Abstract
RNA transcripts without obvious coding potential are widespread in many creatures, including the fruit fly, Drosophila melanogaster. Several noncoding RNAs have been identified within the Drosophila bithorax complex. These first appear in blastoderm stage embryos, and their expression patterns indicate that they are transcribed only from active domains of the bithorax complex. It has been suggested that these noncoding RNAs have a role in establishing active domains, perhaps by setting the state of Polycomb Response Elements A comprehensive survey across the proximal half of the bithorax complex has now revealed nine distinct noncoding RNA transcripts, including four within the Ultrabithorax transcription unit. At the blastoderm stage, the noncoding transcripts collectively span ∼75% of the 135 kb surveyed. Recombination-mediated cassette exchange was used to invert the promoter of one of the noncoding RNAs, a 23-kb transcript from the bxd domain of the bithorax complex. The resulting animals fail to make the normal bxd noncoding RNA and show no transcription across the bxd Polycomb Response Element in early embryos. The mutant flies look normal; the regulation of the bxd domain appears unaffected. Thus, the bxd noncoding RNA has no apparent function.
Keywords: noncoding RNA (ncRNA), Polycomb response element (PRE), bithorax complex, Ultrabithorax, bithoraxoid
IN the genomes of higher animals, only a small fraction of the DNA codes for potential proteins, but RNA expression surveys reveal that nearly all of the nonrepetitive DNA is transcribed (Ponting et al. 2009). Several possible functions have been suggested for the noncoding RNAs (ncRNAs). Some could code for short peptides that would be overlooked in most scans for coding regions (Kondo et al. 2010; Ingolia et al. 2011). Some RNAs could have structural or enzymatic function (Penny et al. 1996; Meller and Rattner 2002). The act of transcription could affect promoters or enhancers within the transcription unit, perhaps by altering nucleosome subunits or modifications along the template (Schwartz and Ahmad 2005; Edmunds et al. 2008). The default possibility is that these ncRNAs have no function, that they represent transcriptional “noise”.
In view of the several possible roles for ncRNAs, it is difficult to determine their functions, and doubly difficult to show that such an RNA has no function. If the RNA structure is important, it may be difficult to disturb with point mutations. Deleting some or all of the transcription unit risks removing enhancers or other regulatory sequences for nearby genes. Inserting a premature termination signal is more attractive, but designing a transcription terminator that approaches 100% efficiency is challenging. RNA interference (RNAi) against the target ncRNA has multiple complications. The knockdown might not be complete, there may be off-target effects, and RNAi should not block transcription-coupled modifications of the template. A further issue is that RNAi can induce transcriptional gene silencing, sometimes involving DNA methylation or histone modification (Castel and Martienssen 2013).
The Drosophila melanogaster bithorax complex (BX-C) presents a striking example of the ncRNA conundrum. The gene cluster spans ∼310 kb, but it codes for only three homeobox transcription factors [Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B)] and one sugar transporter (Glut3). The exons of the mRNAs for these four proteins (including alternative splicing forms) add up to ∼16.5 kb, or only ∼5% of the DNA sequence in the cluster (Martin et al. 1995). However, the majority of the remaining DNA is transcribed.
The BX-C can be divided into nine DNA domains, each defined by mutations that affect a different segment [or parasegment (PS)] of the fly, from the third thoracic (PS5) through the eighth abdominal (PS13). The domains are named for the mutant classes found in each region: bithorax (bx), bithoraxoid (bxd), and infraabdominal-2 (iab-2) through infraabdominal-8 (iab-8). These domains are aligned along the chromosome in the order of the segments that they affect (Lewis 1978). ncRNAs have been discovered in several of these DNA domains, primarily by RNA in situ hybridizations to embryos. Lipshitz et al. (1987) first discovered such ncRNAs, by sequencing cDNA clones homologous to the bxd (PS6) regulatory region. These RNAs are made early in embryonic development and appear in blastoderm embryos in PS6 and more posterior parasegments (Akam et al. 1985; Rank et al. 2002; Petruk et al. 2006). Cumberledge et al. (1990) found cDNA clones representing a noncoding RNA expressed in PS8 and more posterior parasegments. Additional noncoding RNAs from the iab-3 through iab-8 domains between the abd-A and Abd-B genes have been detected by RNA in situ hybridization (Sánchez-Herrero and Akam 1989; Bae et al. 2002; Rank et al. 2002), although most of these have not yet been defined by cDNA clones. Most of these ncRNAs have two features in common: they first appear in blastoderm embryos, at the time when the BX-C domains receive their segmental addresses from the gap and pair rule genes (Maeda and Karch 2009), and the ncRNAs coming from a particular DNA domain appear in the body segments regulated by that domain.
The times and positions of their appearance suggest that these noncoding RNAs could be involved in the establishment of the BX-C domains. RNA transcription across Polycomb Response Elements (PREs) has been shown to antagonize repression by Polycomb group genes (Bender and Fitzgerald 2002; Hogga and Karch 2002; Rank et al. 2002; Schmitt et al. 2005), but this function has not yet been demonstrated for native transcripts in the BX-C. A few more subtle functions for BX-C ncRNAs have been discovered. Two transcripts are known to encode microRNAs from the iab-3 region (Aravin et al. 2003; Bender 2008; Stark et al. 2008; Tyler et al. 2008), but no other microRNAs or likely precursor hairpins have been identified in the BX-C. The iab-8 ncRNA appears to repress abd-A in the eighth abdominal segment by transcriptional suppression (Gummalla et al. 2012), and readthrough transcripts of the bxd ncRNA crossing the Ubx promoter appear to repress early transcription from that promoter (Petruk et al. 2006).
The survey for embryonic noncoding RNAs of Bae et al. (2002) was comprehensive for the iab-2 through iab-8 regulatory regions, which regulate abd-A and Abd-B expression. No comparable screen for noncoding RNAs has been reported for the bx (PS5) and bxd (PS6) domains in the proximal half of the BX-C, responsible for the regulation of Ubx. In particular, the introns of the 75-kb Ubx transcription unit have been ignored, in part because the Ubx transcripts themselves might mask the presence of less abundant noncoding RNAs. We have surveyed the proximal half of the BX-C for noncoding RNAs, to complete the census, and we have ablated the bxd ncRNA to test for its function.
Materials and Methods
Sequence coordinates
The D. melanogaster sequence coordinates follow the SEQ89E numbering of Martin et al. (1995) (GenBank accession no. U31961). Base 1 of SEQ89E corresponds to base 12,809,162 in Release 5.37 of the D. melanogaster genome; SEQ89E numbering proceeds from distal (Abd-B) to proximal (Ubx); the assembled genome proceeds proximal to distal. The genome sequence includes a 6134-bp insertion of the Diver retroposon in the bxd domain, indicated by the small triangle below the DNA line in Figure 1 and Figure 3. The Diver insertion is associated with a 4-bp target duplication of bases 220,924–220,927 in SEQ89E.
Figure 1.
