Abstract
The heme ATP binding cassette (ABC) transporter, ShuUV, of Shigella dysenteriae has been incorporated into proteoliposomes. Functional characterization of ShuUV revealed that ATP hydrolysis and transport of heme from the periplasmic binding protein, ShuT, to the cytoplasmic binding protein, ShuS, are coupled. Site-directed mutagenesis of ShuT residues proposed to be required for stabilization of the complex abolished heme transport. Furthermore, residues His-252 and His-262, located in the translocation channel of ShuU, were required for the release of heme from ShuT and translocation to ShuS. The initial functional characterization of an in vitro heme uptake system provides a platform for future spectroscopic studies.
Gram-negative bacterial pathogens have evolved sophisticated mechanisms for utilizing the hosts’ heme-containing proteins as a source of iron (1, 2). In Shigella dysenteriae, active transport of heme into the periplasm is facilitated by coupling the proton motive force of the cytoplasmic membrane through TonB to the outer membrane receptor, ShuA (3). Heme is then sequestered in the periplasm by the periplasmic binding protein (PBP), ShuT. Translocation of heme into the cytoplasm is facilitated by a heme-specific ABC transporter, ShuUV. ABC transporters make up a diverse family of proteins that use energy from hydrolysis of ATP to pump ligands against a concentration gradient. They are involved in nutrient uptake, osmotic regulation, toxin excretion, and multidrug efflux (4-6). Several ABC transporters with homology to the heme uptake systems have been structurally characterized, including the vitamin B12–BtuCD complex (7-11). The heme ABC transporter consists of two membrane-spanning domains (MSDs) of ShuU, which form a translocation pathway, and two nucleotide-binding domains (NBDs) of ShuV.
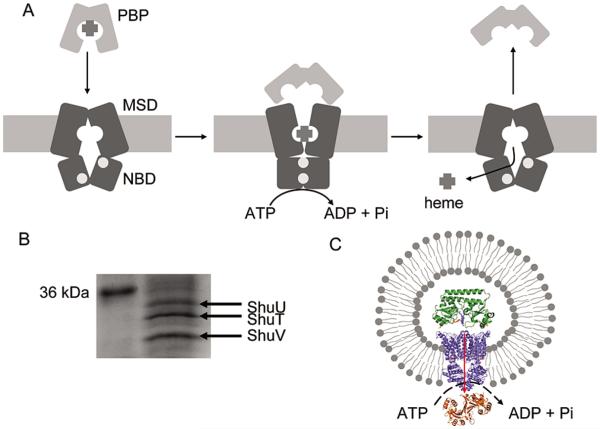
A concerted mechanism for translocation of vitamin B12 by BtuCD has been proposed largely on the basis of structural studies (5, 10-12). In the resting state, the translocation channel of the MSD is closed to the periplasm and open to the cytoplasm (Figure 1A). Interaction of the PBP with the transporter triggers the closing of the NBD interface, generating the open conformation of the PBP such that the affinity for the ligand is reduced. This conformational change in ShuUV opens the translocation channel and brings the NBDs together. Following ATP hydrolysis, the PBP is released, and the NBD dimer reopens, releasing the substrate and resetting the transporter.
FIGURE 1.
Structure and mechanism of the ABC transport system. (A) Proposed mechanism for translocation of heme by ShuUV. (B) Sodium dodecyl sulfate–polyacrylamide electrophoresis of the proteoliposomes with ShuT in the lumen. (C) Schematic of the proteoliposome system indicating the orientation of transport from ShuT in the lumen to ShuS in the external medium.
In this work, we report the first in vitro translocation of heme from the PBP, ShuT, through the ABC transporter, ShuUV, to the cytoplasmic heme-binding protein ShuS. ShuUV was incorporated into 3:1 (w/w) Escherichia coli lipid/L-α-phosphatidylcholine liposomes. On the basis of Sodium dodecyl sulfate–polyacrylamide electrophoresis (SDS–PAGE), we estimate that 50% of the added ShuUV is incorporated into the liposomes (Figure 1B). ShuT was encapsulated within the lumen of the ShuUV proteoliposomes by multiple freeze–thaw cycles (13-15). The heme-bound ShuT protein encapsulated within the proteoliposomes was quantified by pyridine hemochrome assuming a 1:1 heme:protein ratio (16). Approximately 68% of the ShuT added to the proteoliposomes was routinely incorporated into the lumen. The rate of ATP hydrolysis was calculated from a NaH2PO2 calibration curve generated by the modified molybdate method (17). The amount of heme transported from ShuT to apo-ShuS in the external solution was calculated by the pyridine hemochrome method.
