Abstract
Bacteria exposed to bactericidal fluoroquinolone (FQ) antibiotics can survive without becoming genetically resistant. Survival of these phenotypically resistant cells, commonly called “persisters,” depends on the SOS gene network. We have examined mutants in all known SOS-regulated genes to identify functions essential for tolerance in Escherichia coli. The absence of DinG and UvrD helicases and the Holliday junction processing enzymes RuvA and RuvB leads to a decrease in survival. Analysis of the respective mutants indicates that, in addition to repair of double-strand breaks, tolerance depends on the repair of collapsed replication forks and stalled transcription complexes. Mutation in recF results in increased survival, which identifies RecAF recombination as a poisoning mechanism not previously linked to FQ lethality. DinG acts upstream of SOS promoting its induction, whereas RuvAB participates in repair only. UvrD directly promotes all repair processes initiated by FQ-induced damage and prevents RecAF-dependent misrepair, making it one of the crucial SOS functions required for tolerance.
Keywords: SOS response, persisters, tolerance, DNA repair, fluoroquinolones
IN the face of a potentially lethal stress, the presence of phenotypic variants of genetically identical bacterial cells can make a difference between survival of the lineage and extermination. A clonal population of cells of a strain genetically susceptible to an antibiotic can harbor tolerant phenocopies that will survive the antibiotic challenge. These cells are termed “persisters” and are of special interest because of their ability to influence the outcome of antibiotic treatments (Lewis 2010; Mulcahy et al. 2010). Toxins from chromosomally encoded toxin–antitoxin (TA) modules have been shown to play a role in persister formation under certain condition (Keren et al. 2004; Dörr et al. 2010; Gerdes and Maisonneuve 2012) To date, screening of E. coli mutant libraries has yielded no single mutants that completely lack persisters (Spoering 2006; Hansen et al. 2008). These studies have shown that there are many pathways that lead to persister formation (Lewis 2010). We have shown that tolerance to fluoroquinolone (FQ) antibiotics depends on a functional SOS response (Dörr et al. 2009). The SOS-regulated tisAB/istR TA locus contributes to tolerance to ciprofloxacin (Dörr et al. 2010). When overproduced, the membrane peptide TisB decreases proton motive force and intracellular ATP levels (Gurnev et al. 2012). During SOS induction, tisB is overexpressed, which may induce stasis and prevent killing by the antibiotic. However, it influences tolerance only at high ciprofloxacin concentration [100× minimal inhibitory concentration (MIC)]; therefore, it is not the only mechanism of FQ tolerance.
Fluoroquinolones target gyrase and topoisomerase IV (Drlica and Zhao 1997). Upon FQ binding, these enzymes become endonucleases introducing double-strand breaks (DSBs) into the bacterial chromosome (Malik et al. 2006). Cells repair DSBs largely through the DNA-damage-inducible SOS gene network (Radman 1975; Friedberg et al. 2006). The susceptibility to the FQ therefore depends on the cellular concentration of the active gyrase and topoisomerase molecules and the state of induction of the SOS response.
Gyrase and topoisomerase IV are essential during replication and transcription (De Wyngaert and Hinkle 1979), and their maximal amount in a cell is expected during maximal growth rate. Indeed, gyrA and gyrB, structural genes of DNA gyrase, reach maximal expression in the early exponential growth phase and decrease during subsequent growth, attaining the lowest level in stationary phase (Gomez-Gomez et al. 1996; Schneider et al. 1999). The susceptibility to FQs would be expected to parallel this dynamic, and experimental data confirm this: the bactericidal effect of FQs is the strongest in early exponential phase and the weakest in stationary phase (Keren et al. 2004; Dörr et al. 2009).
The SOS gene network is controlled by the LexA repressor, and LexA-regulated genes exhibit heterogeneous expression in a culture not subject to an external SOS-inducing treatment (McCool et al. 2004). The heterogeneity of expression is a result of stochastic factors resulting from the binding affinity of LexA to different SOS boxes and intrinsic DNA damage (Pennington and Rosenberg 2007; Kamensek et al. 2010; Butala et al. 2011). Combined heterogeneity of the expression of the target and the repair mechanism translates into a wide spectrum of phenotypic states and hence varied susceptibility to a FQ.
In this study, we have analyzed knockouts of all known SOS genes (Fernandez De Henestrosa et al. 2000; Courcelle et al. 2001) to identify the SOS functions crucial for tolerance. We have found that the removal of DinG, UvrD, and RuvAB leads to the formation of fewer persisters at high and low concentrations of the antibiotic, confirming that the repair processes that these proteins catalyze are essential for tolerance. In addition, the inactivation of recF leads to an increase in tolerance, identifying the RecFOR recombination pathway as a poisoning mechanism.
Materials and Methods
Tolerance assays
All killing experiments were conducted at 37° in cation-adjusted Mueller Hinton Broth (MHB) (Teknova) buffered with 0.1 M HEPES (pH 7.2) unless otherwise noted. Cultures were treated with 0.1 μg/ml ciprofloxacin, unless otherwise noted. Tolerance was tested by diluting an overnight culture of 1:100 in 3 ml of fresh MHB in 17- × 100-mm polypropylene tubes placed at an angle on a rotating platform and grown for 1.5 hr with shaking (200 rpm) until there were ∼2 × 108 colony-forming units (CFUs) per millimeter. CFU counts were measured by washing cells with 1% NaCl to remove the antibiotic, serially diluting, and plating on Luria–Bertani (LB) agar supplemented with 20 mM MgSO4 to neutralize ciprofloxacin carryover. The colonies were counted after 40 hr of incubation at 37°. For UvrD complementation experiments, 0.1 mM IPTG was added to the diluted culture 30 min prior to ciprofloxacin treatment. All antibiotics and chemicals were purchased from Sigma (St. Louis). For the minimal media persister assays, cultures were grown to stationary phase in minimal medium and diluted into the same medium. Cells were diluted 1:100 into MOPS minimal medium (Neidhardt et al. 1974) with 0.2% glucose as a carbon source and grown for 2 hr at 37° with shaking. Cells were diluted 1:50 into MOPS medium with 0.3% glycerol as a carbon source and grown for 3.5 hr under the same conditions. For experiments done at 28°, cells were diluted 1:100 in buffered MHB and incubated for 12 hr with shaking.
Strain construction
E. coli K-12 MG1655 was used as the parental strain (wild type) for all strain construction. Bacterial strains are listed in Supporting Information, Table S2. P1 transduction was used to move different alleles from the KEIO collection (Baba et al. 2006) into MG1655. The kanamycin cassette was cured using pCP20 when needed. Precise deletions were made in the MG1655 background using the methods described in Datsenko and Wanner (2000).
MG1655 pZS*34uvrD was constructed by cloning the ORF of uvrD into the KpnI/ClaI (in boldface type) cloning site of pZS*34 (Lutz and Bujard 1997) using the primers uvrDkpn (ATTTTAGGTACCAGGAGGCAGCTAATGGACGTTTCTTACCTGCTCGACA GCC) and uvrDClaI (TATTAATCGATTTACACCGACTCCAGCCGGGCGTATG).
