Abstract
Mitochondria play a central role not only in energy production but also in the integration of metabolic pathways as well as signals for apoptosis and autophagy. It is becoming increasingly apparent that mitochondria in mammalian cells play critical roles in the initiation and propagation of various signaling cascades. In particular, mitochondrial metabolic and respiratory states and status on mitochondrial genetic instability are communicated to the nucleus as an adaptive response through retrograde signaling. Each mammalian cell contains multiple copies of mitochondrial genome (mtDNA). A reduction in mtDNA copy number has been reported in various human pathological conditions such as diabetes, obesity, neurodegenerative disorders, aging and cancer. Reduction in mtDNA copy number disrupts mitochondrial membrane potential (Δψm) resulting in dysfunctional mitochondria. Dysfunctional mitochondria trigger retrograde signaling and communicate their changing metabolic and functional state to the nucleus as an adaptive response resulting in altered nuclear gene expression profile and altered cell physiology and morphology. In this review, we provide an overview of the various modes of mitochondrial retrograde signaling focusing particularly on the Ca2+/Calcineurin mediated retrograde signaling. We discuss the contribution of the key factors of the pathway such as Calcineurin, IGF1 receptor, Akt kinase and HnRNPA2 in the propagation of signaling and their role in modulating genetic and epigenetic changes favoring cellular reprogramming towards tumorigenesis.
Keywords: Mitochondrial DNA, Mitochondrial Retrograde Signaling, Epigenetics, Calcineurin, HnRNPA2, Cancer
1. Introduction
Mitochondria in mammalian cells generate most of the cellular energy, ATP which is essential for performing multitude of cellular functions (Sagan, 1967). They are also involved in key cellular functions such as maintaining calcium homeostasis, heme biosynthesis, nutrient metabolism, steroid hormone biosynthesis, removal of NH4+ through urea synthesis, integration of metabolic pathways and integration of cellular signals for apoptosis and autophagy (Cheng and Ristow, 2013;Hammerman, Fox et al. 2004;Marchetti, Castedo et al. 1996;Plas and Thompson, 2002;Renault and Chipuk, 2013). More recent and emerging view is the central role of mitochondria in initiating signals in response to metabolic and genetic stress which affects the nuclear gene expression in a global way, causing changes in cell physiology and function. Mitochondria contain multiple copies of their own DNA genome (mtDNA) which varies from 100–1000 copies per cell depending on the cell and tissue type (Ryan and Hoogenraad, 2007). The human mtDNA is a ~16.5 kbp circular DNA which codes for 13 subunits of the mitochondrial respiratory chain components including 7 subunits of Complex I, 1 subunit of Complex III, 3 subunits of complex IV, and 2 subunits of Complex V in addition to 22 tRNAs, 2 rRNAs, D-loop encoded RNA of unknown function (Vijayasarathy, Biunno et al. 1998; Borst, 1972;Garcia-Rodriguez, 2007) and RNA primers for DNA replication (Bhat, Avdalovic et al. 1989). The limited coding capacity of mtDNA necessitates a major contribution of proteins from the nuclear genome in shaping the tissue and cell specific as well as ubiquitous metabolic reactions and molecular architecture of mitochondria (Westermann and Neupert, 2003). It is estimated that 1500–2000 nuclear encoded and cytoplasmically translated proteins may be imported by mammalian mitochondria. It is therefore apparent that a coordinated expression of the mitochondrial and nuclear genomes is critical in maintaining healthy and functional mitochondria.
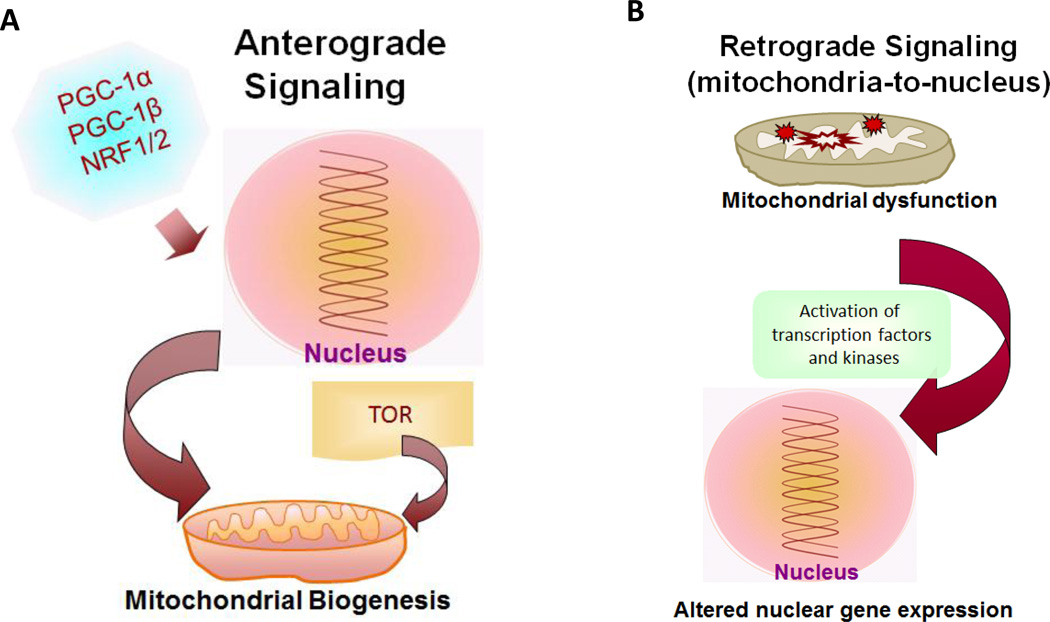
It is becoming increasingly clear that defects in nuclear-encoded mitochondrial proteins, mtDNA-coded proteins and RNAs affecting mitochondrial functions, or altered apoptotic and survival signaling pathways can result in mitochondrial pathologies (Arnold, Sun et al. 2013;Taylor and Turnbull, 2005;Wallace, 2013). Broadly, two distinct signaling pathways exist that modulate mitochondrial structure/function and hence cellular function, causing mitochondrial diseases: 1) Anterograde signaling through activation of nuclear transcription and cytoplasmic mRNA translation which directly regulate mitochondrial biogenesis (See Fig 1A), and 2) Retrograde signaling which is a cellular adaptation to mitochondrial dysfunction induced by low mtDNA copy number, mitochondrial mutations or nuclear mutations which affect mitochondrial function and mitochondrial respiratory defects (Fig. 1B). In particular, the Ca2+/Calcineurin mediated mitochondrial retrograde signaling and its role in cancer progression will be the major focus of this review.
Figure 1. Outline of the two major mitochondrial signaling pathways.
(A) Anterograde signaling is transmitted by activated nuclear transcription factors and cytoplasmic proteins essential for mitochondrial biogenesis. (B) Retrograde signaling originates in dysfunctional mitochondria and communicated to the nucleus resulting in altered expression of nuclear genes.
2. Mitochondrial Anterograde Signaling
Due to an overwhelmingly dominant role of the nuclear genome in regulating mitochondrial structure and functions, most studies are focused on nucleus-to-mitochondria “anterograde” signaling. This signaling controls the flow of proteins and information from the nucleus to mitochondria. As mentioned earlier, mitochondrial biogenesis and functions are tightly regulated by both mitochondrial and nuclear genomes (Garesse and Vallejo, 2001; Attardi and Schatz, 1998).
Mitochondrial biogenesis is a dynamic process which occurs in response to various physiological stimuli and cellular differentiation states. Independent approaches including biochemical, proteomics, bioinformatics and metabolomics have identified a number of nuclear-encoded mitochondrial factors involved in the regulation and coordination of mitochondrial gene expression (Garesse and Vallejo, 2001;Jain, Nilsson et al. 2012;Meisinger, Sickmann et al. 2008;Pagliarini, Calvo et al. 2008;Rensvold, Ong et al. 2013;Rhee, Zou et al. 2013;Westermann and Neupert, 2003). The key proteins required for the replication and expression of the mitochondrial genome including DNA and RNA polymerases, RNA processing enzymes, transcription/mtDNA replication factors, and ribosomal proteins are all nuclear encoded. This includes the mtDNA transcription factors mtTFA (TFAM), mtTFB I and mtTFB II and transcription termination factors MTERF (Aloni and Attardi, 1971;Bestwick and Shadel, 2013;Yakubovskaya, Mejia et al. 2010;Zollo, Tiranti et al. 2012). Additionally, most of the proteins of TCA cycle, fatty acid metabolism, amino acid metabolism, all of the mitochondrial outer membrane, intermembrane space and a vast majority of inner membrane associated proteins are encoded by the nuclear genes.
Two classes of nuclear transcriptional factors/regulators appear critical for the mitochondrial biogenesis in response to various physiological stimuli. The first includes DNA-binding transcription factors, nuclear respiratory factors (NRF)-1 and 2 that regulate nuclear genes involved in maintaining mitochondrial functions and mitochondrial biogenesis (Kelly and Scarpulla, 2004). The second includes transcription coactivators, peroxisome proliferator-activated receptor gamma coactivator 1(PGC-1) alpha, beta and gamma isoforms and PGC-1 related factor, PRC (Puigserver, Wu et al. 1998). The PGC factors are responsive to physiological and nutritional signals mediating thermogenesis, cell proliferation and gluconeogenesis and thereby integrate the action of multiple transcription factors in coordinating nuclear gene expression programs essential to cellular bioenergetics (Meirhaeghe, Crowley et al. 2003). These factors also regulate the expression of mtTFA (TFAM), mtTFB I, mtTFB II and mitochondrial RNA polymerase thus coordinately regulating mitochondrial gene expression. Another important regulation of mitochondrial signaling is by nutrient availability and sensing by the mTOR complex. The TOR kinase acts as nutrient, energy and stress sensor and mTORC2 is activated by growth factor receptor which in turn phosphorylates many kinases like S6Kinase, Akt and PI3 kinase (Laplante and Sabatini, 2012). Since the mTOR signaling is activated in response to the stress signaling pathway, it is a potential regulatory loop at the convergence of mitochondrial anterograde and retrograde signaling.
A large majority of the nucleus-encoded metabolic enzymes necessary for the oxidation of pyruvate, fatty acids (β-oxidation cycle), and acetyl-CoA (tricarboxylic acid cycle), amino acids biosynthesis, and heme synthesis are mitochondrially localized and can regulate mitochondrial functions. Moreover, the steroid and thyroid hormones control mitochondrial functions indicating that mitochondria are responsive to the cellular physiological state (Casas, Daury et al. 2003;Weitzel, Iwen et al. 2003). Cell type-specific regulatory proteins such as the myogenin transcription factor family have a strong influence on some mitochondrial genes in specific cell types (Flynn, Meadows et al. 2010). Mitochondrial localized protein import and transport factors essential for its functions are all nuclear-encoded and dysregulation of these import factors can in turn have profound impact on mitochondrial biogenesis and functions. A new area of research suggests that mtDNA molecules are packaged along with protein components essential for the maintenance and function of mtDNA within nucleoids (Gilkerson, Bravo et al. 2013;He, Cooper et al. 2012;Kaufman, Durisic et al. 2007;Wang and Bogenhagen, 2006); since vast majority of nucleoid proteins are nuclear-encoded, defects in the nucleoid proteins can in turn affect the maintenance of mitochondrial structural integrity and function. Therefore anterograde signaling involving the interplay of various nuclear factors is considered to be a major determinant in the regulation of mitochondrial biogenesis.
3. Mechanisms of mitochondrial retrograde signaling
Mitochondrial retrograde signaling, an emerging area of mitochondrial research, is focused on pathways by which dysfunctional mitochondria communicate with the nuclear genetic compartment for relaying metabolic, oxidative and respiratory conditions prevailing in the mitochondria (Clayton, 1998;Liao and Butow, 1993;Poyton and McEwen, 1996). Defects in mitochondrial respiratory chain components, mtDNA mutations and alterations in mtDNA copy number resulting in the loss of mitochondrial transmembrane potential (Δψm), act as inducers of mitochondrial retrograde signaling. Some models suggest that mitochondrial ROS also induces retrograde signaling. The retrograde signaling is possibly a cellular adaptation mechanism by which dysfunctional mitochondria transmit signals to effect changes in nuclear gene expression leading to metabolic reconfiguration (Amuthan, Biswas et al. 2002;Biswas, Adebanjo et al. 1999;Butow and Avadhani, 2004;Chae, Ahn et al. 2013;Delsite, Kachhap et al. 2002;Guha, Tang et al. 2010;Jazwinski and Kriete, 2012). This signaling was initially reported in Saccharomyces cerevisiae by Butow’s laboratory (Parikh, Conrad-Webb et al. 1989;Liao and Butow, 1993) and has since been observed in mammalian cells of various origins. There are cell-specific differences in factors involved in the propagation of retrograde signaling, although [Ca2+]c is a key molecule involved in both antero- and retrograde mitochondrial-nuclear signaling in response to various stimuli (Amuthan, Biswas et al. 2002;Biswas, Adebanjo et al. 1999;Bononi, Missiroli et al. 2012;Butow and Avadhani, 2004;Cali, Ottolini et al. 2012;Glancy and Balaban, 2012;Hajnoczky and Csordas, 2010;Kelly and Scarpulla, 2004;Le, Mirebeau-Prunier et al. 2011;Wang and Schwarz, 2009). The retrograde signaling pathway interacts with several other stress responsive signaling pathways including metabolic stress, such as target of rapamycin (TOR) and ceramide signaling. The retrograde response is also linked to quality control mechanisms, such as autophagy and mitophagy involving the segregation of dysfunctional mitochondria (Jazwinski and Kriete, 2012). Increasing evidence suggests the involvement of mitochondrial retrograde signaling in various pathological conditions. One of the early observations of mitochondrial retrograde signaling in a pathological setting was reported in MERRF (Myoclonic Epilepsy with Ragged Red Fibers). Low levels of ATP due to inefficient ETC, triggers mitochondria to nuclear signaling resulting in proliferation of defective mitochondria and increased mitochondrial mass in muscle as an attempt to correct the deficiency (Shoffner, Lott et al. 1990). Another report showed the involvement of the retrograde signaling in maternally inherited deafness associated with the A1555G mtDNA mutation. In patient-derived A1555G cells, the methyltransferase mtTFB1-mediated 12S rRNA hypermethylation resulted in ROS generation which in turn activated AMP- kinase and the pro-apoptotic nuclear transcription factor E2F1. In transgenic-mtTFB1 mice ectopic overexpression of E2F1 resulted in apoptosis in the stria vascularis and spiral ganglion neurons of the inner ear, and progressive E2F1-dependent hearing loss (Raimundo, Song et al. 2012;Woo, Green et al. 2012). Mitochondrial retrograde signaling arising from mtDNA mutations and alterations in mtDNA copy number has been implicated as a causal factor in tumorigenesis (Amuthan, Biswas et al. 2002;Biswas, Adebanjo et al. 1999;Brandon, Baldi et al. 2006;Carew and Huang, 2002;Delsite, Kachhap et al. 2002;Guha, Srinivasan et al. 2007;Guo, Zheng et al. 2011;Higuchi, Kudo et al. 2006;Kulawiec, Safina et al. 2008;Park, Sharma et al. 2009;Wallace, 2012;Yu, Shi et al. 2009). Due to reprogramming of the nuclear genome in clinical tumor specimens, it has remained difficult to establish the causal role of the mitochondrial genome defects in driving tumorigenesis. However, a recent report showing mice heterozygous for the mtDNA copy number regulator TFAM which have reduced mtDNA copy number exhibit increased tumor incidence and growth when crossed with the adenomatous polyposis coli multiple intestinal neoplasia (APCMin−/+) mouse model (Woo, Green et al. 2012). In addition, several studies using mtDNA cybrid models (Kaipparettu et al 2013; Fan et al 2012), where nuclear background is unchanged, suggest mitochondrial genome defects and retrograde signaling play a causal role in tumorigenesis.
