Abstract
Pseudomonas fluorescens is a model for the study of adaptive radiation. When propagated in a spatially structured environment, the bacterium rapidly diversifies into a range of niche specialist genotypes. Here we present a genetic dissection and phenotypic characterization of the fuzzy spreader (FS) morphotype—a type that arises repeatedly during the course of the P. fluorescens radiation and appears to colonize the bottom of static broth microcosms. The causal mutation is located within gene fuzY (pflu0478)—the fourth gene of the five-gene fuzVWXYZ operon. fuzY encodes a β-glycosyltransferase that is predicted to modify lipopolysaccharide (LPS) O antigens. The effect of the mutation is to cause cell flocculation. Analysis of 92 independent FS genotypes showed each to have arisen as the result of a loss-of-function mutation in fuzY, although different mutations have subtly different phenotypic and fitness effects. Mutations within fuzY were previously shown to suppress the phenotype of mat-forming wrinkly spreader (WS) types. This prompted a reinvestigation of FS niche preference. Time-lapse photography showed that FS colonizes the meniscus of broth microcosms, forming cellular rafts that, being too flimsy to form a mat, collapse to the vial bottom and then repeatably reform only to collapse. This led to a reassessment of the ecology of the P. fluorescens radiation. Finally, we show that ecological interactions between the three dominant emergent types (smooth, WS, and FS), combined with the interdependence of FS and WS on fuzY, can, at least in part, underpin an evolutionary arms race with bacteriophage SBW25Φ2, to which mutation in fuzY confers resistance.
Keywords: adaptive radiation, phenotypic diversification, mutation spectra, phage–host coevolution
ADAPTIVE radiation—the rapid emergence of phenotypic and ecological diversity within an expanding lineage—is among the most striking of evolutionary phenomena (Darwin 1859; Lack 1947; Dobzhansky 1951; Simpson 1953; Schluter 2000; Kassen 2009; Losos 2010). Fueled by competition and facilitated by ecological opportunity, successive radiations have shaped much of life’s diversity. Of central importance are the phenotypic innovations that fashion the fit between organism and environment (Schluter 2000).
Understanding the nature of these innovations and the pathways by which they emerge and rise to prominence—often in parallel across estranged populations experiencing similar environments—is a central issue (Colosimo et al. 2005; Bantinaki et al. 2007; Conte et al. 2012). How mutational processes generate the variation presented to selection (McDonald et al. 2009; Braendle et al. 2010), how genetic architecture underpinning extant phenotypes determines the capacity of lineages to generate new and adaptive phenotypes (Poole et al. 2003; Wagner and Zhang 2011), and how ecological factors drive phenotypic divergence (Schluter 2009) are questions of seminal interest.
The relative simplicity of microbial systems, their capacity for rapid evolutionary change, and advances in technology that enable detailed genotypic traceability offer a unique opportunity to log moment-by-moment change in experimental populations, with the potential to contribute to understanding of how genotype shapes phenotype, how phenotype interplays with the environment, and how environment may shape genotype (Mortlock 1982; Lenski et al. 1998; Rainey et al. 2000; Beaumont et al. 2009; Buckling et al. 2009; Blount et al. 2012; Hindré et al. 2012). In the context of adaptive radiation, the bacterium Pseudomonas fluorescens SBW25 has served as a useful model (reviewed within MacLean 2005 and Kassen 2009). When propagated in a spatially structured environment, SBW25 rapidly diversifies into a range of niche specialist genotypes. Smooth (SM) genotypes colonize the broth of static broth microcosms; wrinkly spreader (WS) genotypes colonize the air–liquid (AL) interface in the form of a self-supporting biofilm (or “mat”), via the overexpression of an acetylated cellulose-like polymer (ACP); and fuzzy spreaders (FS) form large, spreading colonies and appear to occupy the microcosm floor (Rainey and Travisano 1998). These three dominant types and their variants are maintained over time in serially propagated microcosms by the virtue of negative frequency-dependent interactions: each genotype is fittest when rare and fallible when common.
