Abstract
By combining the high efficiency enrichment via our continuous flow magnetic separation (CFMS) technique with the analytical power of next-generation sequencing, we have enabled the generation of antibody mimetics with a single round of mRNA display. This approach eliminates iterative selection cycles, providing a path to fully automated ligand generation to meet the goal of a renewable, proteome-wide affinity reagent resource.
Keywords: selection methods, antibody mimetics, mRNA display, ligand design, directed evolution
There is a pressing need for new technologies to generate robust, renewable recognition tools against the entire human proteome.[1] Hybridoma-based monoclonal antibodies,[2] the standard for protein reagents, are undesirable for this task because of the number of animals, amount of target, time, and effort required to generate each reagent. Here, we developed a unified approach to solve this problem by integrating four distinct technologies: 1) a combinatorial protein library based on the 10th fibronectin type III domain of human fibronectin (10Fn3),[3] 2) protein library display by mRNA display,[4] 3) selection by continuous flow magnetic separation (CFMS),[5] and 4) sequence analysis by high throughput sequencing (HTS).[6] Next generation sequencing has revolutionized many fields of biology, and is increasingly being utilized to improve ligand design efforts.[7] The result of our integrated approach is the ability to perform selection-based protein design in a single round—an entirely in vitro, highly scalable, multiplexed process. Statistical analysis of input and selected pools reveals the key factors in the success of this first trial were the high uniformity of the input pools and excellent fold enrichment. Our results also demonstrate that highly functional binders (KD ~100 nM or better) are present with a frequency of > 1 in 109 in our library.
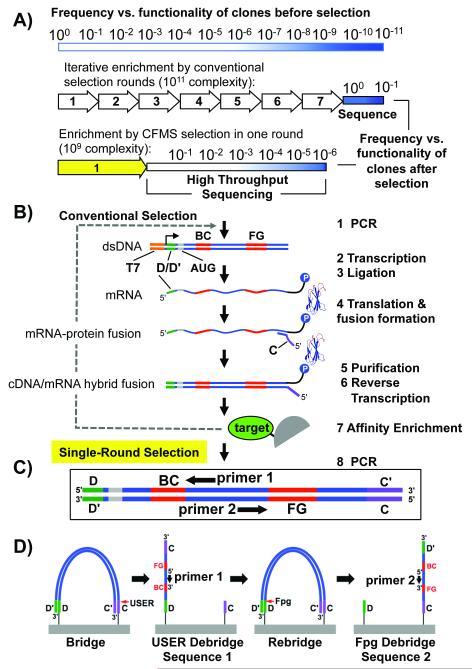
To begin, we needed to devise an appropriate selection protocol and library creation format. Typically, in vitro selections require multiple rounds of modest sequential enrichment, followed by small-scale sequencing of functional clones (Figure 1A). Indeed, the need to generate a target-specific library at each round provides a significant limitation towards parallelizing and accelerating selections. In contrast, for identification of ligands after a single round of CFMS mRNA display (Figure 1A,B), only a single naïve library pool must be synthesized for any number of targets, drastically decreasing the effort needed for ligand discovery.
Figure 1.
Selection scheme. A) Graphical representation of functional sequence enrichment by conventional or single round CFMS mRNA display. Functionality, a combination of specificity and affinity, is depicted by a gradient from white (nonfunctional, more common) to dark blue (high functionality, least common). In conventional selection, many rounds of enrichment are performed until most clones are functional. In our single round selection described here, a lower complexity library (~109) combined with improved enrichment efficiency by CFMS (~1000-fold)[5] enables identification of functional clones >1 in 106 by Illumina sequencing. B) Naïve mRNA display library synthesis steps are illustrated (Steps 1-6). The e10Fn3 library was adapted for Illumina sequencing by inserting one of the annealing regions necessary for bridge amplification (“D”) in the 5′ untranslated region. The second chip-annealing/bridge-amplification region (“C”) is added by the reverse transcription primer (step 4). C) For high-throughput selection we identify ligands after one round of selection by sampling the semi-enriched pools through Ilummina HTS (D) using the incorporated annealing regions for bridge amplification and e10Fn3-specific sequencing primers.