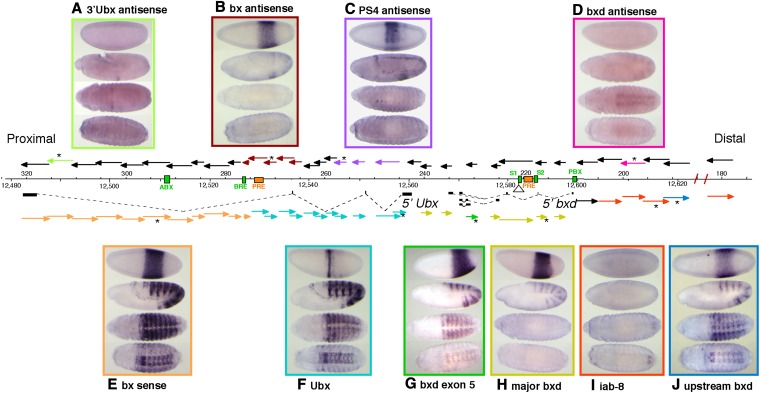
(A–J) Survey of RNA transcripts across the proximal half of the bithorax complex. The arrows above and below the DNA map show the 5′–3′ orientation of strand-specific probes; probes below the map detect distal to proximal transcripts (sense orientation for Ubx mRNA), and those above the map detect proximal to distal (antisense) transcripts. The colored arrows indicate probes that detect RNA products in embryos, and the pictures boxed in corresponding colors show the expression patterns of the different RNAs. Probes marked by an asterisk were the ones used for the displayed in situ results. Several embryonic enhancers are shown in green on the sequence coordinate line, and two mapped PREs are shown in orange. Sequence coordinates (in kilobases) are according to Martin et al. (1995) (above DNA line) and according to Release 5 of the D. melanogaster genome for chromosome 3R (below the DNA line).
Figure 3.
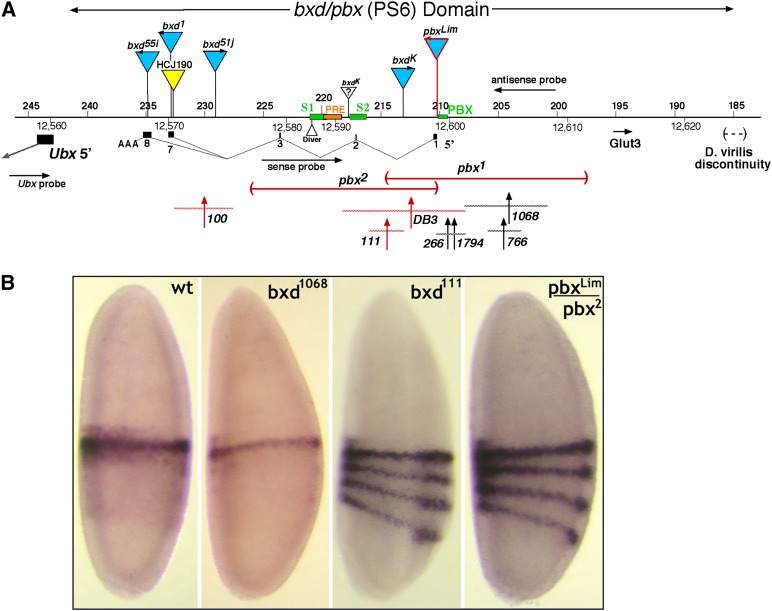
(A and B) Map of the bxd mutant lesions and their effects on early Ubx transcription. (A) Above the DNA line are shown gypsy insertions (blue) and a P-element insertion (yellow). Below the line are diagramed the two large pbx deletions and seven bxd rearrangement breakpoints (vertical arrows with horizontal dashed lines to indicate the limits of uncertainty). The exons of the major bxd RNA (as numbered by Lipshitz et al. 1987) are also shown. The lesions illustrated in red alter the early Ubx RNA pattern. (B) In situs were performed on embryos at 0–3 hr of embryogenesis, using an RNA probe that targets the first exon of Ubx. Early blastoderm embryos from wild type show a single stripe of expression in PS6. Mutations that disrupt the bxd RNA show three additional more posterior stripes, as reported by Petruk et al. (2006).
Whole-mount in situ hybridization
Single-stranded RNA probes were made from subclones of genomic DNA from bacterial phage libraries of the BX-C or from DNA amplified by PCR from Oregon R adults. PCR products were cloned using the Strataclone PCR Cloning Kit (Stratagene, La Jolla, CA). Sense (+) and antisense (−) digoxigenin-labeled RNA probes were made from each clone, using the DIG Northern Starter Kit (Roche). Probes were numbered according to their approximate map coordinates. Probes that detect sense and antisense transcripts over the same region, relative to the orientation of Ubx, were synthesized from the same clone with either T7 or T3 RNA polymerase. The coordinates of each probe pair (+ /−) were as follows: 180, 177,266–182,052; 189, 186,806–191,731; 193, 191,041–195,752; 198, 195,512–200,018; 203, 200,218–205,408; 207, 205,016–208,815; 211, 210,713–212,970; 217, 218,241–215,924; 218, 217,928–219,936; 222, 218,321–225,128; 226, 225,112–227,443; 228, 225,279–230,964; 229, 228,511–230,964; 235, 234,579–236,731; 239, 238,017–240,570; 242, 240,503–243,096; 245, 243,227–246,541; 247, 244,679–249,821; 253, 251,883–254,182; 256, 255,422–257,835; 258, 257,807–259,376; 260, 259,318–261,817; 263, 261,722–264,408; 265, 264,344–266,940; 268, 266,864–269,662; 270, 269,592–271,988; 273, 271,931–275,179; 275, 275,106–275,987; 278, 276,548–279,041; 281, 278,909–284,059; 286, 284,966–287,268; 289, 287,228–291,726; 294, 291,697–296,398; 298, 296,276–300,964; 303, 300,771–305,927; 308, 305,796–310,687; 313, 310,476–315,474; 318, 315,113–320,719.
In situ hybridizations were carried out on 0- to 24-hr Oregon R and (Dp(3;2)P10)2 embryos as described in Nagaso et al. (2001), and stained embryos were photographed at stages 5, 9, 12, and 15. Two-color in situs in Figure 2 were achieved using a biotin-labeled ftz probe that was detected with streptavidin-HRP.
Figure 2.
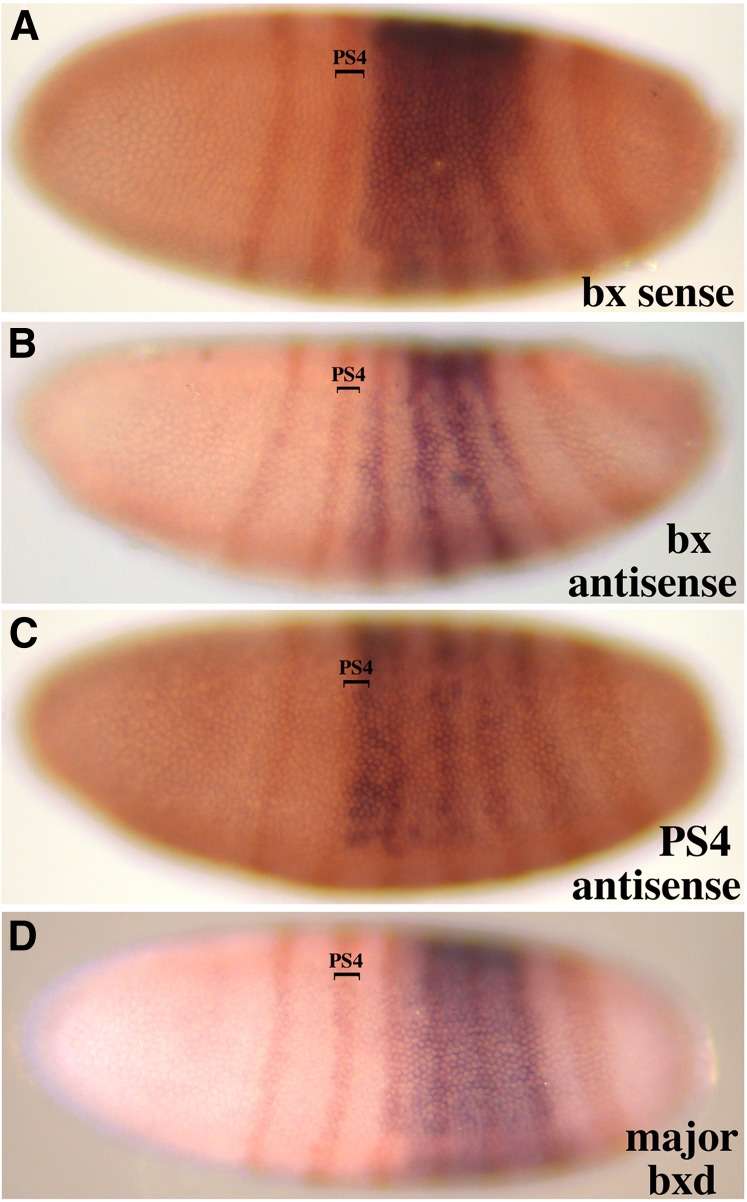
(A–D) Parasegmental limits of early noncoding RNAs. Lateral views of blastoderm embryos are shown with anterior to the left and dorsal toward the top. Two-color RNA in situ hybridizations combined a BX-C probe with one for the FTZ/LACZ fusion mRNA. The “bx sense” and “bx antisense” RNAs (Figure 1, E and B, respectively) show a PS5 anterior limit. The “PS4 antisense” RNA (Figure 1C) begins in PS4, and the “major bxd” RNA (Figure 1H) starts in PS6.