It should be noted that ShuUV may be incorporated into the proteoliposomes right-side-in or, as shown in Figure 1C, inside-out. Although we cannot accurately calculate the ratio of the inside-out to right-side-in forms of ShuUV, on the basis of previous studies of the BtuCD complex it has been shown that the transporters partition in an approximately 50: 50 ratio (13). In this study, we have utilized the inside-out proteins to assess transport of heme to ShuS in the external medium. As shown in Table 1, addition of ATP to the ShuUV proteoliposome preparation gave a basal stimulation of ATP hydrolysis but yielded no further increase over time. In contrast, heme-bound ShuT within the lumen and ShuS in the external medium significantly increases the rate of ATP hydrolysis (Figure 2 and Table 1).
Table 1.
Heme Translocation and ATP Hydrolysis for the Wild-Type and Mutant ShuUV Transportersa
| ShuUV transporter | total amount of heme transferred to ShuS (nmol)d | total amount of ATP hydrolysis (nmol)e |
|---|---|---|
| Wild-Type ShuUV System | ||
| ShuUVc | 2.2 ± 0.1 | 1280 ± 40 |
| ShuUV with ShuT | 2.2 ± 0.1 | 1750 ± 44 |
| ShuUV with heme-bound ShuTb | 16 ± 0.7 | 4353 ± 328 |
| ShuUV with heme-bound Glu-74 Val ShuTb | 1.0 ± 0.04 | 2034 ± 153 |
| ShuUV with heme-bound Glu-207 Val ShuTb | 1.7 ± 0.1 | 1630 ± 33 |
| ShuUV with heme-bound Glu-74/207 Val ShuTb | 2.5 ± 0.35 | 1596 ± 43 |
| ShuUV Mutant System | ||
| His-252 Alac | 3.0 ± 0.1 | 1096 ± 68 |
| His-252 Ala with heme-bound ShuTb | 3.9 ± 0.42 | 1949 ± 78 |
| His-262 Alac | 2.0 ± 0.5 | 460 ± 35 |
| His-262 Ala with heme-bound ShuTb | 2.6 ± 0.15 | 580 ± 74 |
| His-252/262 Alac | 2.0 ± 0.5 | 952 ± 47 |
| His-262 Ala with heme-bound ShuTb | 2.6 ± 0.15 | 895 ± 238 |
The amount of heme-bound ShuT within the lumen was calculated by pyridine hemochrome to be 17 ± 0.6 nmol.
The amount of heme-bound ShuT within the lumen was calculated by pyridine hemochrome to be 17 ± 0.6 nmol.
Basal ATP hydrolysis measured in the absence of ShuT.
The amount of heme transferred to ShuS at the 10 min time point was measured by pyridine hemochrome following removal of the liposomes.
Total ATP hydrolysis at the 10 min time point.
FIGURE 2.
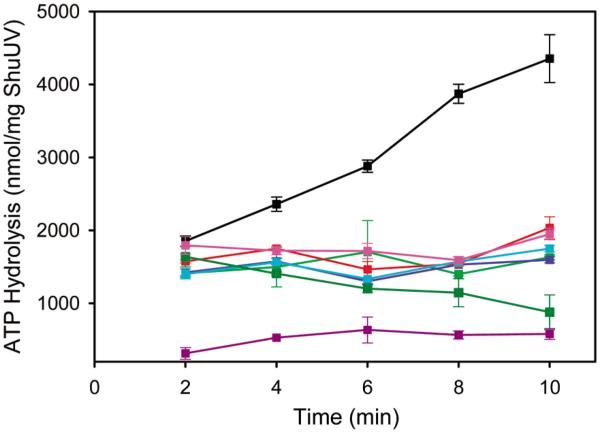
Hydrolysis of ATP over time. Wild-type ShuUV with heme-bound ShuT (black), ShuT (cyan), heme-bound ShuT E74V (red), heme-bound ShuT E207V (light green), heme-bound ShuT E74/207V (blue), ShuU H252A (magenta), ShuU H262A (purple), and ShuU His-252/262 Ala (green) with heme-bound ShuT. All experiments included ShuS in the external medium.