Mitochondrial dysfunction has been correlated with age-associated disorders and most mtDNA defects accumulate with age and are progressive in nature. The direct evidence for the causal role of mitochondrial dysfunction and retrograde signaling in aging was demonstrated in two independent reports using (Polg−/−) mutator mice. These mice with a knockin mutation (D257A) in the exonuclease domain of mtPOLG and have a loss in proofreading function and as a result of a very high amount of mtDNA mutations, these mice exhibit signs of premature aging (Kujoth, Hiona et al. 2005;Trifunovic, Wredenberg et al. 2004).
Several different mechanisms of mitochondrial retrograde signaling have been described that are propagated by different pathways. In this section we provide a mechanistic overview of the different modes of mitochondrial retrograde stress signaling reported thus far. It is important to point out that these different pathways activate many common signaling molecules suggesting a potential convergence of the retrograde signaling independent of the inducing stimuli.
A. Signaling due to oxidative stress induced free radicals
The mitochondrial inner membrane associated electron transport chain is the major site of generation of cellular reactive oxygen species (ROS). Increased production of mitochondrial ROS can damage the electron transport chain (ETC) components resulting in mitochondrial respiratory stress (Kirkinezos and Moraes, 2001). The localization of mtDNA in close proximity to the electron transport chain (ETC) components renders them susceptible to damage by faulty ETC. Therefore, mitochondria play a central role as both producers and sensors of oxidative stress. When the mitochondrial ROS levels exceed that of the antioxidant defense system, it can potentially cause mtDNA damage (Yakes and Van, 1997). Additionally, the mtDNA damage repair machinery is not as regulated, which subjects mtDNA to higher susceptibility to damage than the nuclear DNA. Therefore either ROS itself or by their oxidative damage on mtDNA can impair mitochondrial function and therefore can induce mitochondrial retrograde signaling (Yakes and Van, 1997). MtROS mediated mitochondrial retrograde signaling is also implicated in many pathological conditions (Formentini, Sanchez-Arago et al. 2012). Mitochondrial ROS production is steeply increased under hypoxia, ischemia/reperfusion injury, chemical stress/drug treatment and under many pathophysiological conditions (Srinivasan and Avadhani, 2012). In NARP (Neuropathy, Ataxia and Retinitis Pigmentosa) syndrome, mitochondrial biogenesis has been shown to be regulated by ROS-dependent retrograde signaling. Antioxidant treatment using selenite increased NRF1 and NRF2 levels, thereby modulating mitochondrial biogenesis (Wojewoda, Duszynski et al. 2011). Fan et al have recently reported that mtDNA heteroplasmic mutations in a mouse L cell line resulted in oxidative stress and increased ROS production and the ROS mediated signaling providing survival advantage to these cells (Fan, Lin et al. 2012;Wallace and Fan, 2009). Another example of mitochondrial ROS mediated retrograde signaling in the pathological setting is the mitochondrial localization of the adaptor protein Shc, p66 which regulates ROS production and silencing this gene has been reported to significantly increase the lifespan in mice (Migliaccio, Giorgio et al. 1999). In lung injury, alveolar epithelial cells develop hypoxia, resulting in mitochondrial ROS production and activation of HIF-1α gene targets. This resulted in cellular reprogramming leading to aberrant tissue repair, lung dysfunction and pulmonary fibrosis (Zhou, Dada et al. 2009). Caloric restriction modulates IGF1, TOR, sirtuins and AMP kinase signaling pathways which ameliorates mitochondrial oxidative stress (Raffaello and Rizzuto, 2011). The role of mitochondrial ROS mediated signaling in tumorigenesis has been demonstrated by various groups (Fan, Lin et al. 2012;Formentini, Sanchez-Arago et al. 2012;Hanna, Krishnan et al. 2013;Ishikawa, Takenaga et al. 2008;Pelicano, Xu et al. 2006;Woo, Green et al. 2012). A number of reviews focusing on this aspect of mitochondrial signaling have been published earlier (Naik and Dixit, 2011;Poyton, Ball et al. 2009;Sena and Chandel, 2012). Conflicting views exist on the role of mitochondrial ROS induced HIF-1α activation in the propagation of retrograde signaling. HIF-1α has been implicated as a positive regulator of genes involved in metabolic adaptations and stemness during tumorigenesis (Young and Simon, 2012). While HIF-1α has been shown to be essential in certain cell types for mitochondrial ROS signaling (Majmundar, Wong et al. 2010;Young and Simon, 2012), other studies suggest the involvement of mitochondrial ROS induced JNK/ERK pathway, independent of HIF-1α in initiating retrograde response to mitochondrial dysfunction (Chae, Ahn et al. 2013).
B. Mitochondrial unfolded protein response and generation of regulatory peptides
The mitochondrial unfolded protein response is a mode of retrograde signaling which originates from the efflux of peptides from damaged mitochondrial matrix proteins to the cytosol (Kirstein-Miles and Morimoto, 2010;Loveland, Wang et al. 1990). Mitochondrially generated peptides transported to the cytosol can potentially activate certain nuclear transcription factors resulting in retrograde signaling. For efficient functioning and biogenesis, mitochondria have a number of molecular chaperones to facilitate protein folding and proteases that degrade misfolded proteins. This results in induction of the mitochondrial quality control genes to re-establish the homeostasis within the mitochondrial protein folding machinery. It has been suggested that the mitochondrial protein folding machinery can be damaged by excessive ROS from Complex I and III of the ETC which directly affects protein folding and structure (Runkel, Liu et al. 2013). In yeast, Young et al demonstrated that peptides (6–20 amino acids) derived from proteolysis of inner mitochondrial membrane proteins are exported through the inner member by the ABC transporter, Mdl1 (Young, Leonhard et al. 2001). Recent work from the Haynes group demonstrated that perturbations in the mitochondrial folding machinery leads to protein degradation by the matrix localized ClpXP protease and their efflux by mt UPR component HAF-1 (Haynes, Yang et al. 2010). This mode of peptide signaling process is a stress response mechanism that distinguishes misfolded and damaged mitochondrial proteins and possibly acts as a sensory mechanism for the functional state of the mitochondria. This mode of stress signaling is a rapid response to diverse physiological challenges (Gregersen and Bross, 2010).
C. Mitochondrial Ca2+ Signaling
Calcium is an important signaling molecule involved in the regulation of many cellular functions (Williams, Boyman et al. 2013). Mitochondria maintain a large Ca2+ gradient across their inner membrane and mitochondrial Ca2+ uptake and release play a fundamental role in different physiological processes, such as ATP production and hormone metabolism, while dysregulation of mitochondrial Ca2+ handling triggers the apoptotic cascade through the opening of the permeability transition pore (mPTP) (Rizzuto and Pozzan, 2006;Giacomello, Drago et al. 2007;Bernardi, 1999;Hajnoczky and Csordas, 2010;Zamzami, Maisse et al. 2001;Duchen, 2000;Marchetti, Castedo et al. 1996). Cells largely depend on the mitochondria for Ca2+ buffering within their local environment for the regulation of various metabolic processes. Mitochondrial Ca2+ accumulation is ATP dependent and it utilizes gated channels for Ca2+ uptake and Na+ or H+/Ca2+ exchangers for release (Hajnoczky and Csordas, 2010;Raffaello, De et al. 2012;Rizzuto, De et al. 2012). The driving force for Ca2+ accumulation in the mitochondrial matrix is its membrane potential (ΔΨm) (Rizzuto, De et al. 2012;DELUCA and ENGSTROM, 1961). The exchangers use the Ca2+, H+ and Na+ concentration gradient across the inner membrane to cause the release of Ca2+ back into the cytosol. This ensures the maintenance of a low matrix Ca2+ concentration ([Ca2+]) in resting cells and rapid mitochondrial Ca2+ accumulation when cytosolic Ca2+ is elevated during activation. Hence, when ΔΨm is disrupted, mitochondrial capacity for Ca2+ uptake is impaired resulting in elevated cytosolic Ca2+ levels. It has been recently identified that calcium uptake by mitochondria occurs through the ion channel “uniporter” MCU and its regulatory partner, MICU1 which are conserved across species (Perocchi et al 2010; Mallilankaraman et al, 2012; Baughman et al, 2011; Williams, Boyman et al. 2011). Although details are still unclear, a mitochondrial Ryanodine receptor (RyR) has been suggested as a component of the mCU (Altschafl, Beutner et al. 2007). The activity of MCU is reported to be modulated by protein kinases PKD and PKC mediated phosphorylation (Pinton, Leo et al. 2004;Rizzuto, Duchen et al. 2004). Perturbations in the Ca2+ regulation can potentially offset the Ca2+ balance and trigger a Ca2+ dependent signaling.
Another aspect of calcium signaling of relevance to the mitochondria is its role in regulating mitochondrial motility. Mitochondria are dynamic organelles and their motility within the cell is crucial to meet the constant changing energy and nutrient demands within the cell. Calcium signaling acts as a regulatory input for mitochondrial movement through its binding to Miro thereby rendering the Miro bound kinesin 1 inactive (Saotome, Safiulina et al. 2008;Wang and Schwarz, 2009).
In addition to normal physiological functions, mitochondrial calcium signaling contributes to Ca2+ driven growth and survival pathways in many pathological conditions including cancer. A direct evidence of contribution of mitochondrial dysregulation of Ca2+ comes from a report showing that pharmacologic intervention with an agonist of the mitochondrial Na+/Ca2+ exchanger, CGP-37157, retarded cell growth and increased susceptibility to apoptosis in malignant B16BL6 melanoma cell line while elevated mitochondrial Ca2+ flux which corresponded with high levels of constitutively active protein kinase B/Akt (PKB) (Fedida-Metula, Feldman et al. 2012;Feldman, Fedida-Metula et al. 2010). Another strong evidence implicating Ca2+ dependent signaling in growth factor (EGF) and hypoxia induced epithelial-mesenchymal transition and metastatic reprogramming in breast cancer cells has just been reported by Davis FM et al (Davis, Azimi et al. 2013). It is now established that calcium signaling and mitochondrial response to cellular calcium concentration is an important regulator of cell survival and apoptotic pathways. The link between cytosolic and mitochondrial Ca2+ concentrations and the proximity of mitochondria to Ca2+ release sites makes Ca2+ an important signaling molecule in mitochondrial retrograde signaling pathways.
C (i). Ca2+/ Calcineurin (Cn) mediated Signaling
For more than a decade, research in our laboratory has focused on elucidating the mechanisms of mitochondrial retrograde signaling pathway in various mammalian cells (Amuthan, Biswas et al. 2002;Biswas, Adebanjo et al. 1999;Biswas, Guha et al. 2005;Biswas, Tang et al. 2008;Biswas, Srinivasan et al. 2008;Butow and Avadhani, 2004;Guha, Srinivasan et al. 2007;Guha, Pan et al. 2009;Guha, Tang et al. 2010;Guha, Fang et al. 2010;Srinivasan and Avadhani, 2007;Srinivasan, Koenigstein et al. 2010;Tang, Chowdhury et al. 2012;Amuthan, Biswas et al. 2001). It is now established that disruption of mitochondrial membrane potential ((Δψm) by either genetic (reduction in mtDNA copy number) and pharmacologic interventions (treatment with mitochondrial ionophores) and/or hypoxia-induced mitochondrial dysfunction activate a mitochondrial retrograde signaling pathway. Reduction in mtDNA copy number causes loss of ETC causing a disruption of the membrane potential (Δψm) and lower ATP level. Loss of membrane potential and low ATP disrupts the mitochondrial uptake of Ca2+. Higher leakage of ER calcium due to overexpression of RYR1 channels (discussed later) and lower uptake of Ca2+ by mitochondria results in a net elevated cytosolic Ca2+ in mtDNA depleted cells, which in turn activates the phosphatase, Calcineurin (Cn) (Amuthan, Biswas et al. 2002;Biswas, Adebanjo et al. 1999;Guha, Srinivasan et al. 2007). In the cell lines studied so far, it appears that the Ca2+/Cn mediated retrograde signaling pathway is distinct from the other reported retrograde signaling pathways and propagation of this signaling is dependent on the activation of Cn as an early event at its onset. In skeletal myocytes, the Cn mediated retrograde signaling is an ROS independent event. However, in murine macrophages mitochondrial dysfunction induced by hypoxic insult activates Ca2+/Cn mediated retrograde signaling pathway in which mitochondrial ROS induced membrane damage is a component of the signaling pathway (Srinivasan and Avadhani, 2007;Srinivasan, Koenigstein et al. 2010). Mitochondrial ROS can potentially induce retrograde signaling by alterations in Δψm and activation of Ca2+ and Cn dependent factors suggesting a convergence/overlap of the signaling pathways.
In our early studies, we observed that a reduction in mtDNA copy number to a certain threshold level initiates Ca2+/Cn dependent mitochondrial retrograde signaling resulting in genetic and epigenetic reprogramming. These cellular changes accompany metabolic adaptations and survival advantage by way of resistance to pro-apoptotic stimuli (Biswas et al. 2005). These observations were corroborated by other groups using various tumor cell types (Delsite, Kachhap et al. 2002; Lu, Sharma et al. 2009; Magda, Lecane et al. 2008; Minocherhomji, Tollefsbol et al. 2012). It is important to note that mtDNA depleted cell lines have been used popularly by several groups for characterizing mitochondrial retrograde signaling, some of the studies were carried out with Rho zero (Rho 0) cells that are completely devoid of mtDNA. It is noteworthy that although Rho 0 cells that lack mtDNA cannot maintain a membrane potential by proton pumping, still most mitochondrial proteins encoded by nuclear DNA are imported into mitochondria by a mechanism that requires a membrane potential which is supplied by adenine nucleotide carrier and an incomplete FoF1-ATP synthase in mammalian cells. Moreover, Rho 0 mitochondria maintain an electrochemical potential across the inner membrane by a mechanism coupled to ATP hydrolysis, thus making them net consumers of ATP in cells (Porteous et al. 1998). Rho 0 cells exhibit distinct phenotypic and biochemical properties compared to cells partially depleted of mtDNA which may account for the observed differences in physiological outcomes, including increased apoptosis rates in Rho 0 cells (Cavalli, Varella-Garcia et al. 1997;Liu, Lee et al. 2004). Additionally, Rho 0 state is not clinically relevant as patient tissues are not completely devoid of mtDNA. However, low mtDNA copy number is observed in many pathophysiologic conditions such as mitochondrial depletion syndrome (MDS), diabetes, Alzheimer’s, aging, and many carcinomas (Greaves, Reeve et al. 2012;Taylor and Turnbull, 2005;Wallace, 2013). Moreover reports from many laboratories provide evidence that heteroplasmic, not homoplasmic, mtDNA mutations drive tumorigenesis (He, Wu et al. 2010;Fan, Lin et al. 2012). Therefore, cell models with reduced mtDNA copy number mimic the disease states more closely than Rho 0 cells.
The mitochondrial retrograde signaling is quite diverse with respect to origin, mediators involved, and the phenotypic outcome and this topic has been reviewed by many experts in this field (Ryan and Hoogenraad,2007; Wallace,2012; Westermann and Neupert, 2003;Minocherhomji, Tollefsbol et al. 2012;Modica-Napolitano, Kulawiec et al. 2007;Butow and Avadhani, 2004). We have extensively used immortalized murine skeletal myocytes C2C12 cells to elucidate the mitochondria-to-nucleus stress signaling pathway. Additionally to confirm the commonality of the cellular reprogramming and activation of the signature factors of Ca2+/Cn-mediated mitochondrial stress signaling initially reported in C2C12 cells, we have tested number of different cell lines such as rat cardiomyocytes (H9C2), murine fibroblasts (3T3 and MEFs), human lung adenocarcinoma (A549), murine macrophages (RAW 264.7), MCF7 (Amuthan, Biswas et al. 2002;Biswas, Adebanjo et al. 1999;Guha, Pan et al. 2009;Guha, Fang et al. 2010;Srinivasan and Avadhani, 2007). We have now extended our studies to human breast, pancreatic and colorectal cell lines to delineate the key molecules involved in this signaling.