Measurement of SOS induction
To measure SOS induction by measuring β-galactosidase production of strains carrying recA::lacZ reporter fusion (Casaregola et al. 1982), overnight cultures were diluted 1:100 in 3 ml MHB and grown for 1.5 hr in 17- × 100-mm polypropylene tubes with shaking. An 800-μl aliquot was removed from the culture and the optical density (OD) 600 was measured. Cultures were treated with 0.1 μg/ml ciprofloxacin or 4 μg/ml mitomycin C at 37° for ∼20 min. Both the treated and the culture were measured for recA::lacZ expression using the methods described in Miller (1992) with the exception that β-mercaptoethanol was not added to the Z-buffer stock but was instead added directly to the reaction tube. Miller units were calculated as described in Miller (1992).
Flow cytometry analysis
An E. coli strain with GFP under the sulA promoter (McCool et al. 2004) was used to measure the induction of the SOS response by flow cytometry. Cells were grown as described above and treated with 0.1 μg/ml ciprofloxacin. Prior to the addition of antibiotic, and every 15 min after, 10,000 cells were analyzed using the BD Aria II SORP flow cytometer to measure GFP. Data were analyzed using Flowjo software (Treestar, Inc., San Carlos, CA).
Results
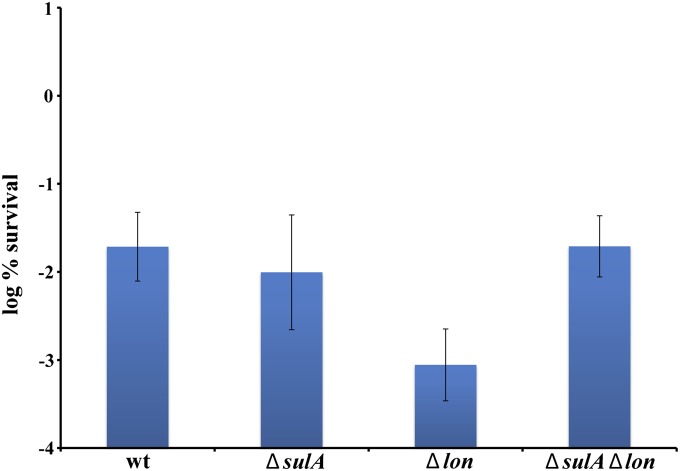
We have previously shown that tolerance to FQs depends on SOS response (Dörr et al. 2009). At high FQ concentrations, the component of the SOS response responsible for tolerance is the tisB/lstR TA module (Dörr et al. 2010). Recently, it has been reported that deletion of the Lon protease, which regulates the antitoxins of 10 TA modules, leads to a decrease in tolerance to a FQ (Maisonneuve et al. 2011). Given that SulA, a cell division inhibitor induced during the SOS response, is also a substrate of the Lon protease (Mizusawa and Gottesman 1983), we decided to reexamine this issue. SulA proteolysis is essential for resuming cell division after repression of the SOS response, which would suggest that decreased tolerance of a lon deletion strain might have resulted from unchecked production of SulA, rather than from the inability of these cells to degrade antitoxins. We measured tolerance to ciprofloxacin in a lon mutant and confirmed that it is indeed low. However, the deletion of sulA in a lon− background restored tolerance to the wild-type level, showing that most of the loss of tolerance to a FQ in a lon− mutant results from SulA-blocked cell division upon SOS induction (Figure 1). This experiment also confirmed that the persister fraction depends on a functional SOS response. The level of persisters surviving treatment with a β-lactam antibiotic was unaffected in lon, sulA, and a double mutant, consistent with the specificity of the SOS-dependent mechanism of tolerance to FQs (data not shown)
Figure 1.
Survival of lon and sulA mutants after ciprofloxacin challenge. Strains were challenged with 0.1 μg/ml ciprofloxacin for 6 hr in exponential phase. CFU counts were determined by plating. The data are averages of three independent experiments, and error bars indicate standard error.
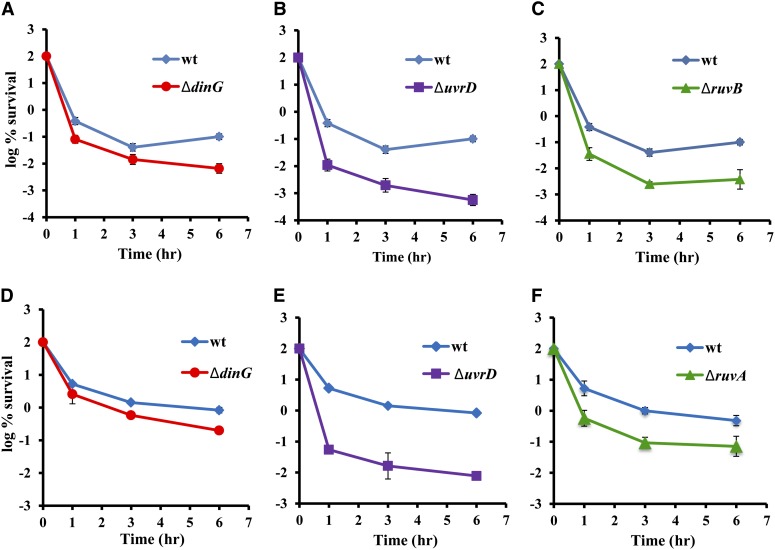
To identify other functions of the SOS response in addition to tisB/lstR, important for FQ tolerance, we compared the survival of deletion mutants of SOS-regulated genes (Fernandez De Henestrosa et al. 2000; Courcelle et al. 2001) to that of the wild type following time-dependent killing in cultures treated with ciprofloxacin at 10× MIC (Table S1). Deletions of dinG, uvrD, and ruvA showed 13-, 434-, and 12-fold decreases in tolerance, respectively (Figure 2, A–C). All of the deletion mutants have the same ciprofloxacin MIC as the wild type. This shows that the decrease in tolerance in these mutants is not due to an intrinsic increase in susceptibility to the antibiotic. The decrease in tolerance is also not growth phase dependent as all three mutants show low persister levels in both exponential and stationary phase (Figure 2, D–F).
Figure 2.
Survival of the wild-type and mutant strains in exponential and stationary phase. (A–C) Exponentially growing cells were treated with 0.1 μg/ml ciprofloxacin for 6 hr. Survival was determined by dilution and plating to count CFU. The graph represents at least 20 independent experiments, and error bars represent standard error. (D–F) Stationary-phase cultures were treated with 1 μg/ml ciprofloxacin. Viable counts were determined by plating. The graphs represent three independent experiments. Standard error is indicated by the error bars. wt, wild type.
