Solutions, Saugus, MA).
Acknowledgements
We thank the microscopy core facility of City of Hope for technical assistance with immunofluorescence staining of MEF cells. We thank D. Finger for technical advice on purification of FEN1 proteins. We thank S. R. da Costa and K. Walker for editorial assistance. All protocols involving animal use were approved by the Research Animal Care Committee of City of Hope National Medical Center and Beckman Research Institute in compliance with the Public Health Service Policy on Use of Laboratory Animals. This work was supported by an NIH grant R01 CA073764 to B.H.S. and a scholarship from China Scholarship Council (CSC) to H.X.
Footnotes
Conflict of Interest: No conflict of financial interest is noted
References
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CP, Jr., et al. Base excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African-Americans. Carcinogenesis. 2009;30:78–87. doi: 10.1093/carcin/bgn261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cistulli C, Lavrik OI, Prasad R, Hou E, Wilson SH. AP endonuclease and poly(ADP-ribose) polymerase-1 interact with the same base excision repair intermediate. DNA Repair. 2004;3:581–591. doi: 10.1016/j.dnarep.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–48. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Figueroa JD, Malats N, Real FX, Silverman D, Kogevinas M, Chanock S, et al. Genetic variation in the base excision repair pathway and bladder cancer risk. Hum Genet. 2007;121:233–42. doi: 10.1007/s00439-006-0294-y. [DOI] [PubMed] [Google Scholar]
- Frank G, Qiu J, Zheng L, Shen B. Stimulation of eukaryotic flap endonuclease-1 activities by proliferating cell nuclear antigen (PCNA) is independent of its in vitro interaction via a consensus PCNA binding region. J Biol Chem. 2001;276:36295–302. doi: 10.1074/jbc.M103397200. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Current Opinion in Genetics & Development. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Gary R, Kim K, Cornelius HL, Park MS, Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J Biol Chem. 1999;274:4354–63. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Guo Z, Zheng L, Dai H, Zhou M, Xu H, Shen B. Human DNA polymerase beta polymorphism, Arg137Gln, impairs its polymerase activity and interaction with PCNA and the cellular base excision repair capacity. Nucleic Acids Res. 2009;37:3431–41. doi: 10.1093/nar/gkp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–95. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- Klungland A, Lindahl T. Second pathway for completion of human DNA base excisionrepair: reconstitution with purified proteins and requirement for DNase IV (FEN1) Embo J. 1997;16:3341–8. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem J. 1997;325(Pt 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi S, Gemignani F, Monnier S, Canzian F. A database of single-nucleotide polymorphisms and a genotyping microarray for genetic epidemiology of lung cancer. Exp Lung Res. 2005;31:223–58. doi: 10.1080/01902140490495624. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, et al. Removal of Oxidative DNA Damage via FEN1-Dependent Long-Patch Base Excision Repair in Human Cell Mitochondria. Mol Cell Biol. 2008;28:4975–87. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beard WA, Shock DD, Prasad R, Hou EW, Wilson SH. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J Biol Chem. 2005;280:3665–74. doi: 10.1074/jbc.M412922200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- Merrick CJ, Jackson D, Diffley JF. Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem. 2004;279:20067–75. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- Ming Y, Huan G, Chen W, Yuefeng H, Dianke Y, Li Z, et al. Functional FEN1 polymorphisms are associated with DNA damage levels and lung cancer risk. Human Mutation. 2009;30:1320–1328. doi: 10.1002/humu.21060. [DOI] [PubMed] [Google Scholar]
- Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 1998;58:222–5. [PubMed] [Google Scholar]
- Pascucci B, Russo MT, Crescenzi M, Bignami M, Dogliotti E. The accumulation of MMS-induced single strand breaks in G1 phase is recombinogenic in DNA polymerase beta defective mammalian cells. Nucleic Acids Res. 2005;33:280–8. doi: 10.1093/nar/gki168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Batra VK, Yang XP, Krahn JM, Pedersen LC, Beard WA, et al. Structural insight into the DNA polymerase beta deoxyribose phosphate lyase mechanism. DNA Repair (Amst) 2005;4:1347–57. doi: 10.1016/j.dnarep.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–41. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- Rotman G, Shiloh Y. ATM: from gene to function. Hum Mol Genet. 1998;7:1555–63. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- Shen B, Singh P, Liu R, Qiu J, Zheng L, Finger LD, et al. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27:717–29. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- Singhal RK, Wilson SH. Short gap-filling synthesis by DNA polymerase beta is processive. J Biol Chem. 1993;268:15906–11. [PubMed] [Google Scholar]
- Sun B, Latham KA, Dodson ML, Lloyd RS. Studies on the catalytic mechanism of five DNA glycosylases. Probing for enzyme-DNA imino intermediates. J Biol Chem. 1995;270:19501–8. doi: 10.1074/jbc.270.33.19501. [DOI] [PubMed] [Google Scholar]
- Vainio H, Boffetta P. Mechanisms of the combined effect of asbestos and smoking in the etiology of lung cancer. Scand J Work Environ Health. 1994;20:235–42. doi: 10.5271/sjweh.1402. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JHJ, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Wang Y, Putnam CD, Kane MF, Zhang W, Edelmann L, Russell R, et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat Genet. 2005;37:750–5. doi: 10.1038/ng1587. [DOI] [PubMed] [Google Scholar]
- Wong D, Demple B. Modulation of the 5′Deoxyribose-5-phosphate Lyase and DNA Synthesis Activities of Mammalian DNA Polymerase β by Apurinic/Apyrimidinic Endonuclease 1. Journal of Biological Chemistry. 2004;279:25268–25275. doi: 10.1074/jbc.M400804200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, Li M, Singh P, Qiu J, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med. 2007;13:812–9. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–36. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]