To integrate CFMS mRNA display with HTS, we adapted a protein scaffold with variable regions that can be easily read by Illumina HTS. Previously, we had designed, created and optimized a high-complexity library termed e10Fn3[5,8]—based on the 10Fn3 scaffold developed by Koide and coworkers.[3] This scaffold contains only two random sequence regions, the BC loop (7 residues) and the FG loop (10 residues) (Figure 1C), which can be read by paired end sequencing using customized primers (Figure 1D; Figure S1). Additionally, the simplicity of the scaffold enables rapid, accurate binder reconstruction using oligonucleotides for validation without the need for cloning into bacteria.
Generally, mRNA display selections utilize large libraries (1012-1014 sequences), with low copy number (3-10 copies) for protein design.[5,8-10] In order to achieve a single round selection, we needed to balance the input diversity with three factors: 1) the number of clones we could sequence, 2) the fold enrichment in a single round of CFMS, and 3) the inherent frequency of functional clones in our library. One lane of an Illumina GAIIx yields 20-30 million sequences, thus we reasoned that clones enriched to greater than 1 in 1 million would be identified with 20-30 copies and therefore would be identifiable above the statistical background. Our previous work indicated that CFMS enrichment was >1,000-fold per round,[5] thus enabling us to identify functional clones occurring at a frequency of ~1 in every 1 billion sequences in the naïve pool (Figure 1A). Prior yeast display work used much smaller libraries to isolate functional clones[11] supporting a complexity of ~109.
We also needed to increase the copy number in our library to detect functional sequences versus background clones by statistics alone. In conventional mRNA display selections with low copy numbers, 1-2 copies of each functional clone are retained by the first enrichment (Figure S2A). However, even if only ~0.01% of the nonfunctional clones are retained due to our CFMS protocol,[5] the individual copy number of these clones (1 copy for each random, sequence-independent carryover) would be indistinguishable from the functional clones. Therefore, here we aimed to achieve a copy number of 1,000 for each sequence, allowing adequate separation of functional from non-functional clones (Figure S2B).
In any random in vitro selection, it is essential that synthesis of the library be as unbiased as possible. This is especially true for our goal of identifying the best binders after one round of selection where the initial frequency cannot be determined for each of the ~1 billion unique library members. After the affinity enrichment step, the observed frequencies of selected clones are dependent on both initial frequency and binding efficiency. Therefore, large skews in clone representation in the initial pool will hinder identification of the clones with the best binding efficiency. Thus, we sequenced a fraction of both the DNA library (step 1, Figure 1B) and purified fusions (step 6, Figure 1B). Using statistical models, we determined the copy number distribution of each library and determined, for the first time, that mRNA display achieves desired target complexities (here ~0.9 × 109 unique purified fusions) with highly uniform clone frequencies (see the Supporting Information, Figures S3-4).
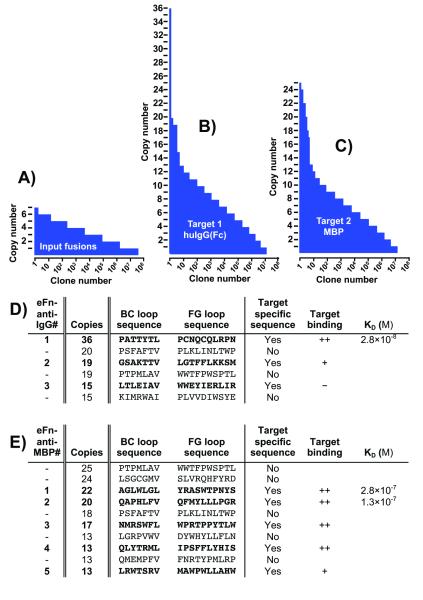
To validate our method, we chose two ubiquitous, highly utilized targets: maltose binding protein (MBP) and human IgG(Fc). MBP is commonly used for purification and solubilization of fusion proteins.[12] Human IgG ligands may be useful for detecting antigen-reactive antibodies in vitro or for purification of IgGs from complex mixtures. Using our CFMS method described previously,[5] we performed affinity enrichment with both targets in parallel (see the Supporting Information). Next, samples were amplified by PCR below saturation prior to performing sequencing on an Illumina GAIIx instrument. Following sequencing, we ranked each unique clone by copy number. Figure 2A demonstrates the clone frequency landscape for the input library (purified fusions prior to selection) and the two semi-enriched pools that had undergone one round of affinity selection versus IgG(Fc) (Figure 2B) or MBP (Figure 2C). The two selected pools contained significantly over-represented clones relative to the frequency distribution of the random carryover as evident by the skewing of the copy number landscape at the highest end. An important feature of our strategy is clones that have high copy numbers but appear in pools selected against different targets are likely to be matrix-binding sequences and can therefore be eliminated from future study (Figure 2D,E).