Both DinG and UvrD are DNA helicases. DinG can remove R-loops and promote replication across transcription units (Voloshin and Camerini-Otero 2007; Boubakri et al. 2010). UvrD catalyzes the unwinding of forked DNA structures (Lestini and Michel 2007) and the dismantling of RecA nucleoprotein filaments (Veaute et al. 2005), allowing various repair processes such as nucleotide excision repair (Husain et al. 1985), mismatch repair (Lahue et al. 1989), and replication fork restart (Florés et al. 2005). Both DinG and UvrD participate in the removal of stalled RNA polymerase (RNAP) (Boubakri et al. 2010). RuvA forms a complex with RuvB and RuvC, which resolves Holliday junctions (HJs) formed during the recombinational repair of damaged DNA (Parsons and West 1993). All of these repair processes involve single-stranded DNA (ssDNA) intermediates. ssDNA is the SOS inducer, so the activities of DinG, UvrD, and RuvA can potentially influence SOS induction (Kuzminov 1999). While the production of these proteins increases during SOS induction, the proteins are all present in an uninduced cell. Therefore DinG, UvrD, and RuvA can promote tolerance either through influencing SOS induction or through the repair functions that they perform, or both. To distinguish among these possibilities, we measured the persister fraction of the respective deletion mutants in a strain where the SOS regulon is derepressed constitutively [lexA(Def)]. In this genetic context, the dinG deletion had no effect on persister levels. When SOS functions are expressed prior to the addition of FQ, DinG is dispensable for tolerance (Figure 3A). This suggests that DinG promotes tolerance mainly through facilitating SOS induction upon FQ treatment. uvrD and ruvA single deletions decreased the persister level even in a lexA(Def) background (Figure 3, B and C). Loss of UvrD decreased tolerance 23-fold in lexA(Def), compared to the ∼430-fold decrease in a lexA+ background. Therefore, UvrD appears to promote tolerance through stimulating SOS induction and by an additional route, likely participating in recombinational repair of DSBs and restarting of stalled replication forks. The loss of RuvA lowered tolerance in both backgrounds, 57- and 12-fold, suggesting that RuvA does not play a role in SOS induction during FQ treatment. These results suggest that RuvA promotes tolerance solely through its role in repair.
Figure 3.
Survival of dinG, uvrD, and ruvAB mutants (A, B, C, respectively) in a strain constitutively expressing SOS. Exponentially growing cells were treated with 0.1 μg/ml ciprofloxacin. Mutants were made in a strain constitutively expressing SOS [lexA(Def)]. Graphs represent 10 independent experiments, and error bars represent standard error.
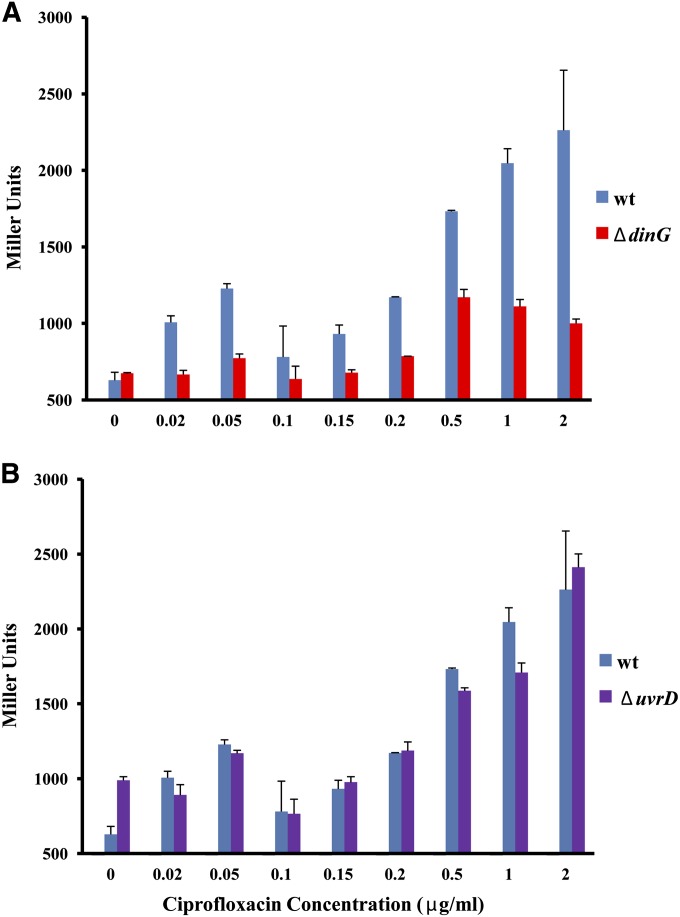
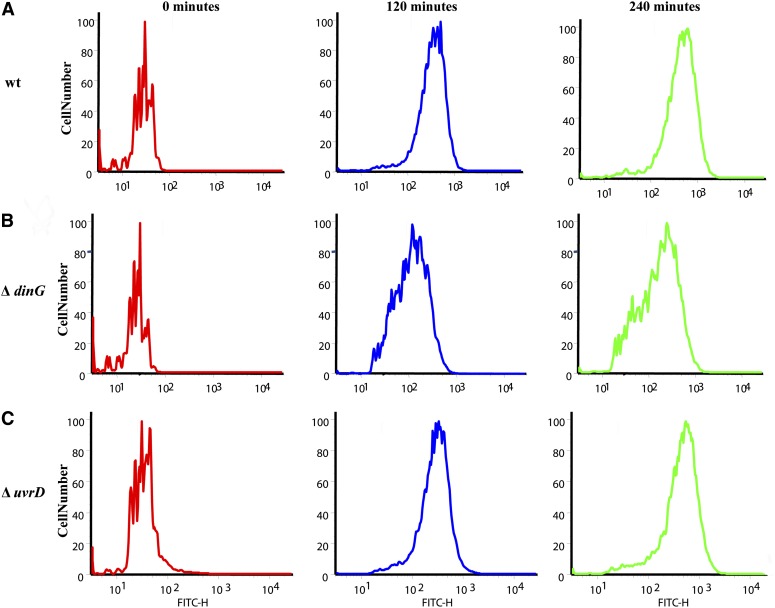
To quantify SOS induction directly, we measured the induction of β-galactosidase under the control of the SOS-inducible recA promoter using various concentrations of ciprofloxacin. A dinG mutant has a deficient SOS response compared to the wild type across a range of ciprofloxacin concentrations (Figure 4A). In contrast, the induction level in a uvrD mutant parallels the wild type (Figure 4B) except for the higher basal expression of the SOS response, which is consistent with previous studies (SaiSree et al. 2000). These measurements report the average SOS induction of the entire population. We next looked into SOS induction in single cells using flow cytometry to probe for differences in the distribution of SOS induction levels across the population in different mutant backgrounds following exposure to ciprofloxacin. We measured the expression of GFP under the control of the sulA promoter, another SOS-inducible gene. In the wild type, the majority of cells exhibited a strong SOS response after 120 min of ciprofloxacin treatment (Figure 5A), reflected in a narrow distribution of fluorescence values. This pattern changed little during the next 120 min. In the dinG knockout after an initial 120 min of treatment, there was a much wider distribution of induction states across the population with fewer highly induced cells compared to the wild type and with many cells showing little or no SOS induction (Figure 5B). The pattern did not change during an additional 120 min, suggesting that the defect in induction is not due to a slower response. These results confirmed that the removal of DinG leads to the impairment of the overall SOS induction, its effect ranging from delaying to abolishing induction.