Mutant lesions
The sites of gypsy mobile element insertions were mapped by inverse PCR. The positions of target site duplications for these gypsy inserts were as follows: bxd55i, 234,906 (TTTG); bxd1, 232,784 (TGTA) (in exon 7); bxd9, 232,247 (TATA); pbxLim, 210,241 (TAAA); and bxdK, 213,109 (TGCA). The bxdK chromosome also carries an unidentified ∼3.4-kb insertion at ∼218,000 (Bender et al. 1985). The pbx2 deletion fusion fragment was cloned by PCR and sequenced; the deletion removes bases 210,404–226,179.
The bxd1794 breakpoint was mapped by genomic Southern blots. The positions of the other rearrangement breakpoints illustrated in Figure 3 were reported in Bender et al. (1985) and Bender and Lucas (2013).
Construction of In(bxd-pro)
Recombination between the two P elements (Figure 4A) used flipase-mediated recombination between FRT sites within each P element. Males of the genotype y, w, [hs-flp]122; Sb, HC154A/HC174B, DlN were heat-shocked as larvae for 1 hr at 37° and mated as adults to cn; ry females. The desired recombinants were recovered as non-Sb, non-Dl, ry− offspring, and the junctions between the genomic edges and the P element were confirmed by PCR.
Figure 4.
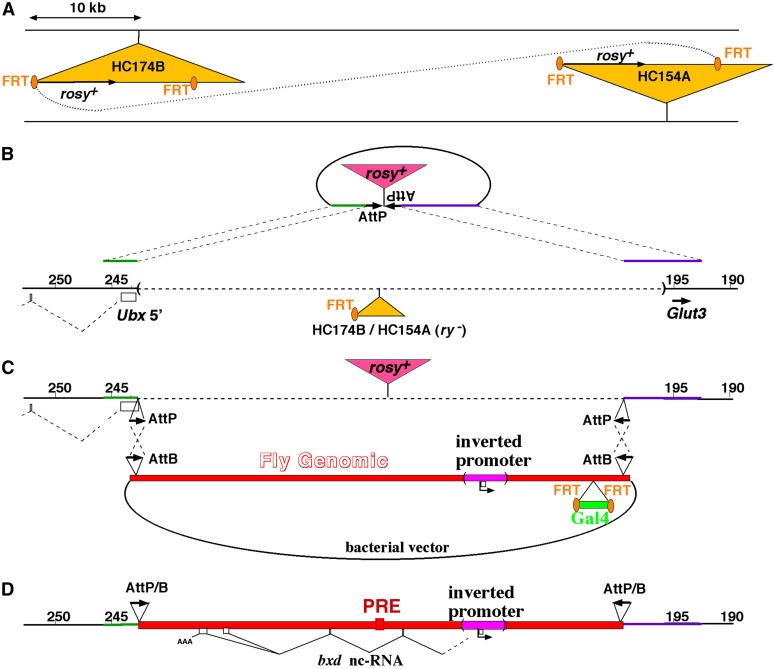
(A–D) Diagram of the recombination-mediated cassette exchange (RMCE) with a BAC covering 45 kb of the bxd domain. (A) The deletion of a 48-kb interval on the chromosome was mediated by flp-induced recombination between two P elements (Bender and Hudson 2000). (B) Two opposing attP sites, separated by a copy of the rosy gene, were inserted by gene conversion, using the remaining P element. This conversion also reduced the deletion interval to match the insert of the BAC [pMB1154 (O’Connor et al. 1989)]. (C) Using recombineering, pMB1154 was modified to carry (i) two opposing attB sites at either end of the domain, (ii) the inversion of the segment containing the bxd RNA promoter and the PBX enhancer, and (iii) the addition of the Gal4 marker, for recovery in flies. After injection of the BAC in a RMCE background (Bateman and Wu 2008), several independent integration lines were recovered. A proper double recombinant was identified, which replaced the 45-kb stretch with the altered bxd domain. (D) The Gal4 marker was then removed with flipase, and embryos homozygous for the integrated BAC [In(bxd-pro)] chromosome were analyzed by RNA in situ hybridization and RT-PCR.
The donor plasmid for gene conversion (Figure 4B) was built in pCR-Blunt (Invitrogen, Carlsbad, CA) with the following sequences: a 3.1-kb genomic fragment from the 5′ Ubx region (246,498–243,392), an AscI site, a 54-bp synthetic AttP site, a 7294-bp genomic HindIII fragment carrying rosy+, an inverted AttP site, an AscI site, and a 6-kb genomic fragment from the distal bxd region (198,832–192,817). This was injected, along with a plasmid encoding the P-element transposase, into preblastoderm eggs of an HC174B-HC154A recombinant/MKRS stock. G0 adults were mated to cn; ry flies, and progeny were scored for integration of the ry+ marker. Junctions of the conversion chromosome were confirmed by PCR.
The Bacterial Artificial Chromosome (BAC) used for replacement of the bxd region (Figure 4C) was pMB1154, from Mike O’Connor (U. Minnesota, Minneapolis, MN); it carries ∼45 kb of BX-C DNA (243,999–198,833). This BAC was modified in four places by recombineering. All the recombineering manipulations were done as described in Warming et al. (2005), using a GalK-based positive/negative selection system in Escherichia coli SW102. For each recombineering site, a donor plasmid was constructed in pCR-Blunt, containing two adjacent DNA segments from the BAC, each 350–700 bp long, separated by an AscI site. The AscI site was used to insert either a copy of the bacterial GalK gene [cloned from the pGalK template (Warming et al. 2005)] or the desired sequence for the final construction.
For the 5′ Ubx site, the AscI site was inserted between bases 243,394 and 243,393. A 50-bp synthetic AttB sequence with an additional 6-bp BglII site was inserted into the AscI site, and its orientation was determined with a BglII restriction digest.
To insert the inverted bxd ncRNA promoter, the initial donor plasmid contained two BX-C genomic fragments spaced ∼1.6 kb apart, joined at an artificial AscI site. The AscI site was flanked by bases 210,614 and 208, 980. The fragment to be inverted (210,613–208,981) was inserted into the AscI site, and its orientation was determined by PCR.