Figure 2.
Clone frequency distribution landscape. (A-C) Sequences are ranked by copy number, illustrating the landscape of variants in the input and selected pools. (D,E) The top sequences are listed for each pool. Target-specific sequences (denoted in bold) indicate clones that were not observed in the other selection pool. Reconstructed clones were analyzed for target binding via western blot analysis. ++ indicates a strong binding, + indicates faint binding and − indicates no binding within the limit of detection. Affinities are from figures S6 and S7.
The clones with the highest copy number that were unique to each target were rapidly validated to bind target. To do this, we developed a strategy for reconstruction, expression and testing entirely in vitro, without the need for cloning into bacteria. We used synthetic oligonucleotides covering the loop regions to amplify and extend each clone from the selected pools. We then used the crude PCR reactions as template for coupled in vitro transcription and translation. Next, the pull- down efficiencies of the expressed binders on beads with or without target were assayed by western blot (Figure S5 and Figure 2E,D). The top two unique, putative IgG(Fc) binders (eFn-anti-IgG1 and 2) and the top five MBP binders (eFn-anti-MBP1-5) were pulled-down specifically by their respective targets with no detectable binding to the beads alone. The frequencies of the best binders validate the predicted outcome that the best IgG(Fc) binder, eFn-anti-IgG1, was estimated to be enriched approximately 1500-fold. Furthermore, even lower frequency binders (e.g., eFn-anti-MBP4-5) displayed greater enrichment than previously achieved using our batch-based affinity enrichment method (see the Supporting Information).[5]
We first attempted to determine the eFn-anti-IgG1 binding constant using surface plasmon resonance (SPR), but were not able to regenerate the chip surface in a timely manner without denaturing the immobilized IgG. As an alternative, we utilized an equilibrium-binding assay to measure pull-down of biotinylated, monomeric eFn-anti-IgG1 onto polystyrene multiwell plates coated with limiting amounts of IgG at room temperature. The amount of eFn-anti-IgG1 bound to IgG coated plates was measured by subsequently adding excess SA-HRP, followed by washing and detection of luminescence (Figure 2D and S6A). Using nonlinear regression, the binding isotherm was fit to a KD of 28 ± 6 nM. The Hill coefficient was 1.005, indicating there was no cooperativity arising from surface or avidity effects. Also, we demonstrated eFn-anti-IgG1 recognizes IgG at the consensus site demonstrated to bind numerous proteins and peptides[13] by competition with protein G (Figure S6B).
We used SPR to determine the binding kinetics and affinity of the top two MBP binders at 25 °C (Figure 2E and S7). MBP binding to eFn-anti-MBP1 immobilized on a SA sensor chip demonstrated a kon = 1.69 × 105 M−1 s−1 and koff = 4.77 × 10−2 s−1 resulting in a KD of 282 nM. MBP binding to eFn-anti-MBP2 demonstrated a kon = 1.01 × 104 M−1 s−1 and koff = 1.30 × 10−3 s−1 resulting in a KD of 129 nM. While eFn-anti-MBP1 displays lower affinity than eFn-anti-MBP2, both display excellent functionality in avidity-enhanced detection assays (see below).