Figure 4.
SOS induction during ciprofloxacin treatment. Induction of SOS response was measured by assaying for β-galactosidase activity in strains carrying lacZ under control of the recA SOS-inducible promoter after 15 min of treatment with ciprofloxacin with the concentration indicated on the x-axis. (A) dinG mutant. (B) uvrD mutant. Graphs are an average of at least three experiments, and error bars represent standard error.
Figure 5.
Single-cell analysis of SOS induction during ciprofloxacin treatment. Induction of SOS response was measured using a flow cytometry by following GFP expression in strains carrying gfp gene under control of the sulA SOS-inducible promoter. Exponential-phase cells were treated with 0.1 μg/ml ciprofloxacin. A total of 10,000 cells were analyzed before ciprofloxacin treatment and again at 120 and 240 min for (A) a wild type (wt), (B) a dinG, and (C) a uvrD mutant.
The difference between the wild type and the uvrD knockout was less pronounced than between dinG and the wild type (Figure 5C). The basal level of SOS induction was higher, as expected (SaiSree et al. 2000). At 120 and 240 min there were more medium-to-highly induced cells compared to the wild type. The ruvA mutant was undistinguishable from the wild type in both assays, further confirming the role of the RuvABC complex in repair processes not influencing the SOS induction (data not shown).
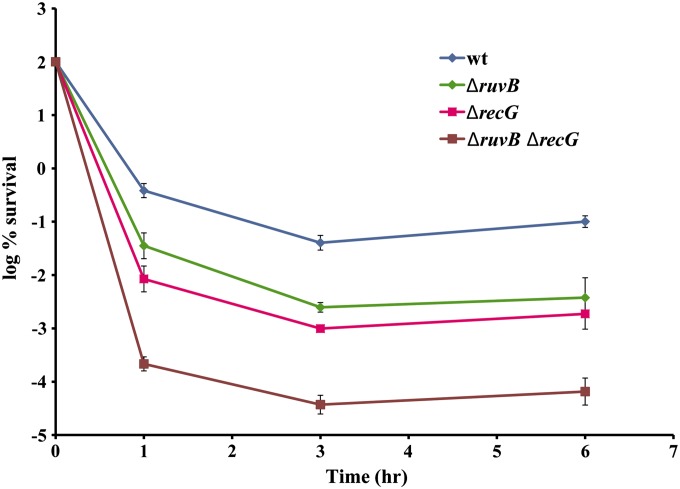
RuvAB catalyzes the late step in recombination, branch migration (Iwasaki et al. 1992), and, together with RuvC, the resolution of HJ (Iwasaki et al. 1991). Therefore, a ruvAB mutant is expected to be as deficient in repair as RecA or RecBC mutants that catalyze the initial steps in recombination, which is not the case. In a ruvAB mutant, HJ can be resolved by an alternative pathway catalyzed by RecG, which is not part of the SOS regulon. We combined ruvAB with a recG mutant, and the resulting strain is indeed as deficient in survival as a recBC mutant, confirming that complete recombinational repair is needed for tolerance (Figure 6). However, it is not clear whether an SOS-induced level of RuvAB is needed for tolerance or whether the basal level is sufficient.
Figure 6.
The role of HJ processing in persistence to fluoroquinolones. Exponentially growing cultures of cells were exposed to 0.1 μg/ml ciprofloxacin. CFU counts were determined by serial dilution and then plating. Error bars represent standard error. The data represent 10 independent experiments.
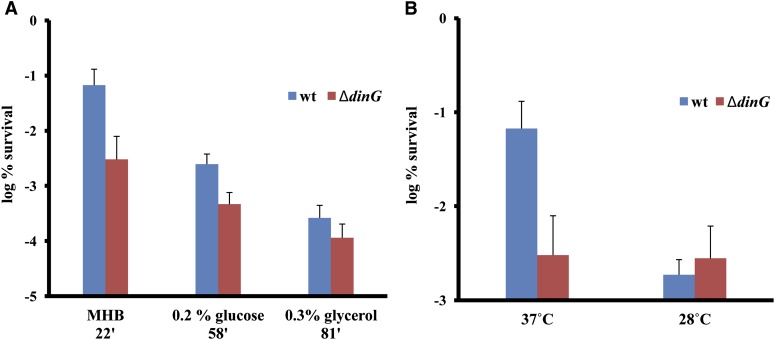
Stalled gyrase and the resulting DSBs create replication and transcription blocks. Transcription blocks cause RNAP to stall, and its removal leads to the formation of R-loops. In turn, R-loops can cause replication stalling (Gan et al. 2011). The DSBs and stalled replication fork cannot be processed until the RNAP dissociates and the resulting R-loop has been removed. By removing RNAP (together with UvrD) and R-loops, DinG (Boubakri et al. 2010) could allow the processing of the break or stalled replication fork, formation of ssDNA, and ultimately SOS induction. If that is the case, the role of DinG in tolerance should correlate with the overall transcription level in the cell. The more transcription complexes there are, the more survival should depend on DinG. To test this, we measured the tolerance in cultures subjected to different growth regimes. The overall transcription level is proportional to the growth rate, as the bulk of transcription takes place at ribosomal operons (Bremer and Dennis 1987). We manipulated the growth rate and hence the transcription levels by growing cells in different media—rich MHB medium and MOPS minimal medium with glucose or glycerol as a sole carbon source—and at different temperatures—37° and 28°. We measured the persister levels in wild type and a dinG deletion mutant. In rich medium, a dinG mutation decreased tolerance 22-fold, compared to 5.2- and 2.2-fold in minimal media with glucose and glycerol, respectively (Figure 7A). Slowing down growth by decreasing temperature from 37° to 28° obviates the need for DinG function in tolerance altogether (Figure 7B). In addition, the effect of a dinG mutation is less pronounced in stationary phase compared to exponential phase (Figure 2, A and D). This is consistent with lower transcription levels in stationary phase. These results are consistent with the role of DinG in allowing DSB processing by RNAP and R-loop removal.
Figure 7.
Role of DinG in survival under different growth conditions. Exponentially growing cultures of dinG mutant and wild type were treated with 0.1 μg/ml of ciprofloxacin for 6 hr, and then CFU counts were determined by serial dilution and plating. (A) Level of persistence in different minimal media; the medium type and generation time in minutes are indicated on the x-axis. (B) Different temperature regimes after 6 hr of treatment. Error bars represent standard error, and graphs represent four independent experiments.