In the following sections, we will discuss the work from our as well as other laboratories on the emerging picture of the mitochondrial retrograde signaling in terms of landmarks of signal propagation and the signature transcription factors involved in modulating nuclear gene expression and tumor progression.
C (ii). Phenotypic hallmarks of mitochondria-to-nucleus signaling
In a series of reports on Cn-mediated mitochondrial retrograde signaling, we have shown that the most prominent change in response to reduced mtDNA copy number in murine skeletal muscle C2C12 cells is a marked increase in cell proliferation rate accompanied by their resistance to etoposide and staurosporin –induced apoptosis (Amuthan, Biswas et al. 2001; Biswas, Adebanjo et al. 1999; Biswas, Guha et al. 2005; Biswas, Anandatheerthavarada et al. 2005; Guha, Srinivasan et al. 2007; Guha, Pan et al. 2009). In human lung adenocarcinoma A549 cell line, which are malignant (form tumors in nude mice) but not invasive, mtDNA depletion renders them highly invasive (Amuthan, Biswas et al. 2002). In C2C12 and A549 cells, mtDNA copy number reduction is accompanied by phenotypic changes characteristic of tumors including altered cell morphology, metabolic shift to glycolysis, apoptosis and increased invasiveness (Biswas, Tang et al. 2008;Biswas, Srinivasan et al. 2008).
The causal role of reduced mtDNA copy number on tumor progression was demonstrated using a xenotransplantation assay, which involves growth of tumor cells inside de-epithelialized rat tracheas, transplanted subcutaneously into SCID mice. Histological examination of transplants showed that parental C2C12 cells grew mostly in the middle of the tracheal transplant with no detectable invasion of the tracheal wall while mtDNA-depleted C2C12 cells invaded the tracheal wall and its cartilage (Amuthan, Biswas et al. 2001). We have demonstrated in a mouse exogenous tumor model, mice injected with mtDNA depleted C2C12 cells had a higher tumor burden (incidence and size) when compared to those injected with parental cells (Tang, Chowdhury et al. 2012). Reports published from other groups using 143B osteosarcoma (Chae, Ahn et al. 2013;Dey and Moraes, 2000), MCF7 and T47D breast carcinoma (Delsite, Kachhap et al. 2002;Yu, Shi et al. 2009), SK-Hep1 (Kim, Kim et al. 2002), hormone dependent LNCaP prostate cancer cells (Moro, Arbini et al. 2009) and in a xenograft mouse model of colorectal cell line (Guo, Zheng et al. 2011) are essentially in agreement with the findings from our laboratory showing reduction in mtDNA copy number initiates cell survival and proliferation pathways conferring resistance to apoptosis and acquired invasive potential in these cells. A recent study published from the Shadel laboratory provides direct in vivo evidence that low mtDNA copy number in Tfam het mouse contribute to increased tumor growth and metastasis in mouse models of intestinal cancer (Woo, Green et al. 2012).
Cells with mtDNA copy number reduction had a distinctly different cellular morphology than the parental cell line. MtDNA depleted C2C12 and A549 cells gain “fibroblast-like” spindle shaped morphology (Amuthan, Biswas et al. 2001;Amuthan, Biswas et al. 2002;Naito, Cook et al. 2008). Pancreatic and breast carcinoma cells devoid of mtDNA have also been shown to undergo cellular transition accompanied by morphological alterations which are characteristics of the metastatic phenotype (Naito, Cook et al. 2008).
A prominent feature of tumor cells is altered metabolism and shift to aerobic glycolysis as was initially observed by Otto Warburg in 1931. Since then, it is well documented that rapidly growing tumors meet their metabolic demands by increased expression of genes encoding glucose transporters and glycolytic enzymes (Lin, 2011; Warburg, 1956; Ward and Thompson, 2012). Also there is increasing evidence to support the notion that mtDNA content directly reflects on the metabolic state of cells. We have observed that cells with reduced mtDNA content below a certain threshold have low ATP levels and meet their energy demands by shifting their cellular metabolism primarily to aerobic glycolysis (Guha, Srinivasan et al. 2007). In the Cn-dependent mitochondrial retrograde signaling, we observed that selective inhibition of IR autophosphorylation and Cn-dependent activation of the insulin-like growth factor 1 receptor (IGF1R). This Cn-dependent IGF1R activation occurs in a growth factor independent manner and IGF1R signaling pathway is the basis for increased glucose utilization and cell proliferation in mtDNA-reduced cells. Interestingly, mitochondrial stress-induced and IGF1R dependent metabolic shift to glycolysis appears to be an important survival factor in these cells, because either inhibiting Cn or IGF1 receptor activity using siRNA expression or pharmacologic inhibitors such as FK506 or Picropodophyllin (PPP) in these cells caused apoptosis. Moreover, cells with reduced mtDNA content had elevated levels of glucose transporters, Glut 4 and Glut 1 as well as higher activity of the glycolytic enzymes, hexokinase, phosphofructokinase, and pyruvate kinase (Guha, Srinivasan et al. 2007).
C (iii). Genetic reprogramming and the nuclear gene signature profile of mitochondrial retrograde signaling
Research from our laboratory on mitochondrial retrograde signaling demonstrated that mitochondrial respiratory stress activates cytosolic Ca2+ which leads to the activation of calcium dependent phosphatase, Cn. This Cn-mediated mitochondrial retrograde signaling culminates in transcription activation/repression of a large set of nuclear genes involved in diverse cellular functions including Ca2+ storage and release, cellular metabolism, mitochondrial transcription and biogenesis, cell survival and apoptosis and cytoskeletal organization (Amuthan, Biswas et al. 2002;Biswas, Tang et al. 2008;Guha, Srinivasan et al. 2007). We have observed activation of Ca2+ dependent transcription factors, kinases and phosphatases which culminates in the transcriptional upregulation of an array of nuclear genes (Guha, Pan et al. 2009;Guha, Tang et al. 2010). We will elaborate on the kinases and phosphatases involved in this pathway in the next section.
Using mouse skeletal myoblast C2C12 as a model cell system we have shown that mitochondrial dysfunction activates Ca2+/calmodulin dependent protein kinase (CAMK) downstream of which Ca2+/Cn dependent transcription factor NFAT, cAMP response elementbinding protein (CREB), CCAAT enhancer binding protein delta (C/EBPδ) and NFκB (cRel:p50) are activated and translocated to the nucleus. The NFκB activation pathway in response to this stress signaling is distinct from the previously reported canonical and non-canonical pathways (Ghosh and Karin, 2002; Madge and May, 2011;Oeckinghaus, Hayden et al. 2011). In mitochondrial retrograde stress signaling, the Rel proteins cRel and p50 form heterodimers and form a complex with PEST domain phosphorylated IkappaB beta. Upon activation, Cn dephosporylates IkappaB beta thereby releasing cRel:p50 complex from IκBβ and in turn promotes nuclear translocation of the Rel factors (Biswas, Anandatheerthavarada et al. 2003;Biswas, Tang et al. 2008). The importance of IκBβ in propagation of stress signaling stems from observations that while mtDNA-depleted C2C12 cells are highly tumorigenic in immunocompromised mice, silencing IκBβ mRNA in these cells abrogates the downstream stress signaling pathway resulting in tumor regression in mice (Biswas, Anandatheerthavarada et al. 2003;Biswas, Tang et al. 2008;Tang, Chowdhury et al. 2012;Wright, Agboke et al. 2012).
Interestingly, alternative NFκB signaling pathways have recently been implicated in cellular metabolism (Bakkar, Ladner et al. 2012) observed that mice with deleted NFκB components IKKα or RelB had reduced mitochondrial content and function. They observed that the alternative NFκB pathway transcriptionally activates regulator for mitochondrial biogenesis, PPAR-γ coactivator 1β (PGC-1β). Regulation of PGC-1β by IKKα/RelB was mammalian target of rapamycin (mTOR) dependent suggesting a possible cross talk between mTOR and NFκB signaling in muscle metabolism (Bakkar, Ladner et al. 2012). More recently it has been shown that mitochondrial ATPase inhibitory factor (IF1) triggers an ROS mediated retrograde signaling which provides cellular proliferative advantage. This involves the phosphorylation of IkBα and NFκB activation for cell survival (Formentini, Sanchez-Arago et al. 2012).
We have reported that the regulation of mitochondrial respiratory stress responsive nuclear genes requires physical interactions and functional synergy between the mitochondrial stress activated transcription factors NFκB (cRel:p50), C/EBPδ, CREB, and NFAT (Biswas, Guha et al. 2005;Guha, Pan et al. 2009;Guha, Tang et al. 2010). A crucial component of this Cn mediated mitochondrial retrograde signaling pathway is the activation of heterogeneous ribonucleoprotein A2 (hnRNPA2), a nuclear RNA-binding protein (Guha, Pan et al. 2009). The functional synergy of these factors in the mitochondrial stress pathway requires coactivation of heterogeneous nuclear ribonucleoprotein A2 (hnRNPA2). Furthermore, we have shown that the oncogenic kinase Akt1 is activated in the nucleus under mitochondrial stress and phosphorylation of hnRNPA2 by Akt1 is essential for the recruitment of hnRNPA2 to the enhanceosome complex of stress target promoters (Guha, Fang et al. 2010). The activation and contribution of hnRNPA2 in the propagation of the Ca2+/Cn mediated mitochondrial retrograde signaling is discussed in a later section.
Although several potential target genes of the mammalian mitochondrial stress pathway have been reported in different cells, the complete genetic footprint of mitochondrial retrograde stress response still remains unknown. Characterization of the genes regulated by the retrograde pathway is central to understanding the putative role of mitochondrial stress in, aging, cancer progression, and neural and bone degenerative diseases and cellular resistance to apoptosis. A number of independent studies have elucidated genome wide transcriptional profiles of cells altered in response to mitochondrial retrograde signaling (Biswas, Tang et al. 2008;Chae, Ahn et al. 2013;Desler, Marcker et al. 2011;Magda, Lecane et al. 2008;Minocherhomji, Tollefsbol et al. 2012;Singh, Rasmussen et al. 2004). Microarray analyses from our laboratory has shown that the transcriptional targets of the mitochondrial retrograde signaling pathway in mammalian cells are diverse and include genes that are involved in mitochondrial biogenesis, cell adhesion, cytoskeletal markers, invasiveness, metabolism, transcription factors, alternative splicing. The cDNA microarray results reported from Singh’s laboratory compared differences in the nuclear gene expression profile between a breast cancer cell line (parental) and its Rho 0 (devoid of mtDNA) derivative with impaired mitochondrial function. Their gene sets are similar to cDNA array results from our laboratory which included higher expression of several nuclear genes involved in cell signaling, cell architecture, energy metabolism, cell growth, apoptosis including general transcription factor TFIIH, v-maf, AML1 in Rho 0 cells. Delsite et al (Delsite, Kachhap et al. 2002) reported downregulation of several genes including phospholipase C, agouti related protein, PKC gamma, protein tyrosine phosphatase C, phosphodiestarase 1A (cell signaling), PIBF1, cytochrome p450, (metabolism) and cyclin dependent kinase inhibitor p19, and GAP43 (cell growth and differentiation). Kulawiec M et al (Kulawiec, Safina et al. 2008) identified two major gene networks, FN1 (fibronectin) and p53 were differentially regulated between parental and Rho 0 epithelial cells. Bioinformatic analyses of FN1 network identified laminin, integrin and 3 of the 6 members of peroxiredoxin whose expression were altered in Rho 0 epithelial cells. In the p53 network, SMC4 and WRN were identified suggesting that this network may affect chromosomal stability. This study reported an increase in DNA double strand breaks and unique chromosomal rearrangements in Rho 0 breast epithelial cells. Further, tight junction proteins claudin-1 and claudin-7 were downregulated in Rho 0 breast epithelial cells suggesting mitochondria-to-nucleus retrograde signaling plays a key role in oncogenic transformation of breast epithelial cells.
Nuclear transcriptome analysis by Magda et al in A549 lung cancer cells and mtDNA-depleted ρ0 counterparts grown in culture and as tumor xenografts in immune-deficient mice identified a specific gene expression profile. Similar to our findings in C2C12 cells, the gene expression profiling analyses of parental A549 and A549 ρ0 cells by Magda et al reported differential expression of transcripts related to immune recognition, autophagy of defective mitochondria, Hif-1mediated gene regulation, the epithelial-mesenchymal transition (EMT), and accumulation of lipophilic molecules which contribute to metabolic advantage to tumor cell proliferation (Magda, Lecane et al. 2008).
A recent study by Chae et al (Chae, Ahn et al. 2013) using systems biology approach further added to our knowledge of the key nuclear transcription factors involved in retrograde signaling pathways. In 143B Rho 0 osteosarcoma cells A3243G mutations in the mt-tRNA Leu induced mitochondrial retrograde signaling involving the retinoid X receptor α, c-JUN N-terminal kinase (JNK), and PGC1α. The results from Chae et al are essentially similar to an earlier report by Ivanov et al (Ivanov, Ghandhi et al. 2011) demonstrating alteration in the nuclear transcriptome in response to mitochondrial retrograde signaling. However, contrary to our results, they observed a downregulation of NFκB signaling and increased susceptibility to apoptosis in their cell types. As mentioned earlier, these differences in the types of genes reported in different studies may reflect cell/tumor specific differences. Another reason may be the use of partial mtDNA depletion in our studies compared to the Rho 0 cells used in other studies. Notably many tumors exhibit partial mtDNA depletion.
The published findings from all the above groups, including ours, are overall quite similar and indicate that reduction in mtDNA levels causes a global upregulation of genes involved in tumorigenic pathways. This raises the possibility that mitochondria can be considered as therapeutic targets in tumor progression.
C (iv). Activation of growth factor receptors and kinases in response to mitochondrial retrograde signaling
Activation of a number of kinases, phosphatases and growth factor receptors are part of the reprograming observed in cells with mitochondrial dysfunction as they transition towards tumorigenesis. It is well established that the high metabolic activity of tumor cells is associated with activation of PI-3K/Akt kinase, which is downstream of IGF1R. Akt activation in response to mitochondrial respiratory stress has been reported in different tumor cell systems (Guha, Fang et al. 2010;Moro, Arbini et al. 2009;Pelicano, Xu et al. 2006). Another kinase, AMP-activated protein kinase (AMPK) is known to play an important role in insulin signaling, glucose uptake, and energy homeostasis. AMPK has also been implicated to be involved in PGC1α-mediated mitochondrial biogenesis and mitochondrial signaling pathways (Mihaylova and Shaw, 2011). In Cn mediated mitochondrial stress signaling pathway, PI3K / Akt kinase is activated downstream of IGF1R activation; however we have not observed AMPK activation in C2C12 cells (Guha, Srinivasan et al. 2007). In one of the earlier reports from our laboratory, we observed activation of P44 and P42 MAP kinases in A459 cells subjected to mitochondrial stress (Amuthan, Biswas et al. 2002). Activation of MAP kinase under mitochondrial stress has also been reported in other tumor cell lines (Naito, Cook et al. 2008).