For the Gal4 marker, the AscI site was inserted between bases 199,802 and 199,801, along with a NotI site. The AscI site was used for the introduction of GalK, and the NotI site was used to insert an FRT/Gal4/FRT cassette. The Gal4 coding sequence, flanked by the Drosophila synthetic core promoter (Pfeiffer et al. 2008) and the hsp70 poly(A) addition site, was cloned by PCR from the pBPGUw plasmid, a gift of Barret Pfeiffer (HHMI Janelia Farm, Ashburn, VA). It was recovered as a NheI fragment and inserted between two 34-bp FRT sites, flanked by NotI sites. The integrity of Gal4 was verified by sequencing.
The distal AttB site was inserted at an AscI site between the distal Drosophila base of the BAC (198,833) and the adjacent plasmid sequence. Its orientation was selected to be inverted relative to the AttB site near the Ubx start.
The final BAC with the inverted bxd RNA promoter was injected into preblastoderm eggs from a cross of w, [nanos-phiC31 integrase]; ry, Fab7/MKRS females with BX-C Dp(3;1)68; ΔHC174B-HC154A > AttP-ry+-AttP homozygous males. The G0 adults were mated to w; P[UAS-GFP]; pbx, Fab7/MKRS, and the progeny were screened as young larvae for GFP fluorescence. Stocks were established for the chromosomes carrying a BAC integration, and these were tested by PCR to determine the orientations of the insertions, and whether the integrations used one or both AttP sites. Double integrations were also recognized by loss of the ry+ marker from the target chromosome.
One double-integration chromosome in the desired orientation was further treated with flipase (as above) to remove the Gal4 marker. A stock homozygous for the final chromosome (Figure 4D) was used for RNA in situ hybridizations (Figure 5 and Figure 6) and examinations of larval and adult cuticles.
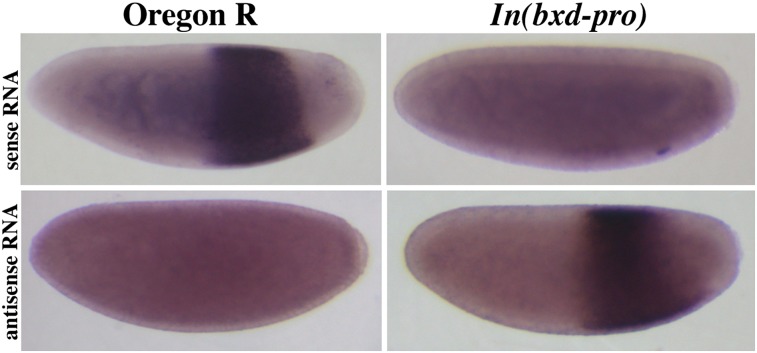
Figure 5.
RNA in situ hybridization of embryos with probes from the bxd region. Embryos of 0–3 hr of embryogenesis were collected from wild-type Oregon R flies and the In(bxd-pro) line. Hybridizations used strand-specific probes to either sense bxd (probe 222) or antisense bxd (probe 203), as illustrated in Figure 3. The normal bxd RNA is gone when the promoter is inverted, and a new proximal-to-distal RNA is detected instead.
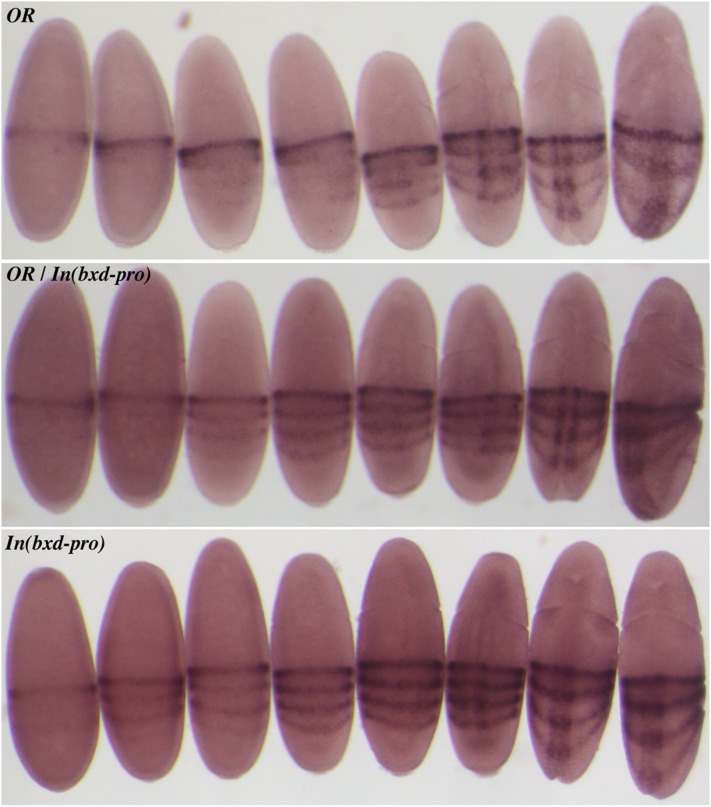
Figure 6.
RNA in situ hybridization of embryos with a probe to the first exon of Ubx (illustrated in Figure 3). The top panel shows embryos of the Oregon R wild type, the bottom panel shows embryos homozygous for In(bxd-pro), and the middle panel shows embryos from a cross between these two strains. The first four embryos in each panel cannot be staged except by staining intensity. The fifth embryo in each panel shows the initial indentation of the cephalic furrow, the first sign of gastrulation. This and subsequent stages of gastrulation can be accurately compared by morphology. The posterior three stripes of Ubx RNA appear prior to gastrulation in the In(bxd-pro) embryos, but in the same pattern as the later expression in wild type. Heterozygous embryos look like In(bxd-pro) homozygotes, with reduced intensity in the posterior three stripes.
Results
RNA in situ hybridizations to embryos were used to discover transcripts from the BX-C. The method is particularly sensitive and reveals the tissue and segment specificity of expression. Strand-specific probes were used to distinguish transcripts from either strand; the probes were designed to give nearly continuous coverage across 143 kb. This DNA segment spans the Ubx transcription unit and its bxd cis-regulatory region (Figure 1). The Ubx domain is bounded on the proximal side by the adjacent modular serine protease gene [at map position 322 kb in Figure 1 (Martin et al. 1995)]. On the distal side, the bxd domain border is defined by loss of sequence homology with D. virilis (at 185 kb in Figure 1) and by the Fub border deletion (Von Allmen et al. 1996; Bender and Lucas 2013). We routinely used embryos from a Drosophila stock containing chromosome II with two copies of Dp(3;2)P10 (Smolik-Utlaut 1990), which covers the entire Ubx domain. The resulting embryos had up to six copies of the left half of the BX-C, which increased the signal strength of the in situ hybridizations. However, most of the ncRNAs reported here were also seen in wild-type (Oregon R) embryos. The positions of the ncRNAs in blastoderm embryos were defined in relation to seven stripes of fushi tarazu (ftz) expression, defined by lacZ RNA expression from a FTZ/LACZ fusion construct on a balancer chromosome (Heemskerk and Dinardo 1994).
Seven new RNA transcripts were detected, in addition to the Ubx mRNA and the bxd and iab-8 ncRNAs. Four of the novel ncRNAs were seen with multiple adjacent probes, all giving the same expression pattern. The hybridization intensities of adjacent probes were comparable; there was no clear indication of splicing that might produce stable exons and unstable introns.
Ubx sense-strand transcripts
The expression patterns for the protein-coding Ubx mRNA have been described (Akam and Martinez-Arias 1985; Casares et al. 1997; Petruk et al. 2006) (Figure 1F). Probes near the 5′ end of the Ubx transcription unit first reveal a single circumferential stripe of Ubx RNA at the cellular blastoderm stage (Figure 1F, top embryo). This stripe marks PS6. Slightly older embryos show a more anterior partial stripe on the dorsal side, and yet older embryos (still blastoderm) show three more posterior circumferential stripes. During germ-band elongation, these three posterior stripes intensify and split, eventually resolving into one stripe for each segment, from the second through the seventh abdominal segments (A2–A7) (Figure 1F, second embryo). From the extended germ-band stage onward, the RNA expression pattern closely resembles the UBX protein pattern.