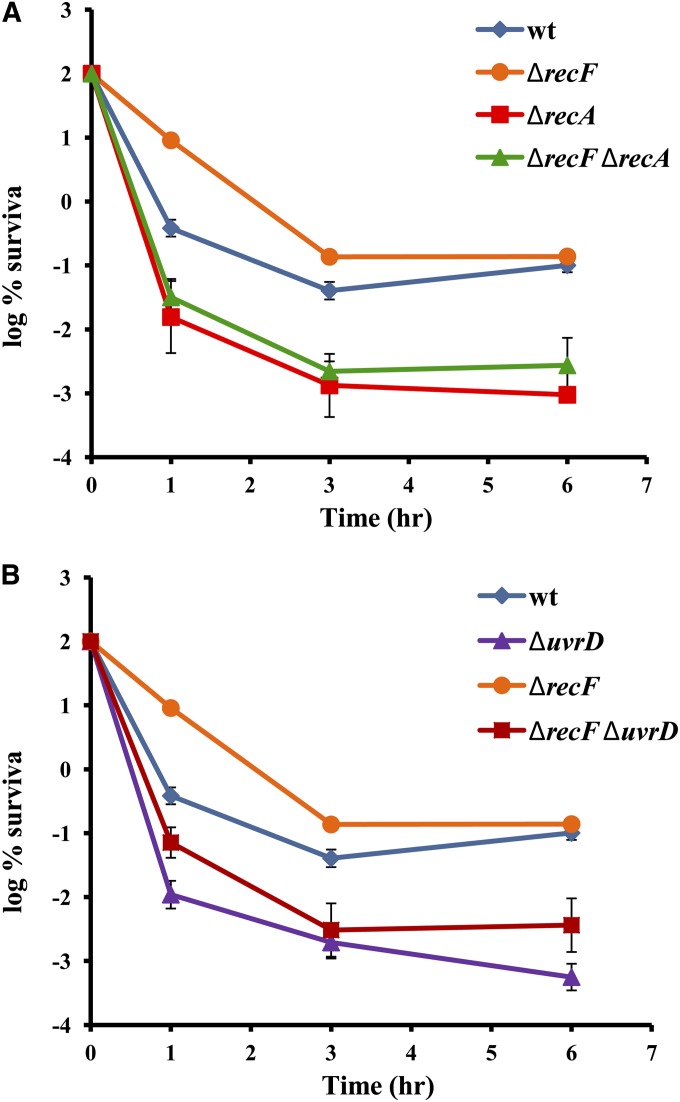
UvrD exerts quality control over the RecA-dependent recombination intermediates and also inhibits the formation of RecF-dependent RecA filaments (Veaute et al. 2005). recF mutation increases survival, suggesting that RecF-dependent recombination is indeed happening during the processing of FQ-induced lesions and is toxic. As with the RecBCD pathway, the RecF pathway depends on RecA. To confirm this, we combined recA with a recF mutation. The survival of the double mutant was as low as that of the recA single mutant, confirming that the RecF effect is due to its interaction with RecA (Figure 8A). In an uvrD background, recF increases the survival, but not to the level of a single recF mutant, confirming that, aside from preventing RecF-dependent toxic recombination, UvrD has an additional role in surviving FQ treatment (Figure 8B). Given the extent of the recF effect, this additional role is stronger than the action of UvrD as a RecF counteractor.
Figure 8.
The role of RecF in survival to ciprofloxacin treatment. Cells were diluted and grown to late exponential phase and then treated with 0.1 μg/ml of ciprofloxacin. Data are from at least 20 independent experiments, and error bars represent standard error.
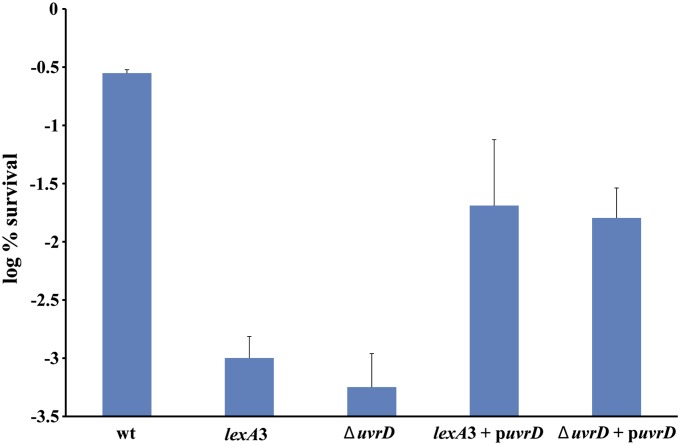
Because it participates at all stages of DSB repair, UvrD could indeed be the single most important SOS function for tolerance to FQs. To test this, we cloned the uvrD gene onto a low copy vector under the control of an IPTG-inducible promoter (Figure 9). First, we introduced this plasmid into an uvrD deletion strain to show that it complements the uvrD defect. Next, we introduced it into a strain carrying the lexA3 mutation. This strain is unable to induce SOS and shows a low tolerance phenotype. In the resulting strain (lexA3 carrying the plasmid with IPTG-inducible uvrD gene), uvrD is the only SOS gene that is overexpressed upon the addition of IPTG into growth medium prior to ciprofloxacin treatment. In this strain, the tolerance was increased to the level of the complemented strain, confirming that UvrD could indeed be the crucial component of the SOS gene network needed for tolerance to FQ antibiotics.
Figure 9.
UvrD overproduction suppresses SOS deficiency. Exponentially growing cultures were treated with 0.1 mM IPTG to induce UvrD vector expression 30 min prior to the addition of 0.1 μg/ml ciprofloxacin. The data are an average of at least five experiments, and error bars represent standard error.
Discussion
Bacteria can survive antibiotic treatments by becoming resistant through a genetic change—mutation or gene acquisition. Mechanisms of resistance have been studied extensively, and most cases are well understood. On the other hand, antibiotic tolerance, involving no genetic change, has only recently been recognized as a potentially important factor influencing the efficiency of existing antibiotic therapies (Mulcahy et al. 2010). The molecular mechanism(s) leading to this epigenetic phenomenon have become a subject of intense research.
In the case of fluoroquinolone antibiotics, tolerance largely depends on the SOS gene network (Dörr et al. 2009), a cellular stress response induced by DNA damage. Mutations resulting in deficiency in SOS induction and repair of DSBs either completely abolish or lead to extremely low tolerance to FQs, but do not affect tolerance to other types of antibiotics. The type I TA module tisAB/istR has a role in FQ tolerance, although only at very high antibiotic concentrations (Dörr et al. 2010). It is a part of the SOS gene network, but its precise role has not been established. In this work, we have identified additional SOS genes needed for FQ tolerance regardless of the antibiotic concentration.
Recently, it was shown that the deletion of the gene encoding Lon protease, the regulator of the antitoxin component of 10 TA modules, results in a low-tolerance phenotype, arguing for the major role of these modules in tolerance (Maisonneuve et al. 2011). We confirmed that tolerance is reduced in the absence of Lon protease. However, inactivation of sulA completely suppresses lon tolerance deficiency during ciprofloxacin treatment (Figure 1). FQs are potent inducers of the SOS response; therefore, during FQ treatment, SulA is produced and blocks cell division during the repair process. The resumption of growth depends on SulA removal by proteolysis. In the absence of the Lon protease, SulA is not degraded, leading to an irreversible cell-cycle checkpoint due to SulA-blocked cell division.