The increase in the level of ATP hydrolysis appears to be coupled to the translocation of heme from heme-bound ShuT in the lumen to apo-Shus in the external medium (Table 1). Indeed, on the basis of the calculated amount of heme-bound ShuT trapped in the lumen of the proteoliposomes, complete transfer of the heme from ShuT in the lumen to apo-ShuS in the external medium is observed (Table 1). However, ShuT within the lumen of the proteoliposmes did not stimulate an increase in the level of ATP hydrolysis and is similar to the initial stimulation observed in the absence of ShuT (Figure 2 and Table 1). These data are consistent with a concerted model, in which the closed conformation of heme-bound ShuT is required for interaction with the ShuUV transporter, thus inducing the conformational change required to bring the NBDs together for ATP hydrolysis. Recent crystallographic studies from our collaborators have confirmed the open and closed forms of ShuT and heme-bound ShuT, respectively (18).
On addition of ATP to the ShuUV proteoliposomes alone or the proteoliposomes in the presence of ShuT, a residual amount of heme is released to ShuS and corresponds to the fraction of heme trapped nonspecifically within ShuUV on purification. Interestingly, upon addition of ATP to the heme-bound ShuT/ShuUV proteoliposome system in the absence of ShuS, no sustained increase in the rate of hydrolysis is observed, indicating that in the absence of ShuS the heme is most likely not released to solvent. These data provide the first evidence that the cytoplasmic heme-binding protein ShuS directly accepts heme from the ABC transporter ShuUV. This is in agreement with previous genetic studies that have shown the ShuS protein is required for efficient heme utilization, and in its absence, high concentrations of heme are toxic to the cell (19). The requirement that ShuS accept heme from ShuUV is not surprising given the ability of free heme to catalyze the production of reactive oxygen species that may cause DNA and lipid damage. These data differ from those of previous studies on the BtuCD/BtuF system in which cobalamin is apparently released into the lumen of the proteoliposomes in the absence of an acceptor protein (13). However, this may be due to the differences in the physiochemical properties of the water soluble corrin ring of vitamin B12 versus the more hydrophobic porphyrin macrocycle.
In the concerted model of translocation, it is proposed that the PBP remains complexed to the ABC transporter on ligand translocation (13). Previous structural studies of the BtuCD and BtuF proteins have suggested that specific Glu residues on the lobes of BtuF (Glu-72 and −202) are required for stabilizing the interaction with the BtuCD ABC transporter (7). To test this hypothesis, the conserved Glu residues on the surface of the lobes of ShuT, Glu-74 and Glu-207, were mutated to Val (see Figure S1 of the Supporting Information). The addition of ATP to the single or double Glu heme-bound ShuT mutants within the ShuUV-reconstituted proteoliposomes resulted in no increase in the rate of ATP hydrolysis (Figure 2), and consequently no translocation of heme to ShuS. Therefore, in addition to requiring the closed conformation of heme-bound ShuT, Glu-74 and Glu-207 are essential for stabilizing the protein–protein interaction and driving the conformational change required for substrate release and ATP hydrolysis.
Furthermore, a sequence alignment of all predicted heme ABC transporters revealed two absolutely conserved His residues. The conserved His residues, which are also present in the vitamin B12 transporter, BtuC, are located in the substrate translocation channel. Therefore, we hypothesized that the corresponding residues in ShuU, His-252 and His-262, are responsible for coordination of heme in the translocation channel. A homology model of ShuUV indicating the location of the conserved His residues is given in the Supporting Information (Figure S1). To determine if His-252 and/or His-262 is required for heme transport, site-directed mutagenesis was utilized to create both the single and double His to Ala ShuU mutants. Proteoliposomes of the His-252 Ala, His-262 Ala, or His-252/262 Ala ShuUV mutants with heme-bound ShuT incorporated into the lumen were tested for their ability to transport heme to ShuS. In the presence of heme-bound ShuT in the lumen, none of the His ShuUV mutants gave an increase in the rate of ATP hydrolysis (Figure 2). Furthermore, the amount of heme transferred to ShuS compares with that calculated to be nonspecifically associated with ShuUV on purification (Table 1). The inability of heme to be translocated from heme-bound ShuT to ShuS when either His-252 or His-262 is mutated suggests that both residues are essential for the transport of heme. Furthermore, the fact that there is no increase in the rate of ATP hydrolysis in the proteoliposomes containing the ShuUV His mutants, and no heme is translocated to ShuS, suggests the heme may be kinetically trapped within ShuT. We therefore hypothesize that His-252 and His-262 provide a high-affinity binding site within the translocation channel of ShuU that is required, in addition to the conformational changes induced on binding of heme-bound ShuT, for the release of heme to the transporter. The functional characterization of ShuT and ShuUV mutants in which we can potentially trap kinetic intermediates provides a platform for future spectroscopic studies.