Previous work has defined the underlying mutational bases of WS genotypes (Spiers et al. 2002; Goymer et al. 2006; Bantinaki et al. 2007; McDonald et al. 2009), identified a set of mutational pathways that evolution repeatedly follows to generate this adaptive type (Rainey and Rainey 2003), and explained why evolution follows a subset of all possible pathways (McDonald et al. 2009). But what of fuzzy spreader? These morphotypes take longer to appear, but like WS, they arise repeatedly (Rainey and Travisano 1998). While WS is well understood, little is known about the ecology and genetics of FS. Here we describe the mutational causes of the FS phenotype, delineate the genetic route from genotype to phenotype to fitness, and unravel the links between FS physiology and the causes of its ecological specialization.
Materials and Methods
Bacterial strains, growth conditions, and manipulation
The ancestral (wild-type) SM strain is P. fluorescens SBW25 (PBR368) that was isolated from the leaf of a sugar beet plant grown at the University Farm, Wytham, Oxford, in 1989 (Rainey and Bailey 1996). The niche-specialist archetypal FS genotype (PBR367) was derived from the ancestral genotype after 7 days of selection in a spatially structured microcosm (Rainey and Travisano 1998). The niche-specialist WS genotype (PBR366; wspF236insGACCGTC) was derived from the ancestral genotype after 3 days of selection in a spatially structured microcosm (Rainey and Travisano 1998). The awsX mutant WST (PBR656; awsXΔ229-261) and the mwsR mutant WSZ (PBR662; mwsRG3055A) were similarly derived (McDonald et al. 2009). SBW25-Tn7-lacZ (PBR760), FS-Tn7-lacZ (PBR761), SBW25-fuzYT443G::Tn7-lacZ (PBR762), SBW25ΔfuzY::Tn7-lacZ (PBR763), and FS-fuzYwt::Tn7-lacZ (PBR764) derivatives used in Figure 2 competition assays carry a chromosomal insertion of mini-Tn7-lacZ (Choi et al. 2005). The strain FS-lacZ (PBR742) used in competition assays has a promoterless ′lacZ inserted within the defective prophage locus of the chromosome (Zhang and Rainey 2007). Both the ′lacZ and Tn7-lacZ markers are neutral with respect to fitness. A full list of strains and plasmids used in this study is given in Supporting Information, Table S1. The 112 independent FS genotypes FS3–FS214b were obtained by propagating the ancestral SM genotype (PBR368) in 214 independent spatially structured microcosms (Table S2). Microcosms were diluted and plated onto King’s medium B (KB) agar after 5 days incubation and (except where otherwise indicated) a single FS genotype was selected at random from each microcosm. These genotypes were streaked for single colonies to check purity and then stored at −80°. In cases where two or more distinct fuzzy-like morphotypes were apparent, representatives of each genotype were selected and stored; these are indicated by “a”, “b”, etc. (Table S2).