In E. coli, FQs target gyrase and topoisomerase IV (Drlica and Zhao 1997). Gyrase is essential for replication and transcription, relaxing superhelical tension ahead of the progressing replication fork and transcription bubble. It cuts the DNA, and the two ends are then twisted around each other and resealed to form supercoils. FQs block the resealing step, and the gyrase is therefore stuck to the cut DNA strands. This reaction intermediate is called the cleavage complex and is essentially a DNA adduct. DSBs can form by direct cleavage of this adduct by XseAB (exonuclease VII) (Nichols et al. 2011) and also by a recombination nuclease initiating repair of the replication fork stalled by the cleavage complex. In both cases, the free DNA ends are released. The multifunctional enzymatic complex RecBCD, essential for the repair of DSBs, initiates the repair process. It loads onto the DNA ends, unwinds the double-stranded DNA, and degrades it until it encounters the regulatory sequence (Chi site), after which it degrades one of the strands and loads RecA (Kowalczykowski et al. 1994). RecA is the main recombinase that catalyzes a DNA synapsis reaction between ssDNA and a complementary region of double-stranded DNA. In the RecA-ssDNA nucleoprotein filament, RecA becomes activated, and this activated form stimulates the autocatalytic cleavage of the LexA repressor, activating the SOS response. The essential nature of these early steps in survival of FQ treatment is reflected by extremely low or totally suppressed survival of RecB and RecA mutants as well as the LexA mutant’s inability to undergo self-cleavage and induce SOS (Dörr et al. 2009). RecBCD is constitutively produced and RecA is under SOS control with a basal level high enough to carry out recombination. The dependence of tolerance on SOS implies that there are other functions that are needed in addition to the repair of DSBs. We have screened the deletion mutants of all known SOS genes (Table S1) and found that tolerance most depended on dinG, ruvAB, and uvrD.
The analysis of the respective mutants and their known roles suggests that FQ treatment leads to replication fork collapse, stalling of transcription complexes, and formation of DSBs. Therefore, survival ultimately depends on the interplay between the repair of DNA breaks, the clearing of the transcription complexes, and the restarting of the replication fork. These processes depend and/or influence each other and also share some proteins, revealing a complex cascade of events initiated by the action of a FQ.
DinG is a DNA helicase that can remove D- and R-loops in vitro (Voloshin and Camerini-Otero 2007). In vivo studies demonstrated its role in clearing stalled transcription complexes and promoting replication across active transcription units (Boubakri et al. 2010).
We found that, in cells with SOS genes derepressed constitutively, DinG is dispensable for tolerance, which means that it acts upstream of SOS induction (Figure 3A). This is confirmed by measuring SOS induction directly and finding that it is severely impaired in a dinG mutant upon exposure to FQ (Figure 4A and Figure 5B). DinG stimulates SOS induction specifically during FQ treatment and has no influence on the induction resulting from other inducers such as mytomicin C (data not shown). This means that it promotes the formation of the SOS-inducing signal, ssDNA during processing of DSBs and has no influence on the induction resulting from the processing of single-stranded gaps. Following DSBs, ssDNA is formed by RecBCD and therefore DinG must directly or indirectly facilitate its action. Given the known roles in vitro and in vivo of both RecBCD and DinG, it is unlikely that DinG directly influences loading of RecBCD onto DNA ends or its helicase and nuclease activities.
DinG clears the stalled replication complexes in vivo by displacing RNAP and removing R-loops. In doing so, it helps in the resolution of the replication/transcription collisions. By clearing the stalled transcription complexes during FQ treatment, DinG may enable RecBCD to process DSBs, release the ssDNA, and hence trigger the induction of the SOS response. This is consistent with the finding that reducing the overall transcription level decreases the role of DinG in tolerance (Figure 7).
UvrD is a multifunctional enzyme: it edits the RecA-formed recombination intermediates, dismantling intermediates of insufficient length, and also prevents the formation of RecF-dependent RecA filaments (Veaute et al. 2005). RecF helps the loading of RecA onto single-stranded gaps and stabilizes the RecA-DNA filaments that form. Even though the RecA-DNA filament is the initial intermediate in recombination that is essential for repair of many types of DNA lesions, out-of-control RecA filamentation can, depending on the substrate, prevent the repair and become a poisoning mechanism. RecF/A filaments can form at stalled replication forks, where, by initiating pairing of single-strand gaps, they prevent the replication restart and the repair of DSBs. UvrD’s ability to prevent this toxic recombination makes it a major player in both restarting collapsed replication forks and DSB repair. The absence of UvrD decreases tolerance to FQ almost as much as RecA, suggesting that quality control of recombination that UvrD exerts is crucial for survival (Figure 8).
We found that inactivation of RecF results in increased tolerance and that this effect is dependent on RecA (Figure 8A). This confirms that RecF/RecA filaments form during FQ treatment and that their formation contributes to the toxicity. The recF mutation increases the survival of the uvrD mutant, showing that part of UvrD’s role in promoting survival is the suppression of RecF toxicity. Single-strand gaps, which are substrates for RecF/RecA, are not introduced directly by FQ-blocked gyrase; therefore, they must result from a different process. Their most likely source is the collapsed replication fork. Active gyrase is located ahead of replisome. Moving at 800 bp/sec, replication forks must run into FQ-blocked gyrase shortly after the formation of the cleavage complex, which leads to their collapse. In a different system Cirz et al. (2005) showed that tolerance to FQ depends on PriA, a protein essential for replication fork restart, which is strong evidence that replication forks do disintegrate during FQ treatment and that their repair/restart is vital for survival.
RuvA and RuvB form a complex with RuvC that then mediates branch migration and resolution of the HJ, the final steps of recombination. SOS induction is not impaired in ruvA and ruvB mutants, but the survival is decreased. It means that RuvAB does not participate in formation of the SOS-inducing signal, but acts downstream of SOS induction. This is consistent with its role in recombinational repair either of gyrase-induced DSBs, collapsed replication fork, or both. Even though resolution of HJ is essential for completing repair, ruvAB mutants survive better than recBC mutants that are unable to initiate recombination. In the absence of RuvAB, RecG can catalyze the resolution of HJ. The double mutant ruvA recG (as well as ruvB recG) is as sensitive as a recBC mutant, confirming that all steps of recombinational repair are needed for tolerance (Dörr et al. 2009).
Taken together, our findings confirm that tolerance to FQs primarily depends on the capacity of the cell to repair the DNA damage resulting from FQ action. RecA is the central protein and it has two roles: (1) sensing the damage allowing for induction of repair proteins through the induction of the SOS network and (2) carrying out recombination. In addition to halting cell division while repair takes place, the SOS response increases the capacity for damage repair and/or tolerance. The capacity to carry out recombination is increased mainly through the elevated levels of RecA and RuvAB. However, it is not clear whether SOS-induced levels of RecA and RuvAB or elevated levels of some other SOS functions are indeed needed for FQ tolerance. Both DSBs and collapsed replication forks are ultimately repaired through recombination, but during the processing of these lesions, there is a considerable potential for improper pairing, which can impede the repair and lead to cell death. The UvrD helicase exerts quality control over recombination and is therefore an essential part of the repair process. We show that increasing the amount of UvrD by overproducing it artificially is sufficient to ensure a wild-type level of survival in the absence of SOS induction (Figure 9). This raises the possibility that UvrD is the crucial SOS function needed for tolerance to FQs, but it is not excluded that overexpression of other SOS genes or combination of genes can have the same effect.