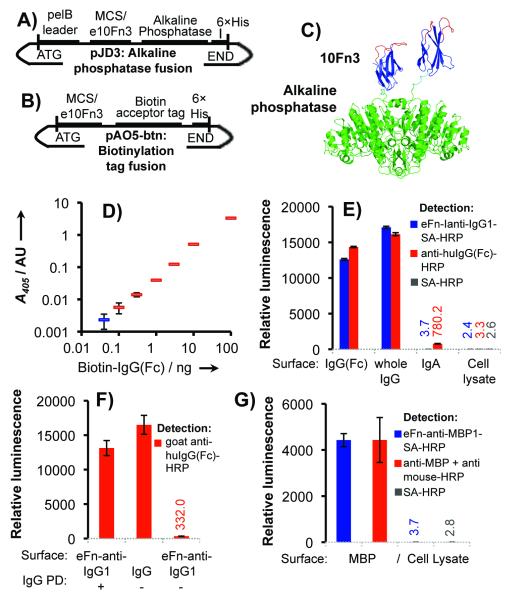
To maximize the utility of an affinity reagent, it is valuable to be able to introduce modifications that enable simple detection of targets in vitro or in vivo. Previously, we have demonstrated utility of mRNA display selected ligands similar to and beyond that of antibodies in inhibition or detection assays both intracellularly and in vitro.[8,9,14] Here, we demonstrate two methods for detection in vitro by linking two validated IgG ligands (eFn-anti-IgG1 and 2) and five validated MBP ligands (eFn-anti-MBP1-5) to alkaline phosphatase (AP) or horseradish peroxidase (HRP). To link our fibronectins with AP, we generated direct, genetic fusions as demonstrated previously,[15,16] using a new vector designed to simplify cloning (Figure 3A, Figure S8). Alternatively, for detection via HRP, we fused the C-terminus of our e10Fn3s with a short peptide that can be specifically biotinylated by the E. coli biotin ligase, BirA (Figure 3B, Figures S9-S10). This construct can then be detected using streptavidin-conjugated HRP (SA-HRP). Since AP forms a stable dimer (Figure 3C) and SA a tetramer, the multivalent display of fibronectins may enhance target detection through avidity.
Figure 3.
Validation of binders for use in enzyme-linked detection assays. (a) pJD3 vector for creating e10Fn3-alkaline phosphatase fusions and (b) pAO5-btn vector for generating fusions to a biotin acceptor peptide tag. (c) Structural model of 10Fn3-AP fusions. The 10Fn3 scaffold is blue, randomized loops are red, and alkaline phosphatase is green. 10Fn3 domains (PDBID: 1FNF and 1FNA) are joined to AP (PDBID: 1ALK) by a GSGGSG linker (cyan). (d) Mock sandwich antibody-free ELISA assay for IgG detection. (Blue is the no-IgG control). (e) Comparison of eFn-anti-IgG-SA-HRP to anti human IgG-HRP. (f) eFn-anti-IgG1 efficiently captures IgG (Pull-down, “PD”) for detection by polyclonal anti-huIgG-HRP (g) Comparison of eFn-anti-MBP1-SA-HRP to a commercially available monoclonal. Binding assays were performed in triplicate.
We first tested our enzyme-linked e10Fn3s for target detection using ELISA-like assays (Figure S11). eFn-anti-IgG1 displayed saturated detection with no background while eFn-anti-IgG2 was less efficient with significant background in a p-nitrophenyl phosphate (PNPP) reaction assay (Figure S11A). We subsequently used eFn-anti-IgG1-AP in a mock sandwich assay, similar to antigen-reactive IgG detection assays, using SA-coated polystyrene multiwell plates to pull down various concentrations of biotinylated IgG(Fc). eFn-anti-IgG1-AP was able to detect IgG(Fc) at a wide range of concentrations spanning three orders of magnitude using a single experimental condition (Figure 3D).