The “bx sense” ncRNA can be distinguished from the Ubx mRNA because it appears at cellular blastoderm as a broad and uniform band, rather than a thin stripe (Figure 1E, top embryo). It is detected by probes covering ∼45 kb through the 3′ half of the Ubx transcription unit, and it appears to initiate near the well-characterized BRE enhancer (Qian et al. 1993). This broad band has an anterior edge between the second and third ftz stripes (therefore, at PS5), and its posterior edge coincides with the posterior edge of the sixth ftz stripe (PS12) (Figure 2A). The staining is less intense near the ventral midline (in the cells of the future mesoderm) and is especially weak or absent in midline cells of PS5 and PS11–12. At the start of gastrulation and germ-band extension, the staining intensifies in a thin band at about PS6, presumably reflecting readthrough from the Ubx promoter. From early germ-band extension onward, the pattern closely resembles that of Ubx (compare Figure 1, E and F). It is not clear whether the noncoding RNA disappears or is merely hidden by the strong staining from the Ubx transcript.
Ubx antisense transcripts
We detected three distinct transcripts coming from the antisense strand within the Ubx transcription unit. The “3′ Ubx antisense” ncRNA was detected by only a single probe from near the 3′ end of the Ubx transcription unit (Figure 1A). It first appeared in late germ-band-extended embryos (stage 11, according to Campos-Ortega and Hartenstein 1985) as a weak stripe across the middle of the third thoracic segment (Figure 1A, second embryo). As the germ band retracts, the staining is limited to a single ventro-lateral patch in each half of the third thoracic segment (Figure 1A, third embryo). No staining was detected beyond full germ-band retraction (stage 13). This staining was quite weak, just above the limit of our detection using the (Dp(3;2)P10)2 embryos.
The “bx antisense” ncRNA is detected by probes spanning ∼12 kb (Figure 1B). The most proximal probe that detects this RNA (a 0.9-kb fragment) is also the most distal probe that detected the bx sense ncRNA. Thus, the two RNAs appear to come from divergent but nearly coincident promoters, situated just distal to the BRE enhancer. The bx antisense ncRNA appears at blastoderm as a broad band, with the same pattern and extent (PS5–12) as the bx sense ncRNA (Figure 2B). The staining resolves into four stripes at the start of gastrulation; the fourth (most posterior) stripe is limited to the dorsal side of the embryo. The staining fades during germ-band extension. At the time that segmental grooves are first appearing, only a faint band of staining is visible across the third thoracic segment. Later stages are unstained.
The “PS4 antisense” ncRNA is defined by three probes spanning ∼15 kb, covering portions of the first two introns of the Ubx transcription unit (Figure 1). It first appears as a broad band, uniform along its anterior/posterior extent, but absent in the most ventral cells destined to become the mesoderm (Figure 1C). The anterior edge of this band coincides with the posterior edge of the second ftz stripe, so that it includes part of PS4 and extends through PS10 (Figure 2C). The broad band quickly evolves into four stripes, with the anterior stripe wider and more intense than the others (Figure 1C, top embryo, and Figure 2C). During germ-band extension, these stripes fade, with the ventral-most cells of the anterior stripe being the last to disappear (Figure 1C, second embryo). As this pattern fades, a new pattern emerges, an anterior–posterior line of cells along the ventral midline, through all the thoracic and abdominal segments. During germ-band retraction, the line resolves into one cluster of cells per segment, in the developing central nervous system, underneath each intersegmental furrow. The staining fades in older embryos, with the spots at the T1/T2 and T2/T3 segmental borders lasting the longest (Figure 1C, third embryo).
bxd sense transcripts
The cDNA clones homologous to the bxd domain (Lipshitz et al. 1987) defined several alternately spliced products, which together included eight exons, and spanned 23 kb. The RNA initiates just proximal to the “PBX” enhancer (Pirrotta et al. 1995). The embryonic expression patterns of these RNA species have been described (Akam et al. 1985; Rank et al. 2002; Petruk et al. 2006), although there has been no report of different patterns for alternative splicing forms. In our studies, probes spanning the bxd ncRNA transcription unit showed two different patterns. Expression of the “major bxd” ncRNA begins at the blastoderm stage with a band of staining, uniform along the anterior/posterior axis, but weaker in the ventral zone destined to become the mesoderm (Figure 1H). The anterior edge of this band coincides with the anterior edge of ftz stripe 3 (i.e., PS6), and the posterior edge lies slightly posterior to ftz stripe 6 (i.e., PS12) (Figure 2D). At the start of gastrulation, the broad band resolves into six stripes of equal width and intensity, and two weak stripes also appear (strongest near the ventral midline) anterior and posterior to the strong group of six. As the posterior end of the embryo curls to the dorsal side (stage 7), the anterior weak stripe strengthens, to give a seven-stripe pattern. By stage 10, the posterior stripe also intensifies, to give an eight-stripe pattern. When segmental grooves appear (stage 11), the stripes are disappearing, but they appear to coincide with the posterior edges of segments T3–A7. A faint stripe corresponding to posterior A8 also appears transiently. By the beginning of germ-band retraction, no more staining is detected. Two probes downstream of (proximal to) the 3′ exons also showed the patterns described above, although much weaker at every stage. These probes could be detecting readthrough transcripts past the poly(A) sites at the ends of exons 7 and 8 (exons numbered in Figure 3).
One probe (covering exon 5 of Lipshitz et al. 1987) shows the pattern described above in early embryos, but at stage 11, additional stripes appear in between each of the eight stripes of the canonical bxd pattern, and mesodermal expression is also apparent (Figure 1G, second embryo). We call this transcript the “bxd exon 5” ncRNA. From the germ-band retraction stage (stage 12) onward, the pattern superficially resembles that of the Ubx RNA, except that there is no staining in PS5. At stage 13, there is epidermal staining in PS6–12 plus mesodermal staining in PS7–12 and weak staining in PS13 (Figure 1G, third embryo). As the central nervous system (CNS) condenses, the staining weakens, with a full pattern in the PS6 CNS and narrower bands of staining in PS7–12 (Figure 1G, fourth embryo).
Another RNA, called the “upstream bxd” ncRNA, resembles the exon 5 ncRNA in pattern, although it is distinguishable. It was detected by a single probe ∼20 kb distal to the initiation site of the major bxd ncRNA. At blastoderm, there is again a broad band, weaker along the ventral midline; the staining does not extend as far posteriorly as that of the major bxd ncRNA (Figure 1J, first embryo). At gastrulation, the staining evolves into eight stripes, like those of the major bxd ncRNA. During germ-band elongation, mesodermal staining appears and the epidermal stripes intensify at their lateral edges (Figure 1J, second embryo). It becomes clear by about stage 10 that the most anterior staining is in the third thoracic segment (PS5), although the pattern here is less intense than that of PS6–12. At germ-band shortening, the pattern again resembles that of Ubx, with prominent staining in PS6–12 and weaker staining in PS5 (Figure 1J, third embryo). When the CNS condenses, the PS6 section of the nerve chord is broadly stained, with more sparse staining in PS7–12 (Figure 1J, fourth embryo). There is also faint CNS staining in the anterior third thoracic segment.