Mutations in all gene functions that promote and suppress survival affect the bulk as well as the persister fraction. The initial rate of killing is increased in recA, recB, ruvAB, dinG, and uvrD mutants and is slowed down in a recF mutant. When repair is completely suppressed (such as in recBC, ruvA recG, and priA backgrounds), there are no survivors. We have shown before that the tolerance to FQs is a result of an induced, active process (Dörr et al. 2009). Here we confirm that all the cells are damaged and also show that the same repair mechanism takes place in the entire population. In studies of survival of γ-irradiated cells, it was estimated that E. coli K-12 can successfully repair up to four simultaneous DSBs (Krisch et al. 1976). Therefore, the persister fraction likely consists of the cells that received between one and four DSBs and managed to repair them. Restoration of the collapsed replication fork is an essential part of successful repair, indicating that at the time of the FQ treatment future persister cells are actively replicating. Cells growing exponentially with the minimal generation time (20 min in rich LB medium), have four to eight chromosomes (Akerlund et al. 1995) and very high transcription activity, the bulk of it at ribosomal RNA operons (Bremer and Dennis 1987). Those cells have four to eight replication forks and multiple transcription bubbles, all relying on gyrase and topoisomerase IV to relieve the negative supercoil accumulation. They collapse when they collide with FQ-induced DNA-gyrase or DNA-topoisomerase IV adducts. The amount of DSBs formed due to these collisions is beyond the cell’s repair capacity. On the other hand, cells that slow down and have fewer replication forks and reduced transcription activity experience few breaks and are able to repair them and survive, forming a persister fraction.
The question remains as to what happens after the completion of the repair. The medium is not exhausted and can support growth, and the antibiotic is still present. Therefore, the surviving cells could resume growth and become vulnerable again, yet there is no significant loss of viability for an extended period of time. The induction of the SOS-controlled TisB toxin dissipates the proton motive force (Gurnev et al. 2012) and depletes the cellular ATP pool (Unoson and Wagner 2008), possibly preventing further damage by effectively shutting down the cell after the repair of the original lesions. However, this mechanism would operate in only a fraction of cases as the survival is TisB dependent only at high concentrations of FQ. Upon SOS induction, an uncharacterized SOS-controlled mechanism halts respiration (Swenson et al. 1978; Swenson and Norton 1983), which would be another way of slowing down all cellular processes and preventing further damage.
Even in the absence of this type of damage avoidance by SOS-induced slowdown or complete dormancy, the repair can probably be completed during the SOS-imposed division block. Broken chromosomes lose superhelicity in the large domains around the break, so there will be a window of time during the repair process and shortly after its completion during which gyrase and topoisomerase IV are not active, thus rendering cells insensitive to FQ. When the repair is completed, SOS is shut off, and superhelicity is restored, the cells then become susceptible again. However, the damage would be repairable because it would occur before multiple replication forks and transcription bubbles form, leading to a few lesions only, not exceeding the repair capacity. In this scenario, the persistent state would consist of successive rounds of low amounts of damage followed by repair.
In addition to FQ antibiotics, ionizing irradiation and starvation for the DNA precursor dTTP (leading to thymineless death) also result in multiple DSBs, and in both cases survival depends on RecABCD/RuvAB repairing the breaks and UvrD counteracting RecAF toxic recombination, revealing a common survival mechanism following chromosome fragmentation (Sargentini and Smith 1986; Fonville et al. 2010, 2011; Kuong and Kuzminov 2010).
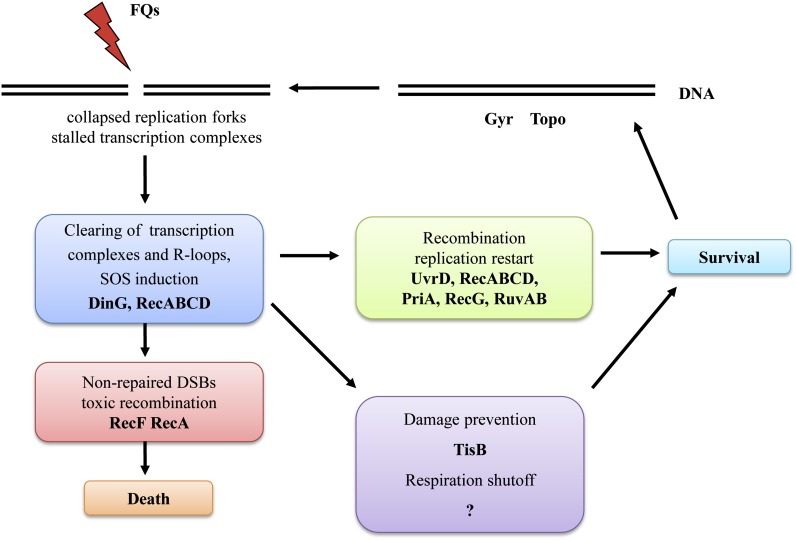
The data presented identify the main functions controlled by the SOS response in tolerance to FQ antibiotics (Figure 10). Upon exposure to a FQ, gyrase and topoisomerase IV are blocked and form DNA adducts that lead to the collapse of the replication forks and stalling of the transcription complexes. Processing of these lesions leads to the formation of DSBs, which in turn results in the induction of the SOS gene network and hence the elevated levels of the proteins needed for repair. The repair consists of clearing the stalled transcription complexes, restoring replication forks, and repairing the DSBs. Recombination plays a central role in the repair process, but, because of the type of lesions, some recombination functions can interfere with each other and exacerbate cell death. The quality control of the recombination process is therefore one of the main contributors to survival. The intrinsic property of FQ-induced damage, its sensing, the induction of the SOS response, and the ensuing repair process make cells refractory to the FQs, rendering them tolerant without genetic change.
Figure 10.
Model of cellular events following exposure to fluoroquinolones. See Discussion for detailed explanation.
Supplementary Material
Acknowledgments
We thank members of the Lewis Lab and Veronica Godoy for critical reading of the manuscript; the E. coli Genetic Stock Center and National BioResource Project (National Institute of Genetics, Japan) for E. coli; the Radman Lab, the Walker Lab, the Sandler Lab, Bénédicte Michel, and Peter McGlynn for strains; and Sonja Hansen for constructing the lon deletion mutant.