IGF1R activation by external cues during growth factor signaling has been widely reported in tumor cells. We have reported a novel mode of its activation by Ca2+/ Cn as part of the mitochondrial retrograde signaling in a growth factor independent manner in mtDNA depleted C2C12 cells (Guha, Srinivasan et al. 2007). Growth factor independent activation of IGF1Receptor is a feature of many actively proliferating tumors (Nair, De Armond et al. 2001). The IGF1R signaling is propagated downstream through activation of phosphatidylinositol 3-kinase, Akt, and protein kinase C ultimately leading to activation of genes involved in glucose uptake and utilization, cell growth and proliferation. Downstream activation of Akt kinase in mitochondrial retrograde signaling is crucial for the full stress response, which culminates in altered expression of nuclear gene targets. Phosphorylation of the mitochondrial stress activated coactivator, hnRNPA2 by Akt is essential for the maximal stress response (Guha, Tang et al. 2010;Guha, Fang et al. 2010) and we believe that a complex transcriptional and posttranscriptional regulatory circuit exists in which hnRNPA2 and Akt1 function interdependently. Moreover ROS-dependent Akt1 activation has been reported in mitochondrial Complex I dysfunction induced retrograde signaling (Sharma, Fang et al. 2011).
Other kinases have been reported to be activated by mitochondrial retrograde signaling in other cell types. Retrograde signaling involving increased cytosolic free [Ca2+]c in rat phecromocytoma cells treated with the uncoupler carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone (FCCP) correlated with activation of ERK1 and ERK2 kinases (Luo, Bond et al. 1997). In ρ° human fibrosarcoma 143B cells and MERRF (myoclonic epilepsy with ragged red fibers) cybrid cell line with mitochondrial tRNALys mutation (A8344G), respiratory deficiency and the associated increase in cytosolic free Ca2+ induced the activation of CaMKIV, which in turn activated CREB by protein phosphorylation (Arnould, Vankoningsloo et al. 2002). Additionally elevated mitochondrial H2O2 leading to mitochondrial stress, also induced MAP kinases in HeLa cells (Nemoto, Takeda et al. 2000).
The activation of different kinases reflects cell-specific differences and also possibly the nature of mitochondrial stress initiators. Removal of free Ca2+ by specific chelators, or knock down of CnAα mRNA, abrogated the activation of these various Ca2+responsive factors and the induced transcription of nuclear target genes thus confirming the role of Ca2+ in retrograde signaling.
C (v). HnRNPA2 activation as a critical landmark of mitochondrial retrograde signaling in the context of tumorigenesis
Heterogeneous ribonucleoproteins hnRNP proteins are abundant RNA-binding proteins and expressed in most human tissues. Additional functions of these proteins in post splicing events, such as mRNA trafficking, and replication and transcription of cytoplasmic RNA viruses, telomere biogenesis have also been reported. HnRNP A2/B1 is a member of this family of proteins which shuttles between the nucleus and cytoplasm (Dreyfuss, Matunis et al. 1993) and known to be involved in regulating RNA biogenesis, localization, and metabolism of certain mRNAs (Weighardt, Biamonti et al. 1996). In recent years there is increasing evidence that in addition to their established function in mRNA processing, hnRNP proteins in general, and hnRNP A2 in particular, regulate multiple cellular processes involved in tumorigenesis by transcriptional and posttranscriptional gene regulations (Martinez-Contreras, Cloutier et al. 2007;Moran-Jones, Wayman et al. 2005). Since there are a number of reviews on hnRNP proteins here we will focus on the role of hnRNPA2 in mitochondrial stress induced cancer and its specific contribution in the propagation of the mitochondrial retrograde signaling (Richard, 2010).
In cells with mitochondrial dysfunction due to either reduced mtDNA copy number or drug induced disruption of Δψm, we have observed transcriptional upregulation and overexpression and nuclear accumulation of hnRNPA2. As mentioned earlier, cells with low mtDNA content are reprogrammed for tumorigenic transformation and hnRNPA2 is overexpressed in these cells. The overexpression/activation of nuclear hnRNPA2 is a specific response to mitochondrial retrograde signaling (Guha, Pan et al. 2009;Guha, Fang et al. 2010). To our knowledge, hnRNPA2 is specifically overexpressed in response to mitochondrial stress signaling, and we have not observed any alteration in levels of other closely related hnRNP members either hnRNPA1 or HnRNP DL (Guha, Pan et al. 2009). The significance of activation of hnRNPA2 in the mitochondrial retrograde signaling pathway stems from the established role of this protein in controlling different aspects of cancer cell metabolism, invasion and cancer progression which are discussed below.
HnRNP A2/B1 overexpression and upregulation has been widely implicated in lung, pancreas, liver and brain cancer (glioblastoma, oligodendrogioma, and astrocytoma) (He, Brown et al. 2005;Patry, Bouchard et al. 2003;Yamaoka, Imajoh-Ohmi et al. 2006). Clinical reports suggest that hnRNPA2/B1 is an early marker of lung epithelial transformation and carcinogenesis (Zhou, Mulshine et al. 1996). A pilot study of archival sputum specimens from a high-risk cohort identified a monoclonal antibody (mAb) to hnRNPA2/B1 that specifically reacted with “normal”-appearing bronchial epithelial cells from individuals who subsequently developed lung cancer, indicating hnRNPA2 could be a sensitive marker for early detection of lung cancer (Tockman, Gupta et al. 1988). Lung epithelial cells overexpressing hnRNPA2 are phenotypically different and subcellular localization of hnRNPA2 could be an important factor in lung cancer progression (Man et al., 2000). Further reports from the Mulshine laboratory suggests that hnRNPA2 plays a role in epithelial-mesenchymal transition during lung cancer metastasis (Tauler, Zudaire et al. 2010).
An interesting observation was that hnRNP-A2/B1 expression is controlled by cell proliferation rate in normal bronchial epithelial cell primary cultures, but not in SV40-transformed bronchial epithelial cells or tumor cell lines and lung tumor cells undergoing clonal expansion upregulate hnRNPA2 (Zhou et al., 2001). Overexpression of hnRNP A2 is essential for the growth and viability in cervical, colon, breast, ovarian, and brain cancer cell lines and silencing this protein leads to apoptosis in these cells whereas it did not affect normal cells (Patry, Bouchard et al. 2003). Silencing hnRNP A2/B1 in glioblastoma cells inhibited tumor formation in mice and overexpression of hnRNP A2/B1 in immortal cells led to malignant transformation, suggesting that hnRNPA2 is a putative proto-oncogene (Golan-Gerstl, Wallach-Dayan et al. 2012). As a component of mitochondrial retrograde signaling pathway, hnRNPA2 plays a similar oncogenic role in that, it is essential for the proliferation, survival and invasiveness of the cells with low mtDNA content and silencing hnRNPA2 in these cells leads to apoptosis (Guha, Pan et al. 2009).
Using promoter pull down coupled with proteomic analysis we identified hnRNPA2 in the nuclear protein complex of mtDNA reduced cells. Combinatorial approaches of promoter activation and chromatin IP show hnRNPA2 functions as a mitochondrial stress activated transcriptional coactivator which is essential for the up-regulation of the mitochondrial stress-target genes Cathepsin L, RyR1, Akt1 and Glut-4. Moreover, we have been unable to show direct binding of hnRNPA2 to double stranded DNA from the promoter regions of stress-target genes. We therefore believe its association with the promoter DNA and with the enhanceosome complex occurs mostly through protein–protein interactions (Guha, Pan et al. 2009). However, a report by Torosyan et al (Torosyan, Dobi et al. 2010) shows that hnRNP A2 directly binds to the Annexin A7 promoter DNA and is involved in androgen independent prostate cancer progression.
HnRNPA2, activated in response to mitochondrial stress, is crucial for the activation of target genes either due to its recruitment of DNA binding factors, NFκB (c-Rel/p50), C/EBPδ, CREB, and NFATc to the promoter sites or in the overall stability of the enhanceosome complex of mitochondrial respiratory stress-responsive genes (Guha, Pan et al. 2009). At this time, the exact mechanism of its recruitment to the enhanceosome complex is not clearly understood. Our observation that hnRNPA2 is upregulated in response to mitochondrial retrograde signaling is significant at two levels: first, it adds to the growing body of literature implicating this protein in driving tumorigenesis; second, since mitochondrial dysfunction due to low mtDNA content is common in many cancers, activation of hnRNPA2 in response to this signaling could be used as pharmacologic target in cancers with mtDNA defects. A proposed mechanism of Ca2+/Cn activated retrograde signaling and the transcription coactivator function of hnRNPA2 is presented in Figure 2.
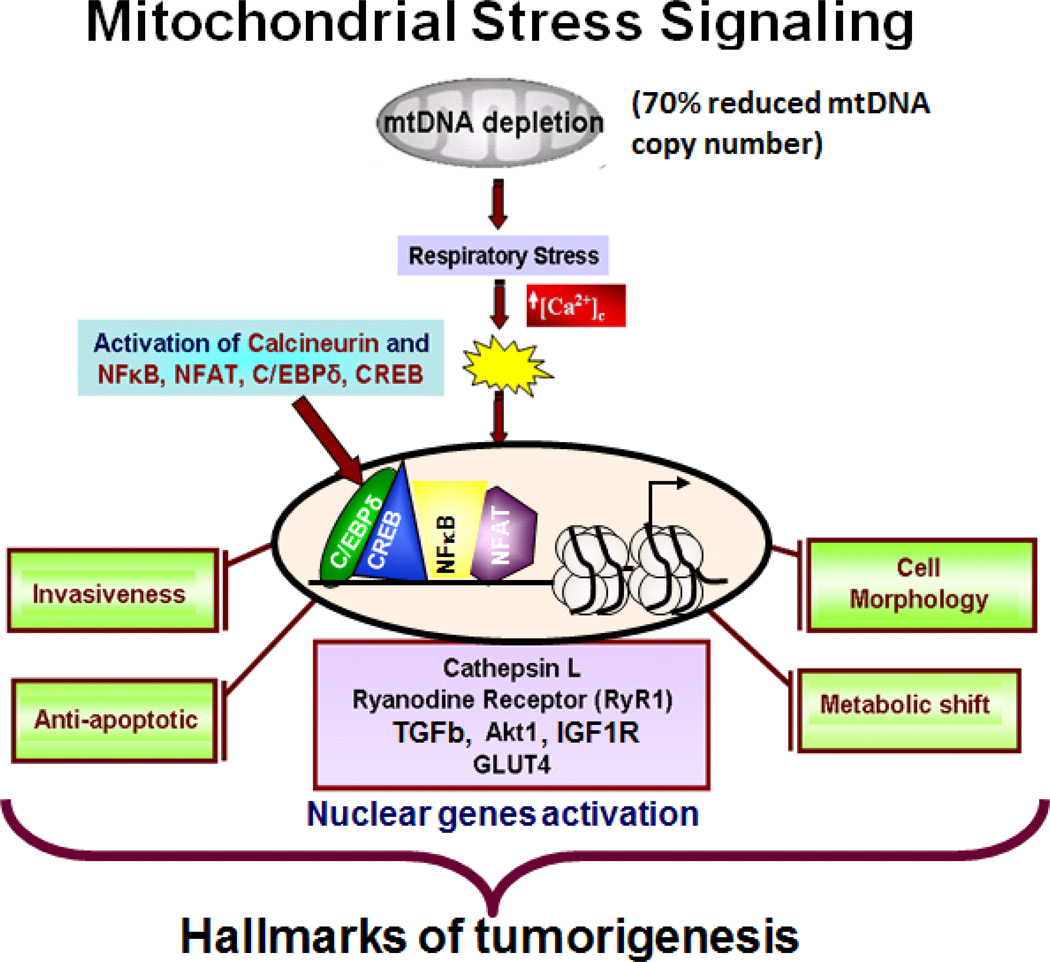
Figure 2. Schematic outline of the reduced mtDNA copy number mediated mitochondrial retrograde signaling in C2C12 myocytes.
Reduction of mtDNA content below 70% disrupts ΔΨm which results in elevated cytosolic Ca2+ and activation of Calcineurin. This initiates a signaling pathway involving a number of kinases and transcription factors along with a mitochondrial stress activated transcriptional coactivator, hnRNPA2. The resultant of this signaling is an upregulation of nuclear genes leading to cellular reprogramming towards tumorigenesis.
Another mechanism by which many hnRNP proteins in general, and hnRNPA2 in particular, regulate tumorigenesis is by regulating oncogenes by alternative splicing (Martinez-Contreras, Cloutier et al. 2007). In cancer cells, the process of alternative splicing is widely misregulated and recent reports suggest that many hnRNP proteins act as splicing repressors by either antagonizing SR proteins or through the recognition of exonic and intronic splicing silencer elements (Black and Grabowski, 2003;Cartegni, Chew et al. 2002;Mayeda and Krainer, 1992;Rooke, Markovtsov et al. 2003). It is now suggested that hnRNP A2 regulates many of the fundamental events in cancer cell migration and aerobic glycolysis by regulating alternative splicing of many key oncogenes. One such example is the tumor protein p53 inducible nuclear protein 2 (TP53INP2). Using exon-tiling microarrays it was identified that hnRNPA2 mediated alternative splicing of TP53INP2 is essential for cells to invade the three dimensional extracellular matrix (Moran-Jones, Wayman et al. 2005).
HnRNPA2 modulates the expression of many genes involved in tumor metabolism. In cell lines we have studied, hnRNPA2 is essential for the glycolytic shift observed in cells with low mtDNA content. The regulatory role of hnRNPA2 in tumor cell metabolism in cells with mitochondrial dysfunction is a consequence of its function as a transcriptional coactivator of genes involved in cancer cell metabolism (Glut4, IGF1R, Akt). Interestingly, the regulation by hnRNPA2 of Akt kinase at the transcriptional level is a positive feedforward loop since activation of hnRNPA2 under mitochondrial stress is dependent on Akt phosphorylation (Guha, Pan et al. 2009;Guha, Fang et al. 2010). The different functional domains of hnRNPA2 and the Akt phosphorylation sites of the protein are shown in Figure 3.
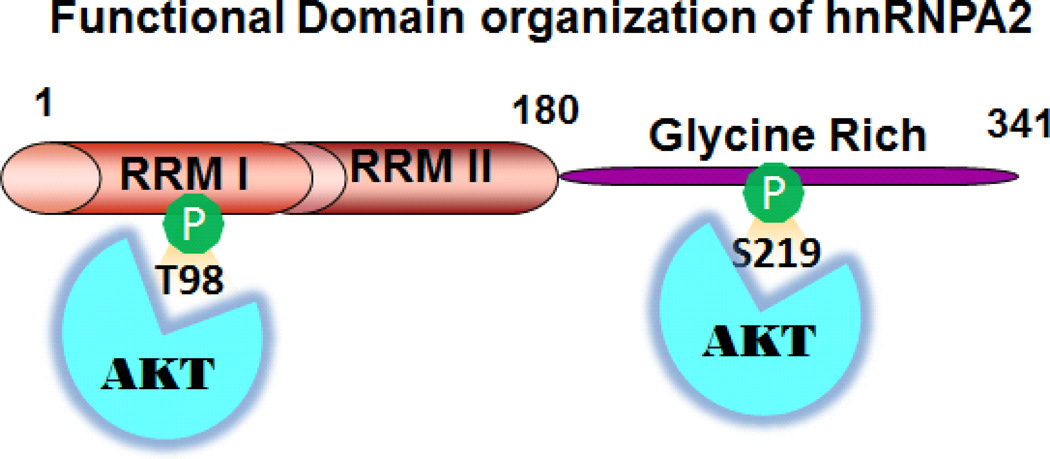
Figure 3. Functional Domains of hnRNPA2.
A cartoon of the different functional domains of human hnRNPA2 identified till date. The residues 1–180 are the two RNA recognition motifs (RRM) which are involved RNA binding and transcriptional activation; residues 181–341 is the glycine rich domain involved in protein-protein interaction. The Akt phosphorylation sites Thr 98 and Ser 213 are indicated in green.