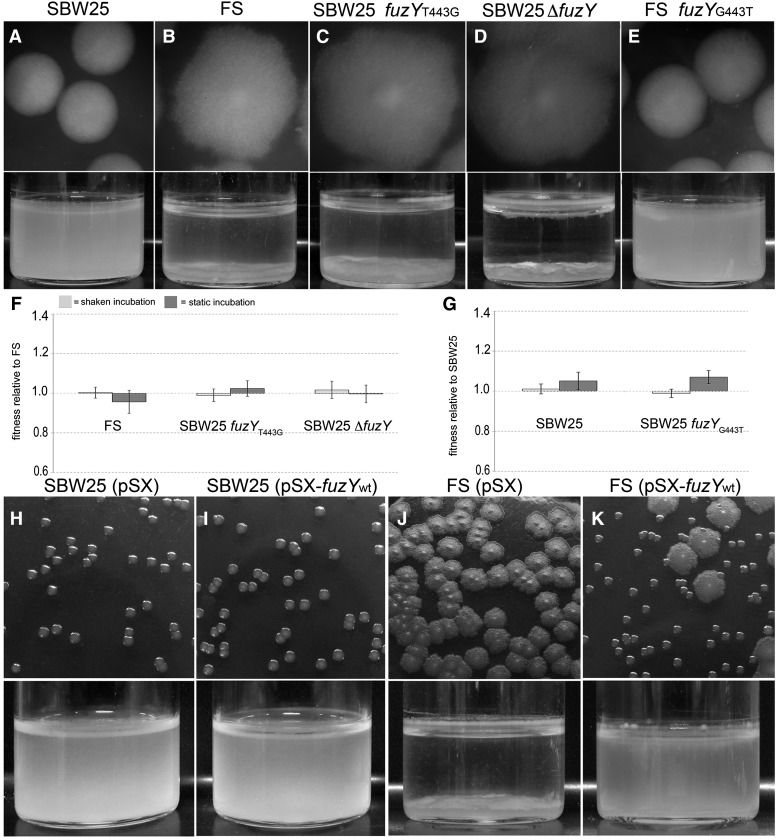
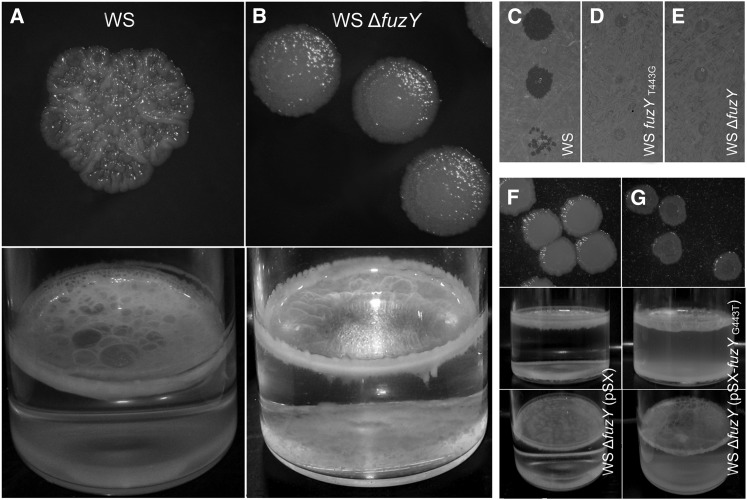
Figure 2.
Mutation of fuzY causes the fuzzy spreader (FS) phenotype. (B–D) fuzYT443G and ΔfuzY mutations reconstructed in SBW25 gave a phenotype indistinguishable from FS. (A and E) Reconstruction of the wild-type fuzY sequence in FS gave a phenotype indistinguishable from SBW25. Top panels show 48-hr colonies on KB plates. Bottom panels show growth after 48 hr of incubation in structured KB microcosms. (F and G) Fitness of the reconstructed mutants was measured relative to lacZ-marked derivatives of FS (F) or SBW25 (G) (see Materials and Methods) with either shaken (bars with light shading) or static incubation (bars with dark shading). No statistically significant fitness differences were measured between native and reconstructed FS mutants under any conditions [one-way ANOVA: P = 0.5048 (shaken), P = 0.4406 (static)]. Similarly, fitness differences between native and reconstructed SBW25 were not statistically significant [Dunnett’s test: P = 0.0745 (shaken), P = 0.5727 (static)]. Fitness measures are a ratio of Malthusian parameters and are the mean of six independent replicates; error bars show standard error of the mean. (H–K) fuzYT443G (carried by FS) was complemented by overexpression of the wild-type fuzY gene in trans from the vector pSX.
P. fluorescens strains were cultured at 28° in KB (King et al. 1954). Escherichia coli strains were grown in LB at 37°. Antibiotics and supplements were used at the following concentrations: ampicillin, 100 μg·ml−1; cycloserine, 800 μg·ml−1; gentamicin, 10 μg·ml−1; kanamycin, 100 μg·ml−1; spectinomycin, 100 μg·ml−1; and tetracycline, 20 μg·ml−1. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) was added at a concentration of 60 μg·ml−1. N-[5-nitro-2-furfurylidene]-1-arminutesohy-dantoin was used to counterselect E. coli (100 μg·ml−1 in agar plates). Plasmid DNA was introduced to E. coli by transformation and P. fluorescens by transformation or conjugation, following standard procedures. The helper plasmid pRK2013 was used to facilitate conjugative transfer between E. coli and P. fluorescens (Figurski and Helinski 1979).