We then tested target detection by eFn-anti-IgG1 via SA-HRP. First, we compared the efficiency and specificity of eFn-anti-IgG1-SA-HRP relative to conventional polyclonal anti-human IgG(Fc) in luminescence-based ELISAs (Figure 3E). Detection was comparable to the commercially available reagent with both Fc and full antibody. The background from mammalian cell lysate was equivalent to either polyclonal anti-IgG(Fc) or SA-HRP alone, within error. The eFn-anti-IgG1-SA-HRP background binding to IgA was negligible, similar to the signal from cell lysate, and significantly lower than the commercially available antibody. Figure 3F demonstrates that eFn-anti-IgG1 captures IgG efficiently and is also highly specific, binding well to human IgG but not appreciably to goat IgG. We also demonstrate utility of eFn-anti-IgG1-SA-HRP in western blotting, as IgG in the low nanogram range is detectable after SDS PAGE and transfer to nitrocellulose (Figure S12A).
We similarly conjugated the MBP binders to SA-HRP for luminescence-based detection assays. eFn-anti-MBP1 displayed the highest detection efficiency, (Figure S11B) and exhibited efficiency of detection comparable to a commercially available anti-MBP monoclonal antibody, with background equal to SA-HRP alone, within error (Figure 3G). eFn-anti-MBP1-SA-HRP was similarly effective in antibody-free western blotting as demonstrated by detecting expression of an MBP-fusion protein (Figure S12B).
This study demonstrates approximately 109 sequences were sufficient for selecting an IgG binder with an affinity of 28 nM, roughly equivalent to long established existing reagents (protein A and protein G) targeting this domain.[17] Also, this affinity is typical of selections using high complexity 10Fn3-based libraries.[5,8,9] This indicates that biases in the multistep selection process may preclude selecting higher affinity binders that theoretically exist in higher complexity libraries. Our CFMS platform requires only minimal target material (100 pMoles or 1-10 μg) and enables performing affinity enrichment in parallel. Without further improvements, ~12 single round selections could be analyzed per lane on an Illumina HiSeq instrument (via barcoding, see the Supporting Information). By scaling up the initial naïve pool synthesis and automating the affinity enrichment step, high throughput ligand generation may be attainable. A proteome-wide affinity reagent resource based on the highly validated 10Fn3 antibody mimetic scaffold[18] would greatly impact molecular biology research as well as human health by providing a wealth of diagnostics and potential therapeutics.
Experimental Section
Detailed methods are provided in the Supporting Information section.
Supplementary Material
Acknowledgments
[**] Funding for this work was provided by the NIH, 5R01AI085583-03 (R.W.R., H.T.S., R.S.). C.A.O. was supported by the NCI Cancer Education Grant, R25 CA 098010. J.A.T was supported by the Charlotte Geyer Foundation and the Morgridge Institute for Research.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Dr. C. Anders Olson, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, CA 90095 (USA)
Jeff Nie, Morgridge Institute for Research, Madison, WI 53707 (USA).
Jonathan Diep, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, CA 90095 (USA).
Dr. Ibrahim Al-Shyoukh, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, CA 90095 (USA)
Terry T. Takahashi, Department of Chemistry, Department of Chemical Engineering, University of Southern California, Los Angeles, CA 90089 (USA)
Dr. Laith Q. Al-Mawsawi, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, CA 90095 (USA)
Jennifer M. Bolin, Morgridge Institute for Research, Madison, WI 53707 (USA)
Angela L. Elwell, Morgridge Institute for Research, Madison, WI 53707 (USA)
Scott Swanson, Morgridge Institute for Research, Madison, WI 53707 (USA).
Dr. Ron Stewart, Morgridge Institute for Research, Madison, WI 53707 (USA)
James A. Thomson, Morgridge Institute for Research, Madison, WI 53707 (USA); Department of Cellular & Regenerative Biology, University of Wisconsin, Madison, WI 53706 (USA); Department of Molecular, Cellular & Developmental Biology, University of California, Santa Barbara, CA 93106
H. Tom Soh, Departments of Materials and Mechanical Engineering, University of California, Santa Barbara, CA 93106 (USA)
Richard W. Roberts, Department of Chemistry, Department of Chemical Engineering, University of Southern California, Los Angeles, CA 90089 (USA)
Ren Sun, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, CA 90095 (USA).