Footnotes
Communicating editor: S. Sandler
Literature Cited
- Akerlund T., Nordstrom K., Bernander R., 1995. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J. Bacteriol. 177: 6791–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., et al. , 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubakri H., de Septenville A. L., Viguera E., Michel B., 2010. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 29: 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H., Dennis P. P., 1987. Modulation of chemical composition and other parameters of the cell by growth rate, pp. 1527–1542 in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, edited by Neidhardt F. C. American Society for Microbiology, Washington, DC. [Google Scholar]
- Butala M., Klose D., Hodnik V., Rems A., Podlesek Z., et al. , 2011. Interconversion between bound and free conformations of LexA orchestrates the bacterial SOS response. Nucleic Acids Res. 39: 6546–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaregola S., D’Ari R., Huisman O., 1982. Quantitative evaluation of recA gene expression in Escherichia coli. Mol. Gen. Genet. 185: 430–439. [DOI] [PubMed] [Google Scholar]
- Cirz R. T., Chin J. K., Andes D. R., de Crecy-Lagard V., Craig W. A., et al. , 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 3: e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Khodursky A., Peter B., Brown P., Hanawalt P., 2001. Comparative gene expression profiles following UV exposure in wild type and SOS-deficient Escherichia coli. Genetics 158: 41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L., 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wyngaert M., Hinkle D. C., 1979. Involvement of DNA gyrase in replication and transcription of bacteriophage T7 DNA. J. Virol. 29: 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T., Lewis K., Vulić M., 2009. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 5: e1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T., Vulić M., Lewis K., 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8: e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Zhao X., 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61: 377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez De Henestrosa A. R., Ogi T., Aoyagi S., Chafin D., Hayes J. J., et al. , 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35: 1560–1572. [DOI] [PubMed] [Google Scholar]
- Florés M.-J., Sanchez N., Michel B., 2005. A fork-clearing role for UvrD. Mol. Microbiol. 57: 1664–1675. [DOI] [PubMed] [Google Scholar]
- Fonville N. C., Bates D., Hastings P. J., Hanawalt P. C., Rosenberg S. M., 2010. Role of RecA and the SOS response in thymineless death in Escherichia coli. PLoS Genet. 6: e1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville N. C., Vaksman Z., DeNapoli J., Hastings P. J., Rosenberg S. M., 2011. Pathways of resistance to thymineless death in Escherichia coli and the function of UvrD. Genetics 189: 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., et al. , 2006. DNA Repair and Mutagenesis. American Society for Microbiology Press, Washington, DC. [Google Scholar]
- Gan W., Guan Z., Liu J., Gui T., Shen K., et al. , 2011. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 25: 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Maisonneuve E., 2012. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 66: 103–123. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez J. M., Baquero F., Blazquez J., 1996. Cyclic AMP receptor protein positively controls gyrA transcription and alters DNA topology after nutritional upshift in Escherichia coli. J. Bacteriol. 178: 3331–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurnev P. A., Ortenberg R., Dörr T., Lewis K., Bezrukov S. M., 2012. Persister-promoting bacterial toxin TisB produces anion-selective pores in planar lipid bilayers. FEBS Lett. 586: 2529–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S., Lewis K., Vulic M., 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52: 2718–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I., Van Houten B., Thomas D. C., Abdel-Monem M., Sancar A., 1985. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc. Natl. Acad. Sci. USA 82: 6774–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Shiba T., Nakata A., Shinagawa H., 1991. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 10: 4381–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Nakata A., Shinagawa H., 1992. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 6: 2214–2220. [DOI] [PubMed] [Google Scholar]
- Kamensek S., Podlesek Z., Gillor O., Zgur-Bertok D., 2010. Genes regulated by the Escherichia coli SOS repressor LexA exhibit heterogeneous expression. BMC Microbiol. 10: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I., Shah D., Spoering A., Kaldalu N., Lewis K., 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186: 8172–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M., 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58: 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisch R. E., Krasin F., Sauri C. J., 1976. DNA breakage, repair and lethality after 125I decay in rec+ and recA strains of Escherichia coli. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 29: 37–50. [DOI] [PubMed] [Google Scholar]
- Kuong K. J., Kuzminov A., 2010. Stalled replication fork repair and misrepair during thymineless death in Escherichia coli. Genes Cells 15: 619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A., 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63: 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue R. S., Au K. G., Modrich P., 1989. DNA mismatch correction in a defined system. Science 245: 160–164. [DOI] [PubMed] [Google Scholar]
- Lestini R., Michel B., 2007. UvrD controls the access of recombination proteins to blocked replication forks. EMBO J. 26: 3804–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K., 2010. Persister cells. Annu. Rev. Microbiol. 64: 357–372. [DOI] [PubMed] [Google Scholar]
- Lutz R., Bujard H., 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1–I2 regulatory elements. Nucleic Acids Res. 25: 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E., Shakespeare L. J., Jorgensen M. G., Gerdes K., 2011. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. USA 108: 13206–13211. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Malik M., Zhao X., Drlica K., 2006. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 61: 810–825. [DOI] [PubMed] [Google Scholar]
- McCool J. D., Long E., Petrosino J. F., Sandler H. A., Rosenberg S. M., et al. , 2004. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol. Microbiol. 53: 1343–1357. [DOI] [PubMed] [Google Scholar]
- Miller J., 1992. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Mizusawa S., Gottesman S., 1983. Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc. Natl. Acad. Sci. USA 80: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy L., Burns J., Lory S., Lewis K., 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192: 6191–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F., 1974. Culture medium for enterobacteria. J. Bacteriol. 119: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols R. J., Sen S., Choo Y. J., Beltrao P., Zietek M., et al. , 2011. Phenotypic landscape of a bacterial cell. Cell 144: 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons C. A., West S. C., 1993. Formation of a RuvAB-Holliday junction complex in vitro. J. Mol. Biol. 232: 397–405. [DOI] [PubMed] [Google Scholar]
- Pennington J. M., Rosenberg S. M., 2007. Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 39: 797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M., 1975. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 5A: 355–367. [DOI] [PubMed] [Google Scholar]
- SaiSree L., Reddy M., Gowrishankar J., 2000. lon incompatibility associated with mutations causing SOS induction: null uvrD alleles induce an SOS response in Escherichia coli. J. Bacteriol. 182: 3151–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargentini N. J., Smith K. C., 1986. Quantitation of the involvement of the recA, recB, recC, recF, recJ, recN, lexA, radA, radB, uvrD, and umuC genes in the repair of X-ray-induced DNA double-strand breaks in Escherichia coli. Radiat. Res. 107: 58–72. [PubMed] [Google Scholar]
- Schneider R., Travers A., Kutateladze T., Muskhelishvili G., 1999. A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol. Microbiol. 34: 953–964. [DOI] [PubMed] [Google Scholar]
- Spoering A., 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 188: 5136–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson P. A., Norton I. L., 1983. Respiration shutoff in Escherichia coli K12 strains is induced by ultraviolet radiation and by mitomycin C. Mutat. Res. 139: 107–110. [DOI] [PubMed] [Google Scholar]
- Swenson P. A., Joshi J. G., Schenley R. L., 1978. Regulation of cessation of respiration and killing by cyclic 3′,5′-adenosine monophosphate and its receptor protein after far-ultraviolet irradiation of Escherichia coli. Mol. Gen. Genet. 159: 125–130. [DOI] [PubMed] [Google Scholar]
- Unoson C., Wagner E., 2008. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 70: 258–270. [DOI] [PubMed] [Google Scholar]
- Veaute X., Delmas S., Selva M., Jeusset J., Le Cam E., et al. , 2005. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 24: 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloshin O. N., Camerini-Otero R. D., 2007. The DinG protein from Escherichia coli is a structure-specific helicase. J. Biol. Chem. 282: 18437–18447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.