Several probes in the distal bxd region detect the “iab-8” ncRNA, but only with faint staining in late embryos in the CNS of the eighth abdominal segment (PS13) (Figure 1I, third and fourth embryos). This is apparently a continuation of the transcription unit that begins in the iab-8 domain of the bithorax complex, near the Abd-B coding region (Bae et al. 2002; Bender 2008; Gummalla et al. 2012). The detection of this RNA in the bxd region extends the length of the iab-8 ncRNA transcription unit to >140 kb. The progressive decline in the in situ signal strength with more proximal probes suggests that iab-8 transcripts terminate or abort at multiple positions along this transcription unit.
bxd antisense transcripts
The bxd region was also surveyed for antisense (proximal to distal) transcripts. Only one probe gave a detectable pattern of hybridization (Figure 1D), and this was detected only in the (Dp(3;2)P10)2 stock. The staining first appears at about stage 10 (extended germ band) as a series of eight weak internal stripes, limited to the midventral region (Figure 1D, second embryo). When segmental creases appear, the stripes are seen to be in the anterior edges of the first through eighth abdominal segments. At the germ-band retraction stage, the weak staining appears to mark the developing CNS in these segments (Figure 1D, third embryo). The staining is absent by the time of dorsal closure. The probe giving this pattern lies just upstream of a Glut3 coding region (Martin et al. 1995; Bender and Lucas 2013). The adjacent (more distal) probe covering the predicted coding region did not give any apparent hybridization. The in situ hybridizations of the Berkeley Drosophila Genome Project (http://insitu.fruitfly.org) also failed to detect any Glut3 embryonic expression.
Mutating the bxd RNA
The major bxd sense transcript (hereafter called the bxd RNA) is the best characterized of these ncRNAs, and was the focus of our efforts to assess function. Figure 3 shows a map of the bxd (PS6) regulatory region upstream of the Ubx transcription unit, showing exons of the bxd RNA. We discovered exon 2 of the bxd RNA (bases 217,105–217,229) as an optional exon in ∼50% of spliced transcripts; it was seen in cDNA products amplified with primers in exons 1 and 7. The reported cDNA starting within exon 2 (Lipshitz et al. 1987) likely lacked a complete 5′ end.
Petruk et al. (2006) reported that the pbx1 and pbx2 deletions lack the bxd RNA. The pbx1 deletion removes the first exon of the bxd RNA and the adjacent PBX enhancer; the pbx2 deletion removes half of exon 1, as well as exons 2 and 3 (Figure 3). They also showed that embryos homozygous for these deletions show three additional stripes of Ubx RNA expression in blastoderm embryos. We have confirmed that observation for pbx1 and pbx2 and extended it to several other mutations in the region. As shown in Figure 3, three rearrangement breakpoints downstream of exon 1 (bxd100, bxd111, and bxdDB3) give the four-stripe Ubx RNA pattern, while four breakpoints just upstream of exon 1 (bxd266, bxd1794, bxd766, and bxd1068) give the wild-type pattern. The pbxLim gypsy element, which is inserted 6 bp upstream of the exon 1 start site (Lipshitz et al. 1987), also gives a four-stripe Ubx pattern, but embryos with other gypsy insertions within the bxd transcription unit (bxdK, bxd51j, bxd55i, and bxd1) show wild-type Ubx RNA patterns.
Petruk et al. (2006) argued that the bxd RNA acts in cis to repress Ubx transcription in the positions of the posterior three stripes. They reported that RNAi against the bxd RNA did not change the Ubx pattern and that a transgene with the Ubx promoter but lacking the bxd RNA gives a four-stripe pattern at blastoderm. Ubx could be repressed by transcriptional interference from the bxd RNA if there is readthrough past the bxd RNA poly(A) addition sites, across the Ubx promoter. As noted above, we could detect weak expression of a presumptive readthrough transcript with probes between the Ubx promoter and the bxd poly(A) sites. All of the lesions lacking the bxd RNA (and giving the four-stripe Ubx pattern) have recessive loss-of-function phenotypes. It has not been clear whether these phenotypes might be due to lack of Ubx repression or loss of some other function of the bxd RNA.
We have attempted to alter or remove the bxd RNA with more directed mutations. We first used P-element-mediated gene conversion (utilizing the HCJ190 P element) to mutate the splice acceptor site of exon 7 of the bxd ncRNA (Figure 3), changing the terminal G (base 232,585) of the intron to T. Flies homozygous for this mutation were morphologically normal. However, cDNA recovered from early embryos homozygous for this mutation showed RNAs spliced to new downstream splice acceptor sites within exon 7.
We have no P elements near the bxd RNA transcription start site with which to initiate gene conversion. Therefore, we developed a strategy based on recombination-mediated cassette exchange (RMCE) (Bateman and Wu 2008) to allow modifications across most of the bxd domain (Figure 4). A BAC had been constructed (O’Connor et al. 1989), covering 45 kb of Drosophila genomic DNA, from within the UBX coding region, to a position 11 kb upstream of the bxd RNA start. This could be used as a substrate for modifications, using recombineering in bacteria. We chose not to delete the bxd RNA start, but rather to invert the promoter. It seemed likely that if we deleted the RNA start site, a new start site would emerge; this apparently happened in other experiments designed to delete the divergent bx ncRNA promoters (B. Pease and W. Bender, unpublished results). Deletion of the enhancer and the promoter would more likely ablate the bxd RNA, but any effects might be due to loss of the enhancer. However, an inversion of the bxd RNA promoter would produce only nonsense RNA, since transcription from this promoter appears to be strongly unidirectional (Figure 1).
The RMCE strategy involved several steps, illustrated in Figure 4. A target chromosome was prepared with a deletion of the comparable region, by using flipase-mediated recombination in trans between two P-element insertions (Figure 4A) (Bender and Hudson 2000). The recombinant chromosome was deleted for ∼47 kb, with an ∼5-kb P element remaining at the site of the deletion. The remaining P element was mobilized to facilitate gene conversion (Figure 4B). The donor plasmid for the conversion event carried genomic DNA spanning either end of the deletion interval, with two inverted phiC31 AttP sites, positioned to match the ends of the 45-kb BAC DNA. The left AttP site lies within the 5′-noncoding leader of the Ubx transcription unit, 525 bp downstream of the transcription start, in a region poorly conserved in related Drosophila species. A copy of the rosy gene was inserted between the AttP sites as a marker for conversion. The BAC was then modified by recombineering, using the GalK marker (Warming et al. 2005), at four distinct positions (Figure 4C). Inverted AttB sites were inserted at either end of the Drosophila DNA. The yeast Gal4 gene was inserted, with flanking FRT sites, as a marker for integration. Finally, we inverted a 1.6-kb segment spanning the PBX enhancer and the first exon of the bxd ncRNA. The BAC was integrated by a double-recombination event, and the Gal4 marker was subsequently removed with flipase. The final chromosome (Figure 4D), designated In(bxd-pro), proved to be homozygous viable and fertile.
We verified the loss of the bxd RNA by in situ hybridization to In(bxd-pro) homozygous embryos. A proximal probe to the sense strand (Figure 3) gives a strong bxd RNA signal in wild type, but gives no signal in the inversion homozygotes (Figure 5, top). Conversely, a distal probe to the antisense strand (Figure 3) gives no signal in wild type, but In(bxd-pro) embryos shows a novel RNA with the same timing and pattern as the bxd RNA (Figure 5, bottom). cDNA was prepared from In(bxd-pro) homozygotes and from wild type, using either oligo(dT) or an exon 3 primer. Quantitative polymerase chain reactions on the cDNA products, using primers in exons 2 and 3, showed at least a 100-fold reduction in bxd RNA in In(bxd-pro) homozygotes.