In summary, the in vitro heme uptake system has provided new insight into the mechanism of heme transport in bacterial pathogens. In this study, we have further refined the concerted mechanism of ligand translocation in ABC transporters (Figure 1A) by providing evidence that substrate coordination within the translocation channel of ShuU is directly coupled to ATP hydrolysis and substrate release. Thus, ATP hydrolyis triggers the ligand-gated switch that releases the substrate at the cytoplasmic face, resets the closed conformation at the periplasmic face, and allows release of the substrate free PBP (Figure 1A). The ABC transporter on release of the substrate free PBP is reset to the resting state to undergo another cycle upon binding the substrate-loaded PBP. Future site-directed mutagenesis studies combined with spectroscopic methods will aid in the elucidation of the molecular mechanism of translocation of heme across the cytoplasmic membrane.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Franz St. John for his help in constructing the homology models of ShuUV and ShuS.
Footnotes
This work was supported by NIH Grant AI-48551 to A.W.
SUPPORTING INFORMATION AVAILABLE
Experimental methods for protein purification, proteoliposome preparation, ATP hydrolysis, heme measurements, and homology model construction. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Wilks A, Burkhard KA. Nat. Prod. Rep. 2007;24:511–522. doi: 10.1039/b604193k. [DOI] [PubMed] [Google Scholar]
- 2.Wandersman C, Delepelaire P. Annu. Rev. Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 3.Burkhard KA, Wilks A. J. Biol. Chem. 2007;282:15126–15136. doi: 10.1074/jbc.M611121200. [DOI] [PubMed] [Google Scholar]
- 4.Higgins CF. Res. Microbiol. 2001;152:205–210. doi: 10.1016/s0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- 5.Davidson AL, Maloney PC. Trends Microbiol. 2007;15:448–455. doi: 10.1016/j.tim.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Davidson AL, Chen J. Annu. Rev. Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 7.Borths EL, Locher KP, Lee AT, Rees DC. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16642–16647. doi: 10.1073/pnas.262659699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locher KP, Lee AT, Rees DC. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 9.Locher KP, Borths E. FEBS Lett. 2004;564:264–268. doi: 10.1016/S0014-5793(04)00289-3. [DOI] [PubMed] [Google Scholar]
- 10.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Nature. 2007;450:515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 11.Hvorup RN, Goetz BA, Niederer M, Hollenstein K, Perozo E, Locher KP. Science. 2007;317:1387–1390. doi: 10.1126/science.1145950. [DOI] [PubMed] [Google Scholar]
- 12.Locher KP. Curr. Opin. Struct. Biol. 2004;14:426–431. doi: 10.1016/j.sbi.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Borths EL, Poolman B, Hvorup RN, Locher KP, Rees DC. Biochemistry. 2005;44:16301–16309. doi: 10.1021/bi0513103. [DOI] [PubMed] [Google Scholar]
- 14.Doeven MK, Abele R, Tampe R, Poolman B. J. Biol. Chem. 2004;279:32301–32307. doi: 10.1074/jbc.M404343200. [DOI] [PubMed] [Google Scholar]
- 15.Liu CE, Ames GF. J. Biol. Chem. 1997;272:859–866. doi: 10.1074/jbc.272.2.859. [DOI] [PubMed] [Google Scholar]
- 16.Fuhrop JH. In: Porphyrins and Metalloporphyrins. Smith KM, editor. Elsevier; Amsterdam: 1975. [Google Scholar]
- 17.Chifflet S, Torriglia A, Chiesa R, Tolosa S. Anal. Biochem. 1988;168:1–4. doi: 10.1016/0003-2697(88)90002-4. [DOI] [PubMed] [Google Scholar]
- 18.Ho WW, Li H, Eakanunkul S, Tong Y, Wilks A, Guo M, Poulos TL. J. Biol. Chem. 2007;282:35796–35802. doi: 10.1074/jbc.M706761200. [DOI] [PubMed] [Google Scholar]
- 19.Wyckoff EE, Lopreato GF, Tipton KA, Payne SM. J. Bacteriol. 2005;187:5658–5664. doi: 10.1128/JB.187.16.5658-5664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.