Molecular biology techniques
Standard molecular biology techniques were used throughout (Sambrook et al. 1989). Gateway technology (vector pCR8; Invitrogen, Carlsbad, CA) was used to clone PCR fragments. The fidelity of all cloned fragments was checked by DNA sequencing (Macrogen, Seoul, South Korea). Oligonucleotide primers for allelic replacements and reconstructions were designed on the basis of the SBW25 genome sequence (Silby et al. 2009).
Construction of deletion mutants and allelic replacements
A two-step allelic exchange strategy was used as described previously (Rainey 1999; Bantinaki et al. 2007). For the generation of deletion mutants, oligonucleotide primers were used to amplify ∼1000-nucleotide regions flanking the gene(s) of interest. A third PCR reaction was used to join flanking sequences by splicing overlap extension (Horton et al. 1989). The product was cloned into pCR8 (and sequenced to check for errors); the resulting fragment was excised using BglII or SpeI and introduced into the suicide vector pUIC3 (Rainey 1999). Deletion constructs were mobilized into P. fluorescens by conjugation and allowed to recombine with the chromosome by homologous recombination. Recombinants from which pUIC3 had been lost were identified from LB agar plates after a brief period of nonselective growth in KB broth (Kitten et al. 1998). To generate allelic replacements, PCR fragments (∼3 kb) centered upon the allele of interest were cloned as above and introduced into pUIC3, and the replacement was made by recombination. Deletion and allelic exchange mutants were confirmed by PCR; allelic exchange mutants were further checked by DNA sequencing.
Gene complementation
A DNA fragment encoding fuzY (pflu0478) flanked by NdeI and BamHI restriction sites was amplified by PCR and ligated with pCR8. The resultant plasmid was purified from E. coli transformants and the integrity of the insert confirmed by Sanger sequencing (Macrogen). For expression in P. fluorescens, fuzY was excised from pCR8 and ligated between the NdeI and BamHI sites of expression vector pSX encoding lacIq and an IPTG-inducible tac promoter (Owen and Ackerley 2011). P. fluorescens transformants were purified by streaking to single colonies and the presence of the desired construct was confirmed by PCR.
Fitness of genotypes
Competitive fitness of FS genotypes was determined by direct competition between each FS genotype and FS (PBR367), FS-lacZ (PBR742), or FS-Tn7-lacZ (PBR761). Competitive fitness of SM genotypes was determined by direct competition between each SM genotype and SM (PBR368), SBW25-lacZ (PBR743), or SBW25-Tn7-lacZ (PBR760). Figure 2 fitness assays composed of six independent replicate contrasts used reciprocal strain marking as follows: in three replicate assays the reference strain (SBW25 or FS) was marked with mini-Tn7-lacZ, and in three replicate assays the reconstructed mutant strain was marked with mini-Tn7-lacZ, thus nullifying any effect of the marker on competition. Competitive fitness of WS genotypes was determined by direct competition between each WS genotype and WS mutant LSWS-Tn7-lacZ (wspFA901C; PBR741). All strains were grown overnight in shaken KB broth before introduction into spatially structured or unstructured microcosms (∼105 cells of each competitor) at a ratio of 1:1. Relative fitness was calculated as the ratio of Malthusian parameters of the genotypes being compared (Lenski et al. 1991).
Experimental evolution
Cells from −80° glycerol stocks were streaked onto KB agar plates to produce isolated single colonies. Populations were founded by inoculation of 6 ml KB broth in glass microcosms with a single representative colony from these KB plates; replicate populations were derived from independent single colonies. Populations were incubated at 28° in structured (static) microcosms. Propagation of populations occurred at regular intervals (of duration stated in Results) by transfer of a mixed sample (6 µl) to a fresh microcosm. At the time of each transfer, samples were spread onto KB plates to screen for colonies (∼500–1500) with a different morphology (for example, WS or FS or SM, as stated in Results). Representative colonies of new emergent types were streaked to single colonies on KB plates to confirm heritability and purity prior to propagation in liquid culture and storage at −80°.