HnRNPA2 regulates many of the genes involved in aerobic glycolysis by alternative splicing in response to cellular microenvironmental cues arising from mitochondrial dysfunction. Increasing number of studies in recent years, provide new insights on how extracellular signals regulate alternative splicing, contributing to cellular transformation and development of breast cancer (Pelisch, Khauv et al. 2012). An example of alternative splicing by hnRNPA2 in a cancer specific gene is pyruvate kinase M. Two independent reports demonstrate that hnRNP proteins A1, A2 and PTB modulate the expression of the glycolytic enzyme pyruvate kinase isoform M (PKM) in cancer cells by alternative splicing (Chen, Zhang et al. 2010;Chen, David et al. 2012;David, Chen et al. 2010). The embryonic pyruvate kinase isoform, PKM2, is almost universally re-expressed in cancer, and promotes aerobic glycolysis, whereas the adult isoform, PKM1, promotes oxidative phosphorylation (Christofk, Vander Heiden et al. 2008). These two isoforms result from mutually exclusive alternative splicing of the PKM pre-mRNA, reflecting inclusion of either exon 9 (PKM1) or exon 10 (PKM2). Recently, Chen M et al (Chen, David et al. 2012) reported an interesting finding that concentration-dependent interaction of hnRNP proteins determine the outcome of PKM splicing. At high concentrations of hnRNP A1, A2 and PTB, such as those found in most cancer cells, binding of these proteins through cooperative interaction to two sites in the upstream regulated exon (exon 9) orchestrates exon 9 (PKM1) exclusion and expression of isoform M2. Furthermore, overexpression of three hnRNP proteins, hnRNPA1, hnRNPA2 and PTB, in cancer cells is regulated by the myc oncogene, which also promotes the switch to PKM2 isoform expression.
HnRNPA2 is known to associate with a number of tumor regulators including TDP-43, TOG2, SET, Annexin A7 and VEGF although the precise mechanism of these associations and their functional relevance in tumor progression is not well understood (Buratti, Brindisi et al. 2005;Kosturko, Maggipinto et al. 2005;Vera, Jaumot et al. 2006). It is important to note that many hnRNP family proteins regulate cellular processes under both physiologic and pathological processes. HnRNP L regulates immune response by alternatively splicing the transmembrane protein-tyrosine phosphatase CD45 (Preussner, Schreiner et al. 2012). Another example is the alternative splicing of GTPase Rac 1b by hnRNP A1 which is over-expressed in breast and colorectal cancer cells and is suggested to play key role in a number of different oncogenic signaling pathways, including MMP3 mediated tumorigenic transformation of mammary epithelial cells (Pelisch, Khauv et al. 2012).
4. Mitochondrial retrograde signaling as an epigenetic regulator
Mitochondria provide the energy flux required to meet the cellular energy demands. The mitochondrial metabolic centers, including the TCA cycle, fatty acid oxidation and the electron transfer complexes (ETC) are the sites where calories are converted to NADH, acetyl-CoA and ATP. Smirgalia et al (Smirgalia et al. 2008) reported that mitochondrial defects can lead to epigenetic changes in the nucleus. This possibly explains the lack of a correlation between genotype and phenotype, often observed in patients with mitochondrial disorders. Wallace and his associates observed that the clinical phenotypes of bioenergetic diseases which are strikingly similar to those observed in epigenetic diseases such as Angelman, Rett, Fragile X Syndromes, the laminopathies and cancer led to the theory that mitochondria and bioenergetics are involved in modulating the epigenetic status of the cell (Wallace, 2013). It is postulated that in a calorie – abundant environment, ATP and acetyl-CoA phosphorylate and acetylate chromatin facilitating nuclear DNA transcription and replication. In nutrient and calorie limiting state, chromatin phosphorylation and acetylation are lost and gene expression is suppressed. It is suggested that DNA methylation via S-adenosyl Methionine (SAM) can also be modulated by mitochondrial function (Naviaux, 2008; Wallace and Fan, 2010).
In recent years, an increasing number of epigenetic diseases have been suggested to be associated with mitochondrial dysfunction (Chinnery, Elliott et al. 2012;Jazwinski and Kriete, 2012;Manev, Dzitoyeva et al. 2012;Minocherhomji, Tollefsbol et al. 2012). In a recent study, Bellizi et al reported that blood samples from patients with mtDNA haplogroup J had a higher global DNA methylation compared to other mtDNA haplogroups. The higher global DNA methylation pattern in the mtDNA haplogroup J correlated with the overexpression of the methionine adenosyltransferase-Iα (MAT1A) gene and low ATP and ROS levels (Bellizzi, D'Aquila et al. 2012). They suggested that by possibly affecting oxidative efficiency, mitochondria-nucleus interactions regulate epigenetic changes. Using Restriction Landmark Genomic Scanning (RLGS) approach, Smiraglia DJ et al (Smiraglia, Kulawiec et al. 2008) observed major changes in methylation patterns of the nuclear genome in response to alterations in mtDNA copy number suggesting mitochondrial genome changes causes profound alterations in the epigenetic status of the cell. The role of mitochondria as an epigenetic regulator in human diseases has been reviewed earlier (Lu and Thompson, 2012;Minocherhomji, Tollefsbol et al. 2012;Venneti and Thompson, 2013).
Our observation of hnRNPA2 activation in response to mitochondrial retrograde signaling is important not only in genetic regulation but also from the epigenetic perspective. Our ongoing work suggests that the mitochondrial stress responsive target promoter regions have elevated levels of histone acetylation which is abrogated by silencing hnRNPA2 suggesting an involvement of this protein in chromatin remodeling complex. It is also noteworthy that hnRNP A2 binds to single stranded telomeric repeat TTAGGGn and also acts as a molecular adapter between single-stranded telomeric repeats, or telomerase RNA, and another segment of ssDNA (Ford, Wright et al. 2002;McKay and Cooke, 1992;Moran-Jones, Wayman et al. 2005). HnRNPA2 colocalizes with telomeric chromatin in the subset of PML bodies that are a hallmark of ALT cells, reinforcing the evidence for hnRNPs having a role in telomere maintenance. HnRNP A2/B1 was recently shown to be associated with human telomerase reverse transcriptase (hTERT) and increase telomerase activity in hepatocellular carcinoma which is a new prognostic biomarker (Mizuno, Honda et al. 2012). We therefore believe mitochondrial retrograde signaling induced activation of hnRNPA2 which is a telomere regulatory protein is a possible epigenetic link between mitochondrial functional state and nuclear gene expression and telomere state.
5. Mitochondrial Retrograde Signaling in tumor cell metabolism
Mitochondria are the sites of integration and coordination of metabolic pathways in the cell. Mitochondria are important regulators of cancer cell metabolism, since in addition to generating ATP they are major sites of cellular ROS production and are regulators of apoptosis through the mitochondrial permeability transition pore (mPTP) (Marchetti, Castedo et al. 1996). The mPTP acts as a “sensor” about mitochondrial energy metabolism, increased oxidative stress and Ca2+ overload by its interaction with pro- and anti-apoptotic Bcl2 proteins, the outer membrane protein voltage-dependent ion channel protein (VDAC/ porin), the inner membrane protein adenine nucleotide translocator (ANT) and the Ca2+ sensor cyclophilin D (Kinnally, Peixoto et al. 2011). The involvement of mitochondria in cancer arises from the observation that a number of nuclear “oncogenes” code for proteins involved in mitochondrial energy production.
The Warburg effect has been demonstrated in different types of tumors and the increase in glucose uptake has been exploited clinically in detection of tumors by fluorodeoxyglucose positron emission tomography (FDG-PET). Cancer cell metabolism has become a promising area of cancer therapeutics (Cheong, Lu et al. 2012). One of the first implications of the mitochondrial role in cancer was reported in 1977 by Pedersen (Bustamante and Pedersen, 1977) in hepatocarcinoma cells where it was shown that cellular transformation correlated with an increase in hexokinase II and a subsequent decrease in glucokinase (Pedersen, Mathupala et al. 2002). It is believed that increased HKII bound to the mitochondrial outer membrane traps ATP through its ATP-priming site, oxidizes the available glucose to G6P resulting in rapid conversion to pyruvate and lactate. It is now understood that this induction of hexokinase in cancer cells is controlled at the epigenetic level wherein the hexokinase promoter is transcriptionally activated by demethylation (Goel, Mathupala et al. 2003). The gene expression is regulated by glucose, hypoxia, cAMP (CREB, ATF and CREM), p53 mutation and insulin (Thomson, Herway et al. 2008). Renal cell carcinomas and uterine leiomyomas have been linked to mutations in mitochondrial fumarate hydratase while mutations in succinate dehydrogenase (SDHB, C and D) is linked to paraganglioma (Yang, Valera et al. 2012;Favier, Briere et al. 2005).
The close association with altered metabolism and cancer is evidenced from reports demonstrating the role of oncogenes affecting tumor cell metabolism. A few notable examples: the oncogene Ras when mutated, promotes glycolysis (Telang, Yalcin et al. 2006); Akt kinase, activated during cell proliferation also increases glucose uptake in tumor cells; and the transcription factor Myc upregulates metabolic genes in tumors (Jones and Thompson, 2009). Moreover, inhibition of LDH-A prevents Warburg effect reverting cancer cells to oxidative phosphorylation and attenuated tumor growth (Shim, Dolde et al. 1997). Tumor suppressor p53 alters mitochondrial function by regulating the expression of SCO2 (Matoba, Kang et al. 2006). Another p53 effector TIGAR (TP53-induced glycolysis and apoptosis regulator) inhibits glycolysis by decreasing fructose-2,6-bisphosphate (Bensaad, Tsuruta et al. 2006). Kawauchi et al (Kawauchi, Araki et al. 2008) have suggested that p53-mediated regulation of glucose metabolism is dependent on NFκB. We have shown that in mammalian cells, Ca2+/Cn mediated mitochondrial retrograde signaling upregulates a number of oncogenic factors and kinases involved in metabolic switch to glycolysis such as IGF1 receptor, Glucose transporter-4, NFκB, CREB, Akt, PI-3Kinase along with the activation of glycolytic enzymes (Amuthan, Biswas et al. 2001;Guha, Srinivasan et al. 2007;Guha, Fang et al. 2010). Thus mitochondrial retrograde signaling initiated by dysfunctional mitochondria can induce metabolic switch to glycolysis.
In recent years it has been suggested that nuclear transcription factors such as STAT3, p53 and ERalpha are translocated to the mitochondria which possibly regulate mitochondrial function independent of transcription (Wegrzyn et al 2009; Brown et al, 2010; Tammineni et al 2012). Additionally, mitochondrial deacetylases possibly has a significant role in regulating the dynamics of the mitochondrial biogenesis (Aquilano et al 2010) and mitochondrial acetylome (Anderson et al, 2012; Still AJ et al, 2013) favoring tumor metabolism.
Our current understanding is that tumor cell mitochondria are functional and capable of oxidative phosphorylation but favor metabolic reprogramming to meet the increasing macromolecular demands associated with high proliferation of these cells (Wallace, 2012;Ward and Thompson, 2012). It has been proposed that the activation of many of the oncogenes is primarily for the purpose of metabolic reprogramming in tumor cells. However, alternative hypotheses from Lisanti and colleagues propose that the cancer associated fibroblasts and the tumor stromal cells produce “energy rich” nutrients and there is a parasitic dependence of the tumor cells on their stromal counterpart to meet the energy and nutrient requirements (Pavlides, Vera et al. 2012).
6. Future Perspectives
Dysfunctional mitochondria are the point of origin of the mitochondria-to-nucleus retrograde signaling pathway. Even though there are multiple modes of signaling mechanisms involved, depending on the type of mitochondrial insult and cell types, these pathways could possibly converge downstream through some common factors for the propagation of this signal. In this review we provided a brief account of the mechanisms of retrograde signaling. It remains to be seen if a particular retrograde signaling mechanism is specific for cell types or the type of stress inducer. It is important to note here that even though in most of the cell lines we have studied so far, reduction in mtDNA copy number or treatment with mitochondrial ionophores lead to the activation of the tumorigenic program, it is possible that the outcome of this signaling pathway could result in other cellular programming depending on the cell lineage such as induced differentiation of macrophages to osteoclast-like cells (Srinivasan and Avadhani, 2007;Srinivasan, Koenigstein et al. 2010). Mitochondrial retrograde signaling has also been reported in many other pathological conditions such as MERFF, neurodegenerative diseases (Alzheimer’s and Parkinson’s), myocardial ischemia, maternally inherited deafness, diabetes and progeroid aging (Wallace, 2013).
Evidence attributing mtDNA defects to tumor progression comes from studies with transmitochondrial cell hybrids (cybrids) where the defective mitochondrial respiration of tumor cells is transferred to normally respiring cells when mitochondria, not nuclei, are imported (Bonora, Porcelli et al. 2006). It is noteworthy that heteroplasmic mtDNA mutations are frequently reported in cancers (Arnold, Sun et al. 2013; Fan, Lin et al. 2012; Park, Sharma et al. 2009; Petros, Baumann et al. 2005). A recent report using deep sequencing confirmed previous reports that while normal tissues contained marginal levels of mtDNA heteroplasmy, most of the cancer-specific somatic mtDNA mutations were heteroplasmic rather than homoplasmic (He, Wu et al. 2010). This report identified that in tumors 85% of the heteroplasmic mutations were in protein-coding or RNA-coding regions, whereas in normal tissues only 33% of the heteroplasmic variants were in protein-coding or RNA-coding regions. These reports suggest the possibility that heteroplasmic mutations might have been selected for their tumor-promoting effects rather than a random segregation event. Therefore mtDNA depletion model used by us and other investigators mimic the mtDNA heteroplasmy models.
The altered metabolic shift to glycolysis resulting from altered genetic composition seems to be a critical factor in tumor cell reprogramming. In recent years, mitochondria are gaining center stage as a therapeutic target in cancer. A number of drugs termed “mitocans” targeting different aspects of mitochondrial biology such as mitochondrial metabolic pathways, mtDNA, ETC components and mitochondrial proteins have shown promising results in vivo and their effects in clinics remains to be seen (Neuzil, Dong et al. 2013;Edeas and Weissig, 2013).
Our studies for the past decade have defined the mechanism of nuclear gene regulation by Ca2+/Cn regulated retrograde signaling (Figure 4) and the role of the key signaling components involved such as Calcineurin, NFκB, IGF1 receptor, Akt1 and hnRNPA2 have been confirmed using gene silencing approach. From our studies and previously published reports, we suggest that therapeutic intervention of the retrograde signaling pathway by targeting hnRNPA2 or calcineurin would be beneficial in designing cancer therapy for patients with mitochondrial copy number defects. A recent report (Ermak G et al, 2012) suggests that an isoform of the negative regulator of calcineurin RCAN1 (RCAN1-1L) induces mitochondrial autophagy in neuronal cells resulting in metabolic shift to glycolysis. However, in mtDNA-depleted cells, we have not observed mitochondrial autophagy but it will be interesting to see the effect of RCAN1-1L on hnRNPA2 functions.