Ubx RNA in In(bxd-pro) blastoderm embryos showed a four-stripe pattern (Figure 6, bottom), identical to that seen in embryos with rearrangement breaks interrupting the bxd RNA (Figure 3). The three additional stripes in mutant embryos look identical in shape and position to those seen in wild-type embryos at a slightly later developmental stage (Figure 6, top). After stage 10 (germ-band extension), we did not see any difference in Ubx RNA patterns between In(bxd-pro) homozygotes and wild type. Heterozygous embryos [from a cross between In(bxd-pro) homozygotes and wild type] gave Ubx patterns with the three posterior stripes, but reduced in intensity (Figure 6, middle). This confirms that loss of the bxd RNA has a dominant effect on Ubx, as expected if the RNA acts in cis.
Embryos of Oregon R and In(bxd-pro) homozygous stocks were stained for UBX protein; no differences were seen in the UBX pattern when the protein first appeared or at any later time in embryonic development. The cuticles of In(bxd-pro) homozygous first-instar larvae appear like those of wild type, and adults are nearly normal in morphology. Homozygous adults show a subtle enlargement of the halteres, and In(bxd-pro)/Ubx heterozygotes show a very weak bx phenotype (anterior haltere to wing transformation). We also tested In(bxd-pro)/pbx1 and In(bxd-pro)/pbx2 heterozygotes (both genotypes should lack the bxd ncRNA), and both look completely wild type in adult morphology (Figure 7B). We attribute the haltere phenotype of In(bxd-pro) homozygotes to the hybrid AttB/P site in the 5′ leader of the Ubx transcription unit, a remnant of our integration strategy (Figure 4D). In summary, we see no effect of the loss of the bxd ncRNA on the development of the fly.
Figure 7.
Adult phenotype of In(bxd-pro). (A) A strong bxd loss-of-function phenotype is seen in flies hemizygous for the pbx1 deletion (diagrammed in Figure 3). The transformations of parasegment 6 include posterior haltere to wing (single arrowhead), reduction of the first abdominal tergite (double arrowhead), and the appearance of ectopic notal tissue (triple arrowhead). (B) When the In(bxd-pro) chromosome is heterozygous with the pbx1 deletion, the flies look wild type, indicating that there is no unusual activation or repression of the bxd domain.
Discussion
ncRNAs and enhancers
Many ncRNAs, in flies and mammals, initiate near strong enhancers, in tissues where the enhancers are active (Natoli and Andrau 2012). The early BX-C ncRNAs appear to fit this pattern. We find widespread transcription across the regulatory domains of the bithorax complex, beginning at the blastoderm stage. The ncRNAs appear in active domains, often initiating near enhancers where the gap and pair rule genes bind. The bxd RNA, in particular, initiates immediately adjacent to the PBX enhancer. The bx proximal and bx distal ncRNAs both appear to initiate in a small region adjacent to the BRE enhancer. The iab-8 ncRNA begins within a 2.7-kb fragment that includes the iab-8 enhancer (Zhou et al. 1999; Gummalla et al. 2012). There are not yet well-defined enhancers at the PS4 ncRNA start site, but the RNA starts within an 11-kb SalI restriction fragment that drives PS4 expression from transgenes (M. B. O’Connor and W. Bender, unpublished observations). It will be useful to define more precisely the initiation points of these early ncRNAs, and to map additional enhancers that might respond to the gap and pair rule genes. However, the initial indication is that most of the blastoderm ncRNAs in the BX-C start near blastoderm-specific enhancers.
bxd ncRNA function
The loss of the bxd RNA affects the initial pattern of Ubx transcription, as reported by Petruk et al. (2006) and confirmed by the mutants described here. The evidence is consistent with transcriptional suppression, in that low-level readthrough transcripts were detected downstream of the bxd RNA poly(A) addition sites at exons 7 and 8, and the Ubx RNA pattern change is seen in heterozygotes (Figure 6), consistent with action in cis. However, the four-stripe Ubx pattern appears to be identical to the normal Ubx pattern seen in slightly older embryos, as if the bxd RNA merely delays the appearance of the posterior three stripes. By the gastrulation stage, Ubx transcription is getting stronger, and the early broad bxd RNA expression is fading; any remaining bxd readthrough RNA is apparently insufficient to suppress the Ubx promoter. We could not detect any difference in UBX protein patterns between wild-type and In(bxd-pro) embryos, and the first-instar cuticle pattern looks normal in In(bxd-pro) homozygotes. The four-stripe Ubx pattern is in some sense a “phenotype” of bxd RNA mutants, but there is no evidence that it has any consequence for development.
The bxd mutations that lack the bxd RNA (shown in red in Figure 3) cause dramatic segmental transformations. It was not clear from prior studies whether these phenotypes could be due to loss of the bxd RNA or to the removal of other sequence elements. Petruk et al. (2006) injected double-stranded RNA homologous to the bxd RNA into preblastoderm embryos, to ablate the bxd RNA by an RNAi mechanism. They reported no homeotic phenotypes in the injected flies. Aside from the concern of incomplete knockdown, this RNAi treatment would not affect any cis-acting function of transcription across the bxd region.
The most appealing proposal for the function of the bxd RNA and the other blastoderm-stage ncRNAs of the BX-C is that they set the active or repressed state of their segmental domains. More specifically, transcription across a PRE within a domain might antagonize its ability to impose repression. This effect of transcription on PREs was demonstrated most clearly by Schmitt et al. (2005) and was also seen in two cases of transcripts initiating from ectopic promoters inserted into the BX-C (Bender and Fitzgerald 2002; Hogga and Karch 2002). But in the Bender and Fitzgerald (2002) example, it was clear that the loss of repression was gradual. There was almost no effect of persistent transcription in late embryos (Bender and Fitzgerald 2002), although Polycomb repression is established by the early germ-band elongation stage of embryogenesis (stage 10, ∼5 hr), as judged by the time of initial misexpression of the homeotic genes in Polycomb group mutants (Struhl and Akam 1985; McKeon and Brock 1991; Simon et al. 1992). Breaking through Polycomb group repression apparently requires strong and persistent transcription, but neither quality applies to the BX-C ncRNAs at blastoderm.
If the bxd ncRNA were required to activate the bxd domain, In(bxd-pro) would cause a loss-of-function bxd phenotype. In the embryo, this would be seen as a reduction of the first abdominal setal belt and the appearance of ventral pits and trihairs on the first abdominal cuticle. In the adult, bxd loss of function is dramatic, as illustrated by the pbx1 phenotype (Figure 7A). The transformations include posterior haltere to wing, reduction or loss of the first abdominal tergite, and the development ectopic notal cuticle. Adult bxd mutant flies that are also heterozygous for an abd-A mutation often develop extra legs on the first abdominal segment. In(bxd-pro) homozygotes show none of these phenotypes. It is possible that there is another ncRNA that serves redundantly to activate the region. However, we could not detect any other embryonic transcript across the bxd PRE. The “sense” probe used for the in situ hybridization in Figure 5 covers the bxd PRE (Figure 3), but it gives no signal in early embryos homozygous for In(bxd-pro).
If the bxd ncRNA somehow represses or limits UBX expression in PS6–12, then In(bxd-pro) might cause a gain of function. Extra gene doses of the Ubx transcription unit plus its bxd regulatory region cause only subtle changes in the denticles of the larval third thoracic setal belt and a reduction in the size of the adult haltere (Smolik-Utlaut 1990). No such changes were seen in In(bxd-pro) homozygotes, nor was there any apparent increase in UBX protein levels in embryos, as judged by immunohistochemistry. In summary, there is no indication that the bxd RNA is needed for either activation or repression of Ubx.