Sequencing and sequence analysis
Whole-genome resequencing of the entire 6.7-Mbp genome of the archetypal FS isolate (PBR367) was achieved using Illumina Solexa technology. Paired-end sequencing was performed in two channels for 76 cycles on an Illumina GAIIx with a paired-end module. The data were analyzed using the Illumina OLB pipeline v1.8: the total cluster number was 58,919,811 with 38,328,187 clusters passing filter (65%). Reads were aligned to the P. fluorescens SBW25 genome sequence (Silby et al. 2009), using SOAP2; a single SNP (T443G) was identified after extracting all SNPs from regions covered by >10 sequence reads with a PHRED quality >20 and where 95% of the aligned nucleotides agreed. The presence of the T443G SNP in FS and its absence in SBW25 was confirmed by Sanger sequencing (Macrogen). Analyses of regions upstream and downstream of fuzY, including the identification of orthologous genes in Pseudomonas species, were performed using Artemis 14.0.0 (Rutherford et al. 2000) and the Generic Genome Browser (GBrowse) version 2.37 (Stein et al. 2002) within the Pseudomonas Genome Database (Winsor et al. 2009).
fuzY from 112 independently isolated FS isolates was amplified by PCR and sequenced by the Sanger method (Macrogen). DNA sequences were analyzed using Sequencher 4.10.1 (Gene Codes) or Geneious 5.5.6 (Biomatters) software.
Phage resistance assays
Sensitivity of SBW25 mutants to bacteriophage SBW25Φ2 was determined by pipetting 5-μl droplets of diluted phage lysate onto lawns of bacterial cells overlaid onto KB plates. Bacterial overlays were prepared by mixing 200 μl of bacterial cells from an overnight culture with 3 ml of molten top agar (KB with 0.8% agar). Phage sensitivity was scored as the presence of clear zones or individual plaques on bacterial lawns after 24-hr incubation at 28°.
Imaging
Niche preference in structured microcosms:
Six ml of KB broth in glass microcosms was inoculated with ∼5 × 105 bacteria and incubated at 28° without shaking for 48 hr (unless otherwise indicated). Structured microcosms were photographed with a Canon PowerShot G7 camera.
Movies of bacterial growth in structured microcosms over time were produced:
Six ml of KB broth in glass microcosms was inoculated with ∼5 × 105 bacteria and incubated at 28° without shaking. Bacterial growth was recorded by time-lapse photography, using a Canon PowerShot G7 camera and PSRemote (Version 1.9.1) Camera Control software (Breeze Systems, Camberley, Surrey, UK): photographs were taken at 5- or 10-min intervals over 24- to 72-hr periods and compiled into movies, using Microsoft Movie Maker (Version 5.1).
Colony morphology:
Bacteria from a 106-fold dilution of an overnight KB culture were spread onto KB plates and incubated at 28° for 48 hr. Colony morphology was examined and photographed using a Zeiss Stemi 2000-C dissection microscope fitted with a Canon PowerShot A640 digital camera.
Results
The fuzzy spreader phenotype is caused by a single nonsynonymous mutation in a putative lipopolysaccharide modification gene
The FS morphotype has proved intractable to standard genetic analysis. Repeated attempts to locate the causal loci by suppressor analysis have failed. We therefore sequenced the genome of an archetypal FS mutant (PBR367). Comparison of the FS genome with the SBW25 reference genome identified a single-nucleotide polymorphism within pflu0478 (a T to G transversion at position 443), a gene predicted to encode a putative glycosyltransferase involved in modification of surface lipopolysaccharides (LPS). The SNP caused the substitution of valine for glycine at position 148 (Pflu0478V148G). pflu0478 is located in a predicted operon of five genes (pflu0475–pflu0479) that are likely transcribed as a single unit (Figure 1). These genes are hereafter designated as fuzVWXYZ for “fuzzy spreader locus”. The predicted functions of these gene products relate to LPS modification: fuzV (537,073–538,830) encodes a predicted carbamoyl transferase (NodU family), fuzW (538,831–539,727) encodes a hypothetical protein with a predicted acetyltransferase domain (NAT_SF superfamily, E-value: 2.31e-10) with significant similarity to the Mig-14 family of antimicrobial resistance proteins (E-value: 2.03e-150), fuzX (539,697–541,124) encodes a predicted deacetylase, fuzY (541,184–542,332) encodes a predicted β-glycosyltransferase (GT1 group), and fuzZ (542,329–543,210) encodes a predicted α-glycosyltransferase (GT2 group). The fuz locus was previously identified in a suppressor analysis of WS types and shown to be required for expression of the WS phenotype (McDonald et al. 2009), indicating a genetic link between WS and FS phenotypes. A summary of the diguanylate cyclase (DGC)-encoding pathways underpinning the evolution of the WS phenotype is given in Figure S1.