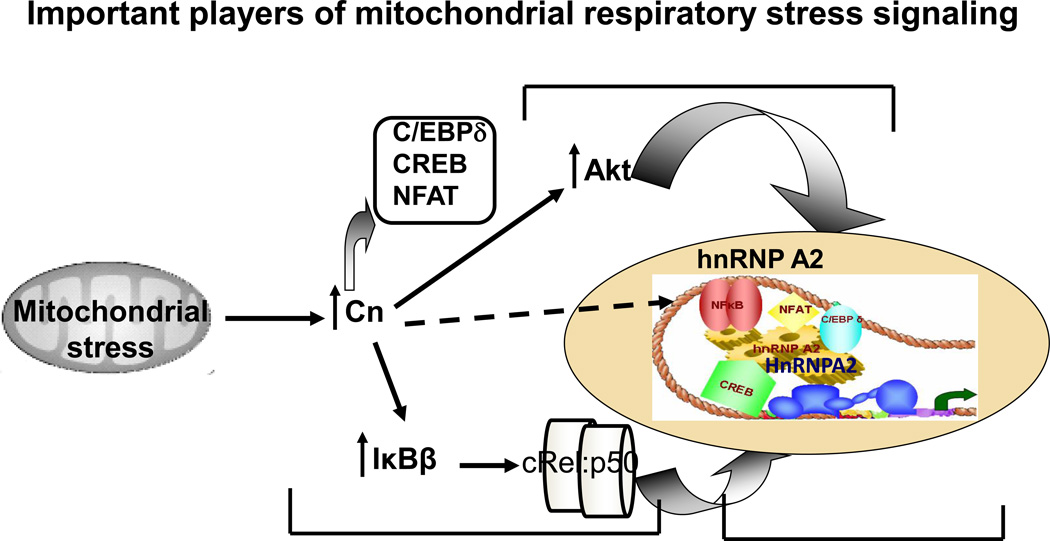
Figure 4. Schematic outline of the key aspects of the Calcineurin mediated mitochondrial retrograde signaling.
The essential components of the Cn-mediated retrograde signaling pathway. In response to this stress signal, NFκB (cRel-p50) and the other signature transcription factors (as indicated) alongwith hnRNPA2 and Akt kinase are activated and translocated to the nucleus. Nuclear Akt phosphorylates hnRNPA2 which associates with the signature transcription factors by protein-protein interaction and stabilizes the enhanceosome complex. The transcriptional coactivator function of hnRNPA2 is essential for activation of the stress responsive nuclear genes Cathepsin L, RyR1, Glut4 and Akt1.
It will be interesting to investigate potential crosstalk between the Cn mediated retrograde signaling reported by us and other reported retrograde signaling pathways such as ROS-induced TFGβ signaling, HIF1α, Ras, p53 signaling (Jain, Rivera et al. 2013;Hage-Sleiman, Esmerian et al. 2013). Identifying a convergence point of these various signaling modes will help in elucidating the global picture of mitochondrial retrograde signaling.
7. Summary
Mitochondrial genome defects such as deleterious mutations, deletions and altered copy number affect normal mitochondrial functions. Mitochondrial retrograde signaling (mitochondria-to-nucleus) is induced as a cellular adaptive response to mitochondrial dysfunction and has been reported in many pathological conditions. We and others have shown that mitochondrial retrograde signaling reprograms cells towards tumorigenesis. In this review, we have compiled the different modes of the propagation of this signaling as reported till date, focusing on the Ca2+/Calcineurin-mediated signaling pathway characterized by us. In this pathway, reduction in mitochondrial genome copy number results in increased cytosolic Ca2+ and Cn is activated. In response to this downstream signature transcription factors NFκB (cRel-p50), NFAT, CREB, C/EBPδ are activated. An RNA-binding protein, hnRNPA2 is also activated with acts as a transcriptional coactivator for the stress responsive nuclear genes Cathepsin L, RyR1, Glut4 and Akt1. IGF1 receptor and Akt kinase are also activated as part of this signaling. The cumulative effect (shown in Figure 4) of the activation of these kinases, growth factors and transcription factors is the transformation of non- tumorigenic genes to a tumorigenic phenotype.
Highlights.
Mitochondrial retrograde signaling from mitochondria to nucleus is an adaptive cellular stress response to dysfunctional mitochondria.
There are broadly three mechanisms of propagation of mitochondrial retrograde signaling.
Mitochondrial retrograde signaling in response to reduced mtDNA copy number is mediated by calcineurin (Cn).
HnRNPA2, Akt, IGF1receptor are essential for propagation of the Cn-mediated retrograde signaling pathway.
Mitochondrial retrograde signaling causes profound changes in nuclear gene expression profile.
Acknowledgements
This work is supported by NIH grant CA-22762 and an Endowment from the Harriet Ellison Woodward Trust to NGA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Conflict of Interest: The authors declare no conflict of interest related to this work.
References
- Aloni Y, Attardi G. Symmetrical in vivo transcription of mitochondrial DNA in HeLa cells. Proc. Natl. Acad. Sci. U. S. A. 1971;68:1757–1761. doi: 10.1073/pnas.68.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschafl BA, Beutner G, Sharma VK, Sheu SS, Valdivia HH. The mitochondrial ryanodine receptor in rat heart: a pharmaco-kinetic profile. Biochim. Biophys. Acta. 2007;1768:1784–1795. doi: 10.1016/j.bbamem.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Amuthan G, Biswas G, Ananadatheerthavarada HK, Vijayasarathy C, Shephard HM, Avadhani NG. Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells. Oncogene. 2002;21:7839–7849. doi: 10.1038/sj.onc.1205983. [DOI] [PubMed] [Google Scholar]
- Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001;20:1910–1920. doi: 10.1093/emboj/20.8.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KA, Hirschey MA. Mitochondrial protein acetylation regulates metabolism. Essays Biochem. 2012;52:23–35. doi: 10.1042/bse0520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold RS, Sun Q, Sun CQ, Richards JC, O'Hearn S, Osunkoya AO, Wallace DC, Petros JA. An inherited heteroplasmic mutation in mitochondrial gene COI in a patient with prostate cancer alters reactive oxygen, reactive nitrogen and proliferation. Biomed. Res. Int. 2013:239257. doi: 10.1155/2013/239257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould T, Vankoningsloo S, Renard P, Houbion A, Ninane N, Demazy C, Remacle J, Raes M. CREB activation induced by mitochondrial dysfunction is a new signaling pathway that impairs cell proliferation. EMBO J. 2002;21:53–63. doi: 10.1093/emboj/21.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1998;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Aquilano K, Vigilanza P, Baldelli S, Pagliei B, Rotilio G, Ciriolo MR. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285:21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkar N, Ladner K, Canan BD, Liyanarachchi S, Bal NC, Pant M, Periasamy M, Li Q, Janssen PM, Guttridge DC. IKKalpha and alternative NF-kappaB regulate PGC-1beta to promote oxidative muscle metabolism. J. Cell Biol. 2012;196:497–511. doi: 10.1083/jcb.201108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, Koteliansky V, Mootha VK. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;19(476):341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi D, D'Aquila P, Giordano M, Montesanto A, Passarino G. Global DNA methylation levels are modulated by mitochondrial DNA variants. Epigenomics. 2012;4:17–27. doi: 10.2217/epi.11.109. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Bernardi P. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol Rev. 1999;79:1127–1155. doi: 10.1152/physrev.1999.79.4.1127. [DOI] [PubMed] [Google Scholar]
- Bestwick ML, Shadel GS. Accessorizing the human mitochondrial transcription machinery. Trends Biochem. Sci. 2013;38:283–291. doi: 10.1016/j.tibs.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KS, Avdalovic N, Avadhani NG. Characterization of primary transcripts and identification of transcription initiation sites on the heavy and light strands of mouse mitochondrial DNA. Biochemistry. 1989;28:763–769. doi: 10.1021/bi00428a052. [DOI] [PubMed] [Google Scholar]
- Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, Anandatheerthavarada HK, Avadhani NG. Mechanism of mitochondrial stress-induced resistance to apoptosis in mitochondrial DNA-depleted C2C12 myocytes. Cell Death. Differ. 2005;12:266–278. doi: 10.1038/sj.cdd.4401553. [DOI] [PubMed] [Google Scholar]
- Biswas G, Anandatheerthavarada HK, Zaidi M, Avadhani NG. Mitochondria to nucleus stress signaling: a distinctive mechanism of NFkappaB/Rel activation through calcineurin-mediated inactivation of IkappaBbeta. J. Cell Biol. 2003;161:507–519. doi: 10.1083/jcb.200211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, Guha M, Avadhani NG. Mitochondria-to-nucleus stress signaling in mammalian cells: nature of nuclear gene targets, transcription regulation, and induced resistance to apoptosis. Gene. 2005;354:132–139. doi: 10.1016/j.gene.2005.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, Srinivasan S, Anandatheerthavarada HK, Avadhani NG. Dioxin-mediated tumor progression through activation of mitochondria-to-nucleus stress signaling. Proc. Natl. Acad. Sci. U. S. A. 2008;105:186–191. doi: 10.1073/pnas.0706183104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, Tang W, Sondheimer N, Guha M, Bansal S, Avadhani NG. A distinctive physiological role for Ikappa Bbeta in the propagation of mitochondrial respiratory stress signaling. J Biol Chem. 2008;283(18):12586–12594. doi: 10.1074/jbc.M710481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL, Grabowski PJ. Alternative pre-mRNA splicing and neuronal function. Prog. Mol. Subcell. Biol. 2003;31:187–216. doi: 10.1007/978-3-662-09728-1_7. [DOI] [PubMed] [Google Scholar]
- Bononi A, Missiroli S, Poletti F, Suski JM, Agnoletto C, Bonora M, De ME, Giorgi C, Marchi S, Patergnani S, Rimessi A, Wieckowski MR, Pinton P. Mitochondria-associated membranes (MAMs) as hotspot Ca(2+) signaling units. Adv. Exp. Med. Biol. 2012;740:411–437. doi: 10.1007/978-94-007-2888-2_17. [DOI] [PubMed] [Google Scholar]
- Bonora E, Porcelli AM, Gasparre G, Biondi A, Ghelli A, Carelli V, Baracca A, Tallini G, Martinuzzi A, Lenaz G, Rugolo M, Romeo G. Defective oxidative phosphorylation in thyroid oncocytic carcinoma is associated with pathogenic mitochondrial DNA mutations affecting complexes I and III. Cancer Res. 2006;66:6087–6096. doi: 10.1158/0008-5472.CAN-06-0171. [DOI] [PubMed] [Google Scholar]
- Borst P. Mitochondrial nucleic acids. Annu. Rev. Biochem. 1972;41:333–376. doi: 10.1146/annurev.bi.41.070172.002001. [DOI] [PubMed] [Google Scholar]
- Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- Bustamante E, Pedersen PL. High aerobic glycolysis of rat hepatoma cells in culture: role of mitochondrial hexokinase. Proc. Natl. Acad. Sci. U. S. A. 1977;74:3735–3739. doi: 10.1073/pnas.74.9.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Cali T, Ottolini D, Brini M. Mitochondrial Ca(2+) as a key regulator of mitochondrial activities. Adv. Exp. Med. Biol. 2012;942:53–73. doi: 10.1007/978-94-007-2869-1_3. [DOI] [PubMed] [Google Scholar]
- Carew JS, Huang P. Mitochondrial defects in cancer. Mol. Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Casas F, Daury L, Grandemange S, Busson M, Seyer P, Hatier R, Carazo A, Cabello G, Wrutniak-Cabello C. Endocrine regulation of mitochondrial activity: involvement of truncated RXRalpha and c-Erb Aalpha1 proteins. FASEB J. 2003;17:426–436. doi: 10.1096/fj.02-0732com. [DOI] [PubMed] [Google Scholar]
- Cavalli LR, Varella-Garcia M, Liang BC. Diminished tumorigenic phenotype after depletion of mitochondrial DNA. Cell Growth Differ. 1997;8:1189–1198. [PubMed] [Google Scholar]
- Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, Hwang D, Park KS. A systems approach for decoding mitochondrial retrograde signaling pathways. Sci. Signal. 2013;6:rs4. doi: 10.1126/scisignal.2003266. [DOI] [PubMed] [Google Scholar]
- Chen M, David CJ, Manley JL. Concentration-dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. Nat. Struct. Mol. Biol. 2012;19:346–354. doi: 10.1038/nsmb.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhang J, Manley JL. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010;70:8977–8980. doi: 10.1158/0008-5472.CAN-10-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ristow M. Mitochondria and Metabolic Homeostasis. Antioxid. Redox. Signal. 2013;19(3):240–242. doi: 10.1089/ars.2013.5255. [DOI] [PubMed] [Google Scholar]
- Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat. Biotechnol. 2012;30:671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery PF, Elliott HR, Hudson G, Samuels DC, Relton CL. Epigenetics, epidemiology and mitochondrial DNA diseases. Int. J. Epidemiol. 2012;41:177–187. doi: 10.1093/ije/dyr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Nuclear-mitochondrial intergenomic communication. Biofactors. 1998;7:203–205. doi: 10.1002/biof.5520070307. [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FM, Azimi I, Faville RA, Peters AA, Jalink K, Putney JW, Jr, Goodhill GJ, Thompson EW, Roberts-Thomson SJ, Monteith GR. Induction of epithelial-mesenchymal transition (EMT) in breast cancer cells is calcium signal dependent. Oncogene. 2013:187. doi: 10.1038/onc.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsite R, Kachhap S, Anbazhagan R, Gabrielson E, Singh KK. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol. Cancer. 2002;1:6. doi: 10.1186/1476-4598-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca HF, Engstrom GW. Calcium uptake by rat kidney mitochondria. Proc. Natl. Acad. Sci. U. S. A. 1961;47:1744–1750. doi: 10.1073/pnas.47.11.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desler C, Marcker ML, Singh KK, Rasmussen LJ. The importance of mitochondrial DNA in aging and cancer. J. Aging Res. 2011;2011:407536. doi: 10.4061/2011/407536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R, Moraes CT. Lack of oxidative phosphorylation and low mitochondrial membrane potential decrease susceptibility to apoptosis and do not modulate the protective effect of Bcl-x(L) in osteosarcoma cells. J. Biol. Chem. 2000;275:7087–7094. doi: 10.1074/jbc.275.10.7087. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Duchen MR. Mitochondria and Ca(2+) in cell physiology and pathophysiology. Cell Calcium. 2000;28:339–348. doi: 10.1054/ceca.2000.0170. [DOI] [PubMed] [Google Scholar]
- Edeas M, Weissig V. Targeting mitochondria: Strategies, innovations and challenges: The future of medicine will come through mitochondria. Mitochondrion. 2013 doi: 10.1016/j.mito.2013.03.009. pii: S1567-7249(13)00062-7. [DOI] [PubMed] [Google Scholar]
- Ermak G, Sojitra S, Yin F, Cadenas E, Cuervo AM, Davies KJ. Chronic expression of RCAN1-1L protein induces mitochondrial autophagy and metabolic shift from oxidative phosphorylation to glycolysis in neuronal cells. J Biol Chem. 2012;287:14088–14098. doi: 10.1074/jbc.M111.305342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Lin CS, Potluri P, Procaccio V, Wallace DC. mtDNA lineage analysis of mouse L-cell lines reveals the accumulation of multiple mtDNA mutants and intermolecular recombination. Genes Dev. 2012;26:384–394. doi: 10.1101/gad.175802.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier J, Briere JJ, Strompf L, Amar L, Filali M, Jeunemaitre X, Rustin P, Gimenez-Roqueplo AP. Hereditary paraganglioma/pheochromocytoma and inherited succinate dehydrogenase deficiency. Horm. Res. 2005;63:171–179. doi: 10.1159/000084685. [DOI] [PubMed] [Google Scholar]
- Fedida-Metula S, Feldman B, Koshelev V, Levin-Gromiko U, Voronov E, Fishman D. Lipid rafts couple store-operated Ca2+ entry to constitutive activation of PKB/Akt in a Ca2+/calmodulin-, Src- and PP2A-mediated pathway and promote melanoma tumor growth. Carcinogenesis. 2012;33:740–750. doi: 10.1093/carcin/bgs021. [DOI] [PubMed] [Google Scholar]
- Feldman B, Fedida-Metula S, Nita J, Sekler I, Fishman D. Coupling of mitochondria to store-operated Ca(2+)-signaling sustains constitutive activation of protein kinase B/Akt and augments survival of malignant melanoma cells. Cell Calcium. 2010;47:525–537. doi: 10.1016/j.ceca.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Meadows E, Fiorotto M, Klein WH. Myogenin regulates exercise capacity and skeletal muscle metabolism in the adult mouse. PLoS One. 2010;5:e13535. doi: 10.1371/journal.pone.0013535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford LP, Wright WE, Shay JW. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene. 2002;21:580–583. doi: 10.1038/sj.onc.1205086. [DOI] [PubMed] [Google Scholar]