The In(bxd-pro) chromosome should make the first exon of the bxd RNA; if that has a function independent of the other exons, it might still be intact. Transcription at an enhancer might be required for the enhancer to function, but the nature of the downstream exons might be irrelevant. Transcription below our level of detection might exist and be sufficient to activate the region, or the bxd ncRNA could have a subtle or a redundant function. However, the most likely conclusion is that the bxd ncRNA is transcriptional noise associated with a strong enhancer, a possibility suggested at the time of its discovery (Lipshitz et al. 1987).
Acknowledgments
We are indebted to E. B. Lewis for providing the mutant lines shown in Figure 3. Xiao-Qiang Qin mapped the bxd1794 rearrangement breakpoint. François Karch (U. Geneva, Geneva Switzerland) provided the nanos-phiC31 stock. Kami Ahmad and Guillermo Orsi provided helpful comments on the manuscript. Plasmids and strains for recombineering were obtained from the NCI BRB Preclinical Repository. This work was supported by the Institute of General Medical Sciences of the National Institutes of Health under award R01-GM028630.
Footnotes
Communicating editor: P. K. Geyer
Literature Cited
- Akam M. E., Martinez-Arias A., 1985. The distribution of Ultrabithorax transcripts in Drosophila embryos. EMBO J. 4: 1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akam M. E., Martinez-Arias A., Weinzierl R., Wilde C. D., 1985. Function and expression of Ultrabithorax in the Drosophila embryo. Cold Spring Harb. Symp. Quant. Biol. 50: 195–200. [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., et al. , 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337–350. [DOI] [PubMed] [Google Scholar]
- Bae E., Calhoun V. C., Levine M., Lewis E. B., Drewell R. A., 2002. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. USA 99: 16847–16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Wu C.-T., 2008. A simple polymerase chain reaction-based method for the construction of recombinase-mediated casette exchange donor vectors. Genetics 180: 1763–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W., 2008. MicroRNAs in the Drosophila bithorax complex. Genes Dev. 22: 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W., Fitzgerald D. P., 2002. Transcription activates repressed domains in the Drosophila bithorax complex. Development 129: 4923–4930. [DOI] [PubMed] [Google Scholar]
- Bender W., Hudson A., 2000. P element homing to the Drosophila bithorax complex. Development 127: 3981–3992. [DOI] [PubMed] [Google Scholar]
- Bender W., Lucas M., 2013. The border between the Ultrabithorax and abdominal-A regulatory domains in the Drosophila bithorax complex. Genetics 193: 1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W., Weiffenbach B., Karch F., Peifer M., 1985. Domains of cis-interaction in the bithorax complex. Cold Spring Harb. Symp. Quant. Biol. 50: 173–180. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J. A., Hartenstein V., 1985. The Embryonic Development of Drosophila melanogaster. Springer-Verlag, Berlin. [Google Scholar]
- Casares F., Bender W., Merriam J., Sánchez-Herrero E., 1997. Interactions of the Drosophila Ultrabithorax regulatory regions with native and foreign promoters. Genetics 145: 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel S. E., Martienssen R. A., 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14: 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberledge S., Zaratzian A., Sakonju S., 1990. Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc. Natl. Acad. Sci. USA 87: 3259–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds J. W., Mahadevan L. C., Clayton A. L., 2008. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 27: 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummalla M., Maeda R. K., Castro Alvarez J. J., Gyurkovics H., Singari S., et al. , 2012. abd-A regulation by the iab-8 noncoding RNA. PLoS Genet. 8: e1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk J., Dinardo S., 1994. Drosophila hedgehog acts as a morphogen in cellular patterning. Cell 76: 449–460. [DOI] [PubMed] [Google Scholar]
- Hogga I., Karch F., 2002. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with Polycomb-mediated silencing. Development 129: 4915–4922. [DOI] [PubMed] [Google Scholar]
- Ingolia N. T., Lareau L. F., Weissman J. S., 2011. Ribosomal profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Plaza S., Zanet J., Benrabah E., Valenti P., et al. , 2010. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329: 336–339. [DOI] [PubMed] [Google Scholar]
- Lewis E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- Lipshitz H. D., Peattie D. A., Hogness D. S., 1987. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1: 307–322. [DOI] [PubMed] [Google Scholar]
- Maeda R. K., Karch F., 2009. The bithorax complex of Drosophila: an exceptional Hox cluster. Curr. Top. Dev. Biol. 88: 1–33. [DOI] [PubMed] [Google Scholar]
- Martin C. H., Mayeda C. A., Davis C. A., Ericsson C. L., Knafels J. D., et al. , 1995. Complete sequence of the bithorax complex of Drosophila. Proc. Natl. Acad. Sci. USA 92: 8398–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon J., Brock H. W., 1991. Interactions of the Polycomb group of genes with homeotic loci of Drosophila. Rouxs Arch. Dev. Biol. 199: 387–396. [DOI] [PubMed] [Google Scholar]
- Meller V. H., Rattner B. P., 2002. The roXRNAs encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 21: 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaso H., Murata T., Day N., Yokoyama K. K., 2001. Simultaneous detection of RNA and protein by in situ hybridization and immunological staining. J. Histochem. Cytochem. 49: 1177–1182. [DOI] [PubMed] [Google Scholar]
- Natoli G., Andrau J.-C., 2012. Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet. 46: 1–19. [DOI] [PubMed] [Google Scholar]
- O’Connor M., Peifer M., Bender W., 1989. Construction of Large DNA segments in Escherichia coli. Science 244: 1307–1312. [DOI] [PubMed] [Google Scholar]
- Penny G. D., Kay G. F., Sheardown S. A., Rastan S., Brockdorff N., 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137. [DOI] [PubMed] [Google Scholar]
- Petruk S., Sedkov Y., Riley K. M., Hodgson J., Schweisguth F., et al. , 2006. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127: 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Janett A., Hammonds A. S., Ngo T.-T. B., Misra S., et al. , 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Nat. Acad. USA 105: 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V., Chan C. S., McCabe D., Qian S., 1995. Distinct parasegmental and imaginal enhancers and the establishment of the expression pattern of the Ubx gene. Genetics 141: 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Oliver P. L., Reik W., 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641. [DOI] [PubMed] [Google Scholar]
- Qian S., Capovilla M., Pirrotta V., 1993. Molecular mechanisms of pattern formation by the BRE enhancer of the Ubx gene. EMBO J. 12: 3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank G., Prestel M., Paro R., 2002. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 22: 8026–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Herrero E., Akam M., 1989. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development 107: 321–329. [DOI] [PubMed] [Google Scholar]
- Schmitt S., Prestel M., Paro R., 2005. Intergenic transcription through a Polycomb group response element counteracts silencing. Genes Dev. 19: 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B. E., Ahmad K., 2005. Transcription triggers deposition and removal of the histone variant H3.3. Genes Dev. 19: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J., Chiang A., Bender W., 1992. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114: 493–505. [DOI] [PubMed] [Google Scholar]
- Smolik-Utlaut S., 1990. Dosage requirements of Ultrabithorax and bithoraxoid in the determination of segmental identity in Drosophila melanogaster. Genetics 124: 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Bushati N., Jan C. H., Kheradpour P., Hodges E., et al. , 2008. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 22: 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G., Akam M., 1985. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 4: 3259–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler D. M., Okamura K., Chung W.-J., Hagen J. W., Berezikov E., et al. , 2008. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 22: 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Allmen G., Hogga I., Spierer A., Karch F., Bender W., et al. , 1996. Splits in fruitfly Hox gene complexes. Nature 380: 116. [DOI] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G., 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Ashe H., Burks C., Levine M., 1999. Characterization of the transvection mediating region of the Abdominal-B locus in Drosophila. Development 126: 3057–3065. [DOI] [PubMed] [Google Scholar]