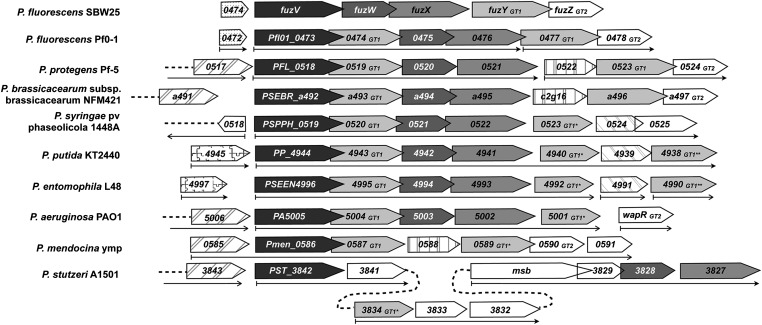
Figure 1.
Comparison of the arrangement of genes at the fuzzy spreader locus in Pseudomonas species. The predicted fuzzy spreader operon in P. fluorescens SBW25 comprises five genes with predicted functions relating to LPS modification: fuzV (537,073–538,830) encodes a predicted carbamoyl transferase (NodU family), fuzW (538,831–539,727) encodes a putative transcriptional regulator (Mig-14 family), fuzX (539,697–541,124) encodes a predicted deacetylase, fuzY (541,184–542,332) encodes a predicted β-glycosyltransferase (GT1 family), and fuzZ (542,329–543,210) encodes a predicted α-glycosyltransferase (GT2 family). Many of the genes within this hypothetical operon have highly conserved orthologs within a range of Pseudomonas species. Orthologous genes were identified by Reciprocal Best Blast Analysis/Ortholuge performed upon data collated in the Pseudomonas Genome Database and are grouped by shade (or pattern). * and ** indicate glycosyltransferases of different GT1 ortholog groups. Open boxes indicate genes with no orthologs within the regions depicted. Overlapping boxes indicate ORFs that initiate within the coding sequence of the upstream ORF. Thin arrows indicate computationally predicted operons from the Database of prOkaryotic OpeRons (DOOR), where available.
Orthologs of fuzVWXYZ were identified across a range of Pseudomonas species (Figure 1). The structure of the 5′ end of the operon (fuzVWX) is highly conserved across all species, except for the more distantly related P. stutzeri. It is likely that the ancestral operon included an additional GT1-group glycosyltransferase between fuzV and fuzW. In contrast, the 3′ end of the operon is more fluid: for most species, orthologs of fuzVWX are coupled with an assortment of glycosyltransferases.
The region upstream of fuzV is poorly conserved. Orthologs of the upstream genes are present in many pseudomonads but only in P. fluorescens SBW25 and Pf0-1 is there juxtaposition of pflu0474—encoding a short hypothetical protein of unknown function—and fuzV (Figure 1); in most other species pflu0474 is linked to the permease component of a putative ABC-type multidrug transport system. This indicates that pflu0474 is not a component of the fuzzy spreader operon.
Loss-of-function mutations in fuzY generate fuzzy spreaders
To test whether the T443G substitution is alone sufficient to cause the FS phenotype, the mutation was reconstructed in a naive SBW25 background. The resultant mutant—SBW25-fuzYT443G (PBR744)—expressed a fuzzy colony morphology on KB plates and exhibited the archetypal fuzzy spreader growth pattern within structured microcosms (Figure 2, A–C). Similarly, reconstruction of the wild-type sequence in FS (FS-fuzYwt, PBR752) generated a mutant with a smooth colony morphology that grew within the broth phase (Figure 2E). To determine whether the FS phenotype is a consequence of loss of function, fuzY was cleanly deleted, taking care to retain the first codon of fuzZ: SBW25ΔfuzY (PBR745) also displayed the typical FS colony morphology and pattern of growth in structured microcosms (Figure 2D). Both SBW25-fuzYT443G and SBW25ΔfuzY were comparable to FS in fitness when competed against a marked FS derivative in both shaken (unstructured) and static (spatially structured) microcosms (Figure 2, F and G).
If the FS phenotype is the simple consequence of a loss-of-function mutation in fuzY, restoration of the ancestral phenotype should be achieved by expression of fuzY in trans. Indeed, complementation of fuzYT443G by overexpression of fuzYwt from vector pSX (PBR753) resulted in restoration of the ancestral phenotype (Figure 2, J and K). Overexpression of fuzYwt in ancestral SBW25 (PBR751) produced no discernible effect on either phenotype or growth (Figure 2, H and I).
A single genetic route confers the fuzzy spreader phenotype through a diverse range of fuzY mutations
To determine whether mutation of fuzY is the sole cause of all fuzzy morphotypes we isolated 112 FS and FS-like morphotypes from independent spatially structured microcosms and classified them on the basis of colony morphology. By visual inspection, two distinct classes of spreading colonies, fuzzy in appearance, were discernible. “Typical” fuzzy spreaders (88 isolates) were rough textured and opaque in appearance, with irregular colony margins. “Atypical” fuzzy spreaders (aFS) (24 isolates) were of a similar form, but differed from the typical class in colony opacity and/or texture. Similarly, an inspection of niche occupancy revealed differences between the classes: FS types failed to grow within the broth phase, appearing as a “volcano-like” biomass on the microcosm floor (as in Figure 2, B and C). In contrast, aFS types grew within the broth phase, with many producing weak mats.
For each of the 112 isolates, we sequenced fuzY: 92 isolates harbored mutations; of these, 88 were of the typical class, and 4 were of the aFS class. The remaining 20 genetically uncharacterized isolates were of the aFS class and phenotypically distinct from FS. These could be further partitioned into two groups on the basis of their susceptibility to bacteriophage SBW25Φ2 (Table S2). Thus, aFS aside, the mutational basis of the FS phenotype appears to be simple, with a diverse range of mutations from SNPs causing nonsynonymous substitutions or nonsense mutations to insertions and deletions (indels) of various lengths causing frameshifts throughout the length of the gene (Table 1, Table S2).
Table 1. Spectrum of fuzY mutations isolated.
| Mutational class (bp) | Unique mutations (no. mutants) |
|---|---|
| SNPs | |
| Transitions | 10 (14) |
| AT > GC | 2 (2) |
| GC > AT | 8 (12) |
| Transversions | 13 (40) |
| AT > CG | 1 (28) |
| GC > TA | 6 (6) |
| AT > TA | 2 (2) |
| GC > CG | 4 (4) |
| Indels | |
| +1 insertions | 5 (6) |
| +2 insertions | 2 (2) |
| −1 deletions | 10 (10) |
| −2 deletions | 4 (4) |
| Small insertions (5–8) | 2 (2) |
| Large insertions (951) | 2 (2) |
| Small deletions (3–6) | 4 (4) |
| Large deletions (24–3373) | 7 (8) |
| Total | 59 (92) |
Of the 112 independent FS and FS-like mutants isolated, 92 harbored mutations within fuzY. The mutations ranged from SNPs causing missense or nonsense mutations to small and large deletions and insertions (indels) causing frameshifts throughout the length of the gene. Numbers not in parentheses indicate the number of unique mutations isolated, while numbers in parentheses indicate the total number of mutants isolated for each class. T443G occurred 28 times (31% of all mutants), C565T occurred twice, C622T occurred three times, G857A occurred twice, and Δ882–947 occurred twice. Of the seven large deletions, two extended into fuzZ and one deleted all of fuzX and fuzY and some of each of fuzW and fuzZ. Of the large insertions, one resulted from the duplication of adjacent sequence and the other from the insertion of a putative transposon encoded elsewhere in the genome. Mutant genotypes are listed in full in Table S2.
Of 92 independent fuzY causal mutations, 54 were SNPs at 23 unique positions (Table 1). Of the 23 unique SNPs, 10 were transitions (predominantly GC > AT) and 13 were transversions (predominantly GC > TA and GC > CG); 16 were missense mutations and 7 were nonsense. Interestingly, more than one-quarter of all the independently isolated fuzzy spreaders were caused by an identical SNP—the same T443G transversion carried by the resequenced “archetypal” isolate—causing valine to be substituted for glycine at position 148 in the protein (V148G). Similar single-site mutation biases have been reported elsewhere and explained by replication template switching within imperfect repeat (quasi-palindromic) sequences during DNA replication (De Boer and Ripley 1984; Mo et al. 1991; Demarini et al. 1998; Viswanathan et al. 2000; Lovett 2004). A comparison of the DNA sequence surrounding the T443G “hotspot” with nearby sequences failed to identify any imperfect palindromic matches likely to template a T > G substitution at position 443.
Of the 92 mutations, 28 were small indels at 27 unique positions. The majority of these can be explained by nascent strand misalignment [slipped-strand mispairing (Levinson and Gutman 1987)] within monotonic nucleotide “runs” (14 of 28), by duplication of adjacent sequence (2 of 28), or by deletion of a directly repeated sequence (2 of 28). How the remaining 12 small indels were created is not clear but, notably, one mutant (FS126a; Table S2) had acquired a single-base-pair insertion producing a perfect 5-bp inverted repeat of the adjacent sequence, potentially a consequence of intramolecular template switching (Lovett 2004). Five of 92 mutants were caused by large deletions within fuzY [Δ629–760, Δ775–896, Δ862–885, and Δ882–947 (which occurred twice)]; two of 92 mutants had deletions extending from the 3′ end of fuzY into fuzZ; and one mutant had a deletion extending from the 3′ end of fuzW to the 5′ half of fuzZ. The remaining mutant was caused by the insertion of a transposon encoded elsewhere in the genome.
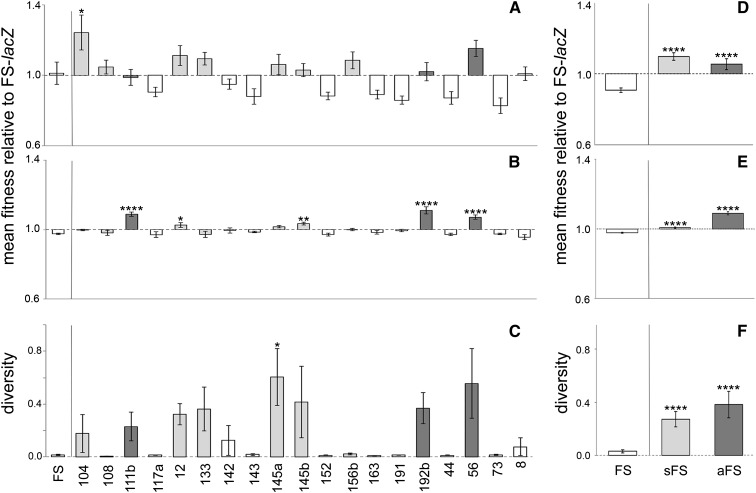
Typical FS isolates are indistinguishable from each other in terms of colony morphology; however, subtle differences in phenotype are conferred by different mutations within fuzY. When the independent FS isolates were screened for niche preference in spatially structured microcosms, most isolates were indistinguishable from the archetypal FS (fuzYT443G; PBR367). However, a subset of seven FS isolates exhibited a small amount of growth within the broth phase in addition to producing biomass on the microcosm floor. The mutations responsible for these “subtle” isolates (sFS) were located in two or three distinct regions of the gene: codons 90–144, codon 221, and codon 271—four were missense substitutions and three were insertions of 1, 8, and 126 nucleotides, respectively. Three of the four aFS isolates with mutations within fuzY carried missense substitutions—two located close together (A316E and G318D) and a third (K213R) located nearby sFS mutant A221