- Formentini L, Sanchez-Arago M, Sanchez-Cenizo L, Cuezva JM. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Mol. Cell. 2012;45:731–742. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Garcia-Rodriguez LJ. Appendix 1. Basic properties of mitochondria. Methods Cell Biol. 2007;80:809–812. doi: 10.1016/S0091-679X(06)80040-3. [DOI] [PubMed] [Google Scholar]
- Garesse R, Vallejo CG. Animal mitochondrial biogenesis and function: a regulatory cross-talk between two genomes. Gene. 2001;263:1–16. doi: 10.1016/s0378-1119(00)00582-5. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Giacomello M, Drago I, Pizzo P, Pozzan T. Mitochondrial Ca2+ as a key regulator of cell life and death. Cell Death. Differ. 2007;14:1267–1274. doi: 10.1038/sj.cdd.4402147. [DOI] [PubMed] [Google Scholar]
- Gilkerson R, Bravo L, Garcia I, Gaytan N, Herrera A, Maldonado A, Quintanilla B. The mitochondrial nucleoid: integrating mitochondrial DNA into cellular homeostasis. Cold Spring Harb. Perspect. Biol. 2013;5:a011080. doi: 10.1101/cshperspect.a011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glancy B, Balaban RS. Role of mitochondrial Ca2+ in the regulation of cellular energetics. Biochemistry. 2012;51:2959–2973. doi: 10.1021/bi2018909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Mathupala SP, Pedersen PL. Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. J. Biol. Chem. 2003;278:15333–15340. doi: 10.1074/jbc.M300608200. [DOI] [PubMed] [Google Scholar]
- Golan-Gerstl R, Wallach-Dayan SB, Zisman P, Cardoso WV, Goldstein RH, Breuer R. Cellular FLICE-like inhibitory protein deviates myofibroblast fas-induced apoptosis toward proliferation during lung fibrosis. Am J Respir Cell Mol Biol. 2012;47:271–279. doi: 10.1165/rcmb.2010-0284RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves LC, Reeve AK, Taylor RW, Turnbull DM. Mitochondrial DNA and disease. J Pathol. 2012;226:274–286. doi: 10.1002/path.3028. [DOI] [PubMed] [Google Scholar]
- Gregersen N, Bross P. Protein misfolding and cellular stress: an overview. Methods Mol. Biol. 2010;648:3–23. doi: 10.1007/978-1-60761-756-3_1. [DOI] [PubMed] [Google Scholar]
- Guha M, Fang JK, Monks R, Birnbaum MJ, Avadhani NG. Activation of Akt is essential for the propagation of mitochondrial respiratory stress signaling and activation of the transcriptional coactivator heterogeneous ribonucleoprotein A2. Mol Biol Cell. 2010;21:3578–3589. doi: 10.1091/mbc.E10-03-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M, Pan H, Fang JK, Avadhani NG. Heterogeneous nuclear ribonucleoprotein A2 is a common transcriptional coactivator in the nuclear transcription response to mitochondrial respiratory stress. Mol Biol Cell. 2009;20:4107–4119. doi: 10.1091/mbc.E09-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M, Srinivasan S, Biswas G, Avadhani NG. Activation of a novel calcineurin-mediated insulin-like growth factor-1 receptor pathway, altered metabolism, and tumor cell invasion in cells subjected to mitochondrial respiratory stress. J Biol Chem. 2007;282:14536–14546. doi: 10.1074/jbc.M611693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M, Tang W, Sondheimer N, Avadhani NG. Role of calcineurin, hnRNPA2 and Akt in mitochondrial respiratory stress-mediated transcription activation of nuclear gene targets. Biochim. Biophys. Acta. 2010;1797:1055–1065. doi: 10.1016/j.bbabio.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zheng L, Liu W, Wang X, Wang Z, Wang Z, French AJ, Kang D, Chen L, Thibodeau SN, Liu W. Frequent truncating mutation of TFAM induces mitochondrial DNA depletion and apoptotic resistance in microsatellite-unstable colorectal cancer. Cancer Res. 2011;71:2978–2987. doi: 10.1158/0008-5472.CAN-10-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage-Sleiman R, Esmerian MO, Kobeissy H, Dbaibo G. p53 and Ceramide as Collaborators in the Stress Response. Int J Mol. Sci. 2013;14:4982–5012. doi: 10.3390/ijms14034982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnoczky G, Csordas G. Calcium signalling: fishing out molecules of mitochondrial calcium transport. Curr Biol. 2010;20:R888–R891. doi: 10.1016/j.cub.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerman PS, Fox CJ, Thompson CB. Beginnings of a signal-transduction pathway for bioenergetic control of cell survival. Trends Biochem Sci. 2004;29:586–592. doi: 10.1016/j.tibs.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Hanna SC, Krishnan B, Bailey ST, Moschos SJ, Kuan PF, Shimamura T, Osborne LD, Siegel MB, Duncan LM, O'Brien ET, III, Superfine R, Miller CR, Simon MC, Wong KK, Kim WY. HIF1alpha and HIF2alpha independently activate SRC to promote melanoma metastases. J Clin. Invest. 2013;123:2078–2093. doi: 10.1172/JCI66715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Cooper HM, Reyes A, Di RM, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, Walker JE, Holt IJ. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 2012;40:6109–6121. doi: 10.1093/nar/gks266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Brown MA, Rothnagel JA, Saunders NA, Smith R. Roles of heterogeneous nuclear ribonucleoproteins A and B in cell proliferation. J Cell Sci. 2005;118:3173–3183. doi: 10.1242/jcs.02448. [DOI] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Kudo T, Suzuki S, Evans TT, Sasaki R, Wada Y, Shirakawa T, Sawyer JR, Gotoh A. Mitochondrial DNA determines androgen dependence in prostate cancer cell lines. Oncogene. 2006;25:1437–1445. doi: 10.1038/sj.onc.1209190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- Ivanov VN, Ghandhi SA, Zhou H, Huang SX, Chai Y, Amundson SA, Hei TK. Radiation response and regulation of apoptosis induced by a combination of TRAIL and CHX in cells lacking mitochondrial DNA: a role for NF-kappaB-STAT3-directed gene expression. Exp Cell Res. 2011;317:1548–1566. doi: 10.1016/j.yexcr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Rivera S, Monclus EA, Synenki L, Zirk A, Eisenbart J, Feghali-Bostwick C, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial reactive oxygen species regulate transforming growth factor-beta signaling. J Biol Chem. 2013;288:770–777. doi: 10.1074/jbc.M112.431973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM, Kriete A. The yeast retrograde response as a model of intracellular signaling of mitochondrial dysfunction. Front Physiol. 2012;3:139. doi: 10.3389/fphys.2012.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaipparettu BA, Ma Y, Park JH, Lee TL, Zhang Y, Yotnda P, Creighton CJ, Chan WY, Wong LJ. Crosstalk from non-cancerous mitochondria can inhibit tumor properties of metastatic cells by suppressing oncogenic pathways. PLoS One. 2013;8(5):e61747. doi: 10.1371/journal.pone.0061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kim YH, Chang I, Kim S, Pak YK, Oh BH, Yagita H, Jung YK, Oh YJ, Lee MS. Resistance of mitochondrial DNA-deficient cells to TRAIL: role of Bax in TRAIL-induced apoptosis. Oncogene. 2002;21:3139–3148. doi: 10.1038/sj.onc.1205406. [DOI] [PubMed] [Google Scholar]
- Kinnally KW, Peixoto PM, Ryu SY, Dejean LM. Is mPTP the gatekeeper for necrosis, apoptosis, or both? Biochim Biophys Acta. 2011;1813:616–622. doi: 10.1016/j.bbamcr.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkinezos IG, Moraes CT. Reactive oxygen species and mitochondrial diseases. Semin Cell Dev Biol. 2001;12:449–457. doi: 10.1006/scdb.2001.0282. [DOI] [PubMed] [Google Scholar]
- Kirstein-Miles J, Morimoto RI. Peptides signal mitochondrial stress. Cell Metab. 2010;11:177–178. doi: 10.1016/j.cmet.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Kosturko LD, Maggipinto MJ, D'Sa C, Carson JH, Barbarese E. The microtubule-associated protein tumor overexpressed gene binds to the RNA trafficking protein heterogeneous nuclear ribonucleoprotein A2. Mol Biol Cell. 2005;16:1938–1947. doi: 10.1091/mbc.E04-08-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van RH, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Kulawiec M, Safina A, Desouki MM, Still I, Matsui S, Bakin A, Singh KK. Tumorigenic transformation of human breast epithelial cells induced by mitochondrial DNA depletion. Cancer Biol Ther. 2008;7:1732–1743. doi: 10.4161/cbt.7.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le PS, Mirebeau-Prunier D, Boutet-Bouzamondo N, Jacques C, Guillotin D, Lauret E, Houlgatte R, Malthiery Y, Savagner F. Nitric oxide and calcium participate in the fine regulation of mitochondrial biogenesis in follicular thyroid carcinoma cells. J Biol Chem. 2011;286:18229–18239. doi: 10.1074/jbc.M110.217521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh-Brown S, Enriquez JA, Odom DT. Nuclear transcription factors in mammalian mitochondria. Genome Biology. 2010;11:215. doi: 10.1186/gb-2010-11-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- Lin M. Molecular imaging using positron emission tomography in colorectal cancer. Discov Med. 2011;11:435–447. [PubMed] [Google Scholar]
- Liu CY, Lee CF, Hong CH, Wei YH. Mitochondrial DNA mutation and depletion increase the susceptibility of human cells to apoptosis. Ann N. Y. Acad Sci. 2004;1011:133–145. doi: 10.1007/978-3-662-41088-2_14. [DOI] [PubMed] [Google Scholar]
- Loveland B, Wang CR, Yonekawa H, Hermel E, Lindahl KF. Maternally transmitted histocompatibility antigen of mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell. 1990;60:971–980. doi: 10.1016/0092-8674(90)90345-f. [DOI] [PubMed] [Google Scholar]
- Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 2009;19:802–815. doi: 10.1038/cr.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Bond JD, Ingram VM. Compromised mitochondrial function leads to increased cytosolic calcium and to activation of MAP kinases. Proc. Natl. Acad. Sci. U. S. A. 1997;94:9705–9710. doi: 10.1073/pnas.94.18.9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madge LA, May MJ. The NFkappaB paradox: RelB induces and inhibits gene expression. Cell Cycle. 2011;10:6–7. doi: 10.4161/cc.10.1.14291. [DOI] [PubMed] [Google Scholar]
- Magda D, Lecane P, Prescott J, Thiemann P, Ma X, Dranchak PK, Toleno DM, Ramaswamy K, Siegmund KD, Hacia JG. mtDNA depletion confers specific gene expression profiles in human cells grown in culture and in xenograft. BMC. Genomics. 2008;9:521. doi: 10.1186/1471-2164-9-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Cárdenas C, Doonan PJ, Chandramoorthy HC, Irrinki KM, Golenár T, Csordás G, Madireddi P, Yang J, Müller M, Miller R, Kolesar JE, Molgó J, Kaufman B, Hajnóczky G, Foskett JK, Madesh M. MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat Cell Biol. 2012;14:1336–1343. doi: 10.1038/ncb2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manev H, Dzitoyeva S, Chen H. Mitochondrial DNA: A Blind Spot in Neuroepigenetics. Biomol. Concepts. 2012;3:107–115. doi: 10.1515/bmc-2011-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti P, Castedo M, Susin SA, Zamzami N, Hirsch T, Macho A, Haeffner A, Hirsch F, Geuskens M, Kroemer G. Mitochondrial permeability transition is a central coordinating event of apoptosis. J Exp Med. 1996;184:1155–1160. doi: 10.1084/jem.184.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- McKay SJ, Cooke H. hnRNP A2/B1 binds specifically to single stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res. 1992;20:6461–6464. doi: 10.1093/nar/20.24.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirhaeghe A, Crowley V, Lenaghan C, Lelliott C, Green K, Stewart A, Hart K, Schinner S, Sethi JK, Yeo G, Brand MD, Cortright RN, O'Rahilly S, Montague C, Vidal-Puig AJ. Characterization of the human, mouse and rat PGC1 beta (peroxisome-proliferator-activated receptor-gamma co-activator 1 beta) gene in vitro and in vivo. Biochem J. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Sickmann A, Pfanner N. The mitochondrial proteome: from inventory to function. Cell. 2008;134:22–24. doi: 10.1016/j.cell.2008.06.043. [DOI] [PubMed] [Google Scholar]
- Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocherhomji S, Tollefsbol TO, Singh KK. Mitochondrial regulation of epigenetics and its role in human diseases. Epigenetics. 2012;7:326–334. doi: 10.4161/epi.19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Honda M, Shirasaki T, Yamashita T, Yamashita T, Mizukoshi E, Kaneko S. Heterogeneous nuclear ribonucleoprotein A2/B1 in association with hTERT is a potential biomarker for hepatocellular carcinoma. Liver Int. 2012;32:1146–1155. doi: 10.1111/j.1478-3231.2012.02778.x. [DOI] [PubMed] [Google Scholar]
- Modica-Napolitano JS, Kulawiec M, Singh KK. Mitochondria and human cancer. Curr Mol Med. 2007;7:121–131. doi: 10.2174/156652407779940495. [DOI] [PubMed] [Google Scholar]
- Moran-Jones K, Wayman L, Kennedy DD, Reddel RR, Sara S, Snee MJ, Smith R. hnRNP A2, a potential ssDNA/RNA molecular adapter at the telomere. Nucleic Acids Res. 2005;33:486–496. doi: 10.1093/nar/gki203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro L, Arbini AA, Yao JL, di Sant'Agnese PA, Marra E, Greco M. Mitochondrial DNA depletion in prostate epithelial cells promotes anoikis resistance and invasion through activation of PI3K/Akt2. Cell Death. Differ. 2009;16:571–583. doi: 10.1038/cdd.2008.178. [DOI] [PubMed] [Google Scholar]
- Naviaux Mitochondrial control of epigenetics. Cancer Biology and Therapy. 2008;7:1191–1193. doi: 10.4161/cbt.7.8.6741. [DOI] [PubMed] [Google Scholar]
- Naik E, Dixit VM. Mitochondrial reactive oxygen species drive proinflammatory cytokine production. J Exp Med. 2011;208:417–420. doi: 10.1084/jem.20110367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair PN, De Armond DT, Adamo ML, Strodel WE, Freeman JW. Aberrant expression and activation of insulin-like growth factor-1 receptor (IGF-1R) are mediated by an induction of IGF-1R promoter activity and stabilization of IGF-1R mRNA and contributes to growth factor independence and increased survival of the pancreatic cancer cell line MIA PaCa-2. Oncogene. 2001;20:8203–8214. doi: 10.1038/sj.onc.1205044. [DOI] [PubMed] [Google Scholar]
- Naito A, Cook CC, Mizumachi T, Wang M, Xie CH, Evans TT, Kelly T, Higuchi M. Progressive tumor features accompany epithelial-mesenchymal transition induced in mitochondrial DNA-depleted cells. Cancer Sci. 2008;99:1584–1588. doi: 10.1111/j.1349-7006.2008.00879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, Takeda K, Yu ZX, Ferrans VJ, Finkel T. Role for mitochondrial oxidants as regulators of cellular metabolism. Mol Cell Biol. 2000;20:7311–7318. doi: 10.1128/mcb.20.19.7311-7318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuzil J, Dong LF, Rohlena J, Truksa J, Ralph SJ. Classification of mitocans, anti-cancer drugs acting on mitochondria. Mitochondrion. 2013;13:199–208. doi: 10.1016/j.mito.2012.07.112. [DOI] [PubMed] [Google Scholar]
- Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh VS, Conrad-Webb H, Docherty R, Butow RA. Interaction between the yeast mitochondrial and nuclear genomes influences the abundance of novel transcripts derived from the spacer region of the nuclear ribosomal DNA repeat. Mol Cell Biol. 1989;9:1897–1907. doi: 10.1128/mcb.9.5.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Sharma LK, Li H, Xiang R, Holstein D, Wu J, Lechleiter J, Naylor SL, Deng JJ, Lu J, Bai Y. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum Mol Genet. 2009;18:1578–1589. doi: 10.1093/hmg/ddp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patry C, Bouchard L, Labrecque P, Gendron D, Lemieux B, Toutant J, Lapointe E, Wellinger R, Chabot B. Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res. 2003;63:7679–7688. [PubMed] [Google Scholar]
- Pavlides S, Vera I, Gandara R, Sneddon S, Pestell RG, Mercier I, Martinez-Outschoorn UE, Whitaker-Menezes D, Howell A, Sotgia F, Lisanti MP. Warburg meets autophagy: cancer-associated fibroblasts accelerate tumor growth and metastasis via oxidative stress, mitophagy, and aerobic glycolysis. Antioxid. Redox. Signal. 2012;16:1264–1284. doi: 10.1089/ars.2011.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen PL, Mathupala S, Rempel A, Geschwind JF, Ko YH. Mitochondrial bound type II hexokinase: a key player in the growth and survival of many cancers and an ideal prospect for therapeutic intervention. Biochim Biophys Acta. 2002;1555:14–20. doi: 10.1016/s0005-2728(02)00248-7. [DOI] [PubMed] [Google Scholar]
- Pelicano H, Xu RH, Du M, Feng L, Sasaki R, Carew JS, Hu Y, Ramdas L, Hu L, Keating MJ, Zhang W, Plunkett W, Huang P. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J Cell Biol. 2006;175:913–923. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisch F, Khauv D, Risso G, Stallings-Mann M, Blaustein M, Quadrana L, Radisky DC, Srebrow A. Involvement of hnRNP A1 in the matrix metalloprotease-3-dependent regulation of Rac1 pre-mRNA splicing. J Cell Biochem. 2012;113:2319–2329. doi: 10.1002/jcb.24103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;16(467):291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinton P, Leo S, Wieckowski MR, Di BG, Rizzuto R. Long-term modulation of mitochondrial Ca2+ signals by protein kinase C isozymes. J Cell Biol. 2004;165:223–232. doi: 10.1083/jcb.200311061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plas DR, Thompson CB. Cell metabolism in the regulation of programmed cell death. Trends Endocrinol. Metab. 2002;13:75–78. doi: 10.1016/s1043-2760(01)00528-8. [DOI] [PubMed] [Google Scholar]
- Porteous WK, James AM, Sheard PW, Porteous CM, Packer MA, Hyslop SJ, Melton JV, Pang CY, Wei YH, Murphy MP. Bioenergetic consequences of accumulating the common 4977-bp mitochondrial DNA deletion. Eur J Biochem. 1998;257:192–201. doi: 10.1046/j.1432-1327.1998.2570192.x. [DOI] [PubMed] [Google Scholar]
- Poyton RO, Ball KA, Castello PR. Mitochondrial generation of free radicals and hypoxic signaling. Trends Endocrinol. Metab. 2009;20:332–340. doi: 10.1016/j.tem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- Preussner M, Schreiner S, Hung LH, Porstner M, Jack HM, Benes V, Ratsch G, Bindereif A. HnRNP L and L-like cooperate in multiple-exon regulation of CD45 alternative splicing. Nucleic Acids Res. 2012;40:5666–5678. doi: 10.1093/nar/gks221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Raffaello A, De SD, Rizzuto R. The mitochondrial Ca(2+) uniporter. Cell Calcium. 2012;52:16–21. doi: 10.1016/j.ceca.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Raffaello A, Rizzuto R. Mitochondrial longevity pathways. Biochim. Biophys. Acta. 2011;1813:260–268. doi: 10.1016/j.bbamcr.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Raimundo N, Song L, Shutt TE, McKay SE, Cotney J, Guan MX, Gilliland TC, Hohuan D, Santos-Sacchi J, Shadel GS. Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell. 2012;148:716–726. doi: 10.1016/j.cell.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault TT, Chipuk JE. Inter-organellar communication with mitochondria regulates both the intrinsic and extrinsic pathways of apoptosis. Commun Integr Biol. 2013;6:e22872. doi: 10.4161/cib.22872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensvold JW, Ong SE, Jeevananthan A, Carr SA, Mootha VK, Pagliarini DJ. Complementary RNA and protein profiling identifies iron as a key regulator of mitochondrial biogenesis. Cell Rep. 2013;3:237–245. doi: 10.1016/j.celrep.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S. Reaching for the stars: Linking RNA binding proteins to diseases. Adv Exp Med Biol. 2010;693:142–157. [PubMed] [Google Scholar]
- Rizzuto R, De SD, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Duchen MR, Pozzan T. Flirting in little space: the ER/mitochondria Ca2+ liaison. Sci. STKE. 2004:re1. doi: 10.1126/stke.2152004re1. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pozzan T. Microdomains of intracellular Ca2+: molecular determinants and functional consequences. Physiol Rev. 2006;86:369–408. doi: 10.1152/physrev.00004.2005. [DOI] [PubMed] [Google Scholar]
- Rooke N, Markovtsov V, Cagavi E, Black DL. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol Cell Biol. 2003;23:1874–1884. doi: 10.1128/MCB.23.6.1874-1884.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel ED, Liu S, Baumeister R, Schulze E. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS Genet. 2013;9:e1003346. doi: 10.1371/journal.pgen.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annu Rev Biochem. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenstrom P, Rizzuto R, Hajnoczky G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20728–20733. doi: 10.1073/pnas.0808953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma LK, Fang H, Liu J, Vartak R, Deng J, Bai Y. Mitochondrial respiratory complex I dysfunction promotes tumorigenesis through ROS alteration and AKT activation. Hum. Mol. Genet. 2011;20:4605–4616. doi: 10.1093/hmg/ddr395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- Singh KK, Rasmussen AK, Rasmussen LJ. Genome-wide analysis of signal transducers and regulators of mitochondrial dysfunction in Saccharomyces cerevisiae. Ann. N. Y. Acad. Sci. 2004;1011:284–298. doi: 10.1007/978-3-662-41088-2_27. [DOI] [PubMed] [Google Scholar]
- Smiraglia DJ, Kulawiec M, Bistulfi GL, Gupta SG, Singh KK. A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol. Ther. 2008;7:1182–1190. doi: 10.4161/cbt.7.8.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Avadhani NG. Hypoxia-mediated mitochondrial stress in RAW264.7 cells induces osteoclast-like TRAP-positive cells. Ann. N. Y. Acad. Sci. 2007;1117:51–61. doi: 10.1196/annals.1402.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Avadhani NG. Cytochrome c oxidase dysfunction in oxidative stress. Free Radic. Biol. Med. 2012;53:1252–1263. doi: 10.1016/j.freeradbiomed.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Koenigstein A, Joseph J, Sun L, Kalyanaraman B, Zaidi M, Avadhani NG. Role of mitochondrial reactive oxygen species in osteoclast differentiation. Ann. N. Y. Acad. Sci. 2010;1192:245–252. doi: 10.1111/j.1749-6632.2009.05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still AJ, Floyd BJ, Hebert AS, Bingman CA, Carson JJ, Gunderson DR, Dolan BK, Grimsrud PA, Dittenhafer-Reed KE, Stapleton DS, Keller MP, Westphall MS, Denu JM, Attie AD, Coon JJ, Pagliarini DJ. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. J Biol Chem. 2013 doi: 10.1074/jbc.M113.483396. (Epub) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammineni P, Anugula C, Mohammed F, Anjaneyulu M, Larner AC, Sepuri NB. The Import of the Transcription Factor STAT3 into Mitochondria Depends on GRIM-19, a Component of the Electron Transport Chain. J Biol. Chem. 2013;288:4723–4732. doi: 10.1074/jbc.M112.378984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Chowdhury AR, Guha M, Huang L, Van WT, Rustgi AK, Avadhani NG. Silencing of IkBbeta mRNA causes disruption of mitochondrial retrograde signaling and suppression of tumor growth in vivo. Carcinogenesis. 2012;33:1762–1768. doi: 10.1093/carcin/bgs190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauler J, Zudaire E, Liu H, Shih J, Mulshine JL. hnRNP A2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res. 2010;70:7137–7147. doi: 10.1158/0008-5472.CAN-10-0860. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telang S, Yalcin A, Clem AL, Bucala R, Lane AN, Eaton JW, Chesney J. Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene. 2006;25:7225–7234. doi: 10.1038/sj.onc.1209709. [DOI] [PubMed] [Google Scholar]
- Thomson DM, Herway ST, Fillmore N, Kim H, Brown JD, Barrow JR, Winder WW. AMP-activated protein kinase phosphorylates transcription factors of the CREB family. J. Appl. Physiol. 2008;104:429–438. doi: 10.1152/japplphysiol.00900.2007. [DOI] [PubMed] [Google Scholar]
- Tockman MS, Gupta PK, Myers JD, Frost JK, Baylin SB, Gold EB, Chase AM, Wilkinson PH, Mulshine JL. Sensitive and specific monoclonal antibody recognition of human lung cancer antigen on preserved sputum cells: a new approach to early lung cancer detection. J. Clin. Oncol. 1988;6:1685–1693. doi: 10.1200/JCO.1988.6.11.1685. [DOI] [PubMed] [Google Scholar]
- Torosyan Y, Dobi A, Glasman M, Mezhevaya K, Naga S, Huang W, Paweletz C, Leighton X, Pollard HB, Srivastava M. Role of multi-hnRNP nuclear complex in regulation of tumor suppressor ANXA7 in prostate cancer cells. Oncogene. 2010;29:2457–2466. doi: 10.1038/onc.2010.2. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly Y, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Venneti S, Thompson CB. Metabolic modulation of epigenetics in gliomas. Brain Pathol. 2013;23:217–221. doi: 10.1111/bpa.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera J, Jaumot M, Estanyol JM, Brun S, Agell N, Bachs O. Heterogeneous nuclear ribonucleoprotein A2 is a SET-binding protein and a PP2A inhibitor. Oncogene. 2006;25:260–270. doi: 10.1038/sj.onc.1209050. [DOI] [PubMed] [Google Scholar]
- Vijayasarathy C, Biunno I, Lenka N, Yang M, Basu A, Hall IP, Avadhani NG. Variations in the subunit content and catalytic activity of the cytochrome c oxidase complex from different tissues and different cardiac compartments. Biochim. Biophys. Acta. 1998;1371:71–82. doi: 10.1016/s0005-2736(97)00278-2. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial bioenergetic etiology of disease. J. Clin. Invest. 2013;123:1405–1412. doi: 10.1172/JCI61398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136:163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, Moh A, Moghaddas S, Chen Q, Bobbili S, Cichy J, Dulak J, Baker DP, Wolfman A, Stuehr D, Hassan MO, Fu XY, Avadhani N, Drake JI, Fawcett P, Lesnefsky EJ, Larner AC. Function of Mitochondrial Stat3 in Cellular Respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weighardt F, Biamonti G, Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- Weitzel JM, Iwen KA, Seitz HJ. Regulation of mitochondrial biogenesis by thyroid hormone. Exp. Physiol. 2003;88:121–128. doi: 10.1113/eph8802506. [DOI] [PubMed] [Google Scholar]
- Westermann B, Neupert W. 'Omics' of the mitochondrion. Nat. Biotechnol. 2003;21:239–240. doi: 10.1038/nbt0303-239. [DOI] [PubMed] [Google Scholar]
- Williams GS, Boyman L, Chikando AC, Khairallah RJ, Lederer WJ. Mitochondrial calcium uptake. Proc. Natl. Acad. Sci. U. S. A. 2013 doi: 10.1073/pnas.1300410110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojewoda M, Duszynski J, Szczepanowska J. NARP mutation and mtDNA depletion trigger mitochondrial biogenesis which can be modulated by selenite supplementation. Int. J. Biochem. Cell Biol. 2011;43:1178–1186. doi: 10.1016/j.biocel.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Woo DK, Green PD, Santos JH, D'Souza AD, Walther Z, Martin WD, Christian BE, Chandel NS, Shadel GS. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(Min/+) mice. Am. J. Pathol. 2012;180:24–31. doi: 10.1016/j.ajpath.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CJ, Agboke F, Muthu M, Michaelis KA, Mundy MA, La P, Yang G, Dennery PA. Nuclear factor-kappaB (NF-kappaB) inhibitory protein IkappaBbeta determines apoptotic cell death following exposure to oxidative stress. J. Biol. Chem. 2012;287:6230–6239. doi: 10.1074/jbc.M111.318246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakes FM, Van HB. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141:982–993. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka K, Imajoh-Ohmi S, Fukuda H, Akita Y, Kurosawa K, Yamamoto Y, Sanai Y. Identification of phosphoproteins associated with maintenance of transformed state in temperature-sensitive Rous sarcoma-virus infected cells by proteomic analysis. Biochem. Biophys. Res. Commun. 2006;345:1240–1246. doi: 10.1016/j.bbrc.2006.04.183. [DOI] [PubMed] [Google Scholar]
- Yang Y, Valera V, Sourbier C, Vocke CD, Wei M, Pike L, Huang Y, Merino MA, Bratslavsky G, Wu M, Ricketts CJ, Linehan WM. A novel fumarate hydratasedeficient HLRCC kidney cancer cell line, UOK268: a model of the Warburg effect in cancer. Cancer Genet. 2012;205:377–390. doi: 10.1016/j.cancergen.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291:2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- Young RM, Simon MC. Untuning the tumor metabolic machine: HIF-alpha: proand antitumorigenic? Nat. Med. 2012;18:1024–1025. doi: 10.1038/nm.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Shi Y, Wei X, Yang Y, Zang F, Niu R. Mitochondrial DNA depletion promotes impaired oxidative status and adaptive resistance to apoptosis in T47D breast cancer cells. Eur. J. Cancer Prev. 2009 doi: 10.1097/CEJ.0b013e32832f9bd6. [DOI] [PubMed] [Google Scholar]
- Zamzami N, Maisse C, Metivier D, Kroemer G. Measurement of membrane permeability and permeability transition of mitochondria. Methods Cell Biol. 2001;65:147–158. doi: 10.1016/s0091-679x(01)65009-x. [DOI] [PubMed] [Google Scholar]
- Zhou G, Dada LA, Wu M, Kelly A, Trejo H, Zhou Q, Varga J, Sznajder JI. Hypoxia-induced alveolar epithelial-mesenchymal transition requires mitochondrial ROS and hypoxia-inducible factor 1. Am. J. Physiol Lung Cell Mol. Physiol. 2009;297:L1120–L1130. doi: 10.1152/ajplung.00007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Mulshine JL, Unsworth EJ, Scott FM, Avis IM, Vos MD, Treston AM. Purification and characterization of a protein that permits early detection of lung cancer. Identification of heterogeneous nuclear ribonucleoprotein-A2/B1 as the antigen for monoclonal antibody 703D4. J. Biol. Chem. 1996;271:10760–10766. doi: 10.1074/jbc.271.18.10760. [DOI] [PubMed] [Google Scholar]
- Zollo O, Tiranti V, Sondheimer N. Transcriptional requirements of the distal heavy-strand promoter of mtDNA. Proc. Natl. Acad. Sci. U. S. A. 2012;109:6508–6512. doi: 10.1073/pnas.1118594109. [DOI] [PMC free article] [PubMed] [Google Scholar]