References
- [1].Stoevesandt O, Taussig MJ. Proteomics. 2007;7:2738. doi: 10.1002/pmic.200700155. [DOI] [PubMed] [Google Scholar]
- [2].Kohler G, Milstein C. Nature. 1975;256:495. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- [3].Koide A, Bailey CW, Huang X, Koide S. J. Mol. Biol. 1998;284:1141. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- [4].Roberts RW, Szostak JW. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12297. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Olson CA, Adams JD, Takahashi TT, Qi H, Howell SM, Wu TT, Roberts RW, Sun R, Soh HT. Angew. Chem. Int. Ed. Engl. 2011;50:8295. doi: 10.1002/anie.201101149. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2011;123:8445. [Google Scholar]
- [6].Bentley DR, et al. Nature. 2008;456:53. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ravn U, Gueneau F, Baerlocher L, Osteras M, Desmurs M, Malinge P, Magistrelli G, Farinelli L, Kosco-Vilbois MH, Fischer N. Nucleic Acids Res. 2010;38:e193. doi: 10.1093/nar/gkq789. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, Ellington AD, Georgiou G. Nat. Biotechnol. 2010;28:965. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]; Cho M, Xiao Y, Nie J, Stewart R, Csordas AT, Oh SS, Thomson JA, Soh HT. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15373. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kupakuwana GV, Crill JE, 2nd, McPike MP, Borer PN. PLoS One. 2011;6:e19395. doi: 10.1371/journal.pone.0019395. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bayrac AT, Sefah K, Parekh P, Bayrac C, Gulbakan B, Oktem HA, Tan W. ACS Chem. Neurosci. 2011;2:175. doi: 10.1021/cn100114k. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hoon S, Zhou B, Janda KD, Brenner S, Scolnick J. Biotechniques. 2011;51:413. doi: 10.2144/000113786. [DOI] [PubMed] [Google Scholar]
- [8].Olson CA, Liao H-I, Sun R, Roberts RW. ACS Chem. Biol. 2008;3:480. doi: 10.1021/cb800069c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Liao HI, Olson CA, Hwang S, Deng H, Wong E, Baric RS, Roberts RW, Sun R. J. Biol. Chem. 2009;284:17512. doi: 10.1074/jbc.M901547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Takahashi TT, Roberts RW. Methods Mol. Biol. 2009;535:293. doi: 10.1007/978-1-59745-557-2_17. [DOI] [PubMed] [Google Scholar]
- [11].Lipovsek D, Lippow SM, Hackel BJ, Gregson MW, Cheng P, Kapila A, Wittrup KD. J. Mol. Biol. 2007;368:1024. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- [12].Kapust RB, Waugh DS. Protein Sci. 1999;8:1668. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].DeLano WL, Ultsch MH, de Vos AM, Wells JA. Science. 2000;287:1279. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- [14].Ishikawa FN, Chang HK, Curreli M, Liao HI, Olson CA, Chen PC, Zhang R, Roberts RW, Sun R, Cote RJ, Thompson ME, Zhou C. ACS Nano. 2009;3:1219. doi: 10.1021/nn900086c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Suzuki C, Ueda H, Suzuki E, Nagamune T. J. Biochem. 1997;122:322. doi: 10.1093/oxfordjournals.jbchem.a021756. [DOI] [PubMed] [Google Scholar]
- [16].Han Z, Karatan E, Scholle MD, McCafferty J, Kay BK. Comb. Chem. High Throughput Screen. 2004;7:55. doi: 10.2174/138620704772884823. [DOI] [PubMed] [Google Scholar]
- [17].Saha K, Bender F, Gizeli E. Anal. Chem. 2003;75:835. doi: 10.1021/ac0204911. [DOI] [PubMed] [Google Scholar]
- [18].Bloom L, Calabro V. Drug Discov. Today. 2009;14:949. doi: 10.1016/j.drudis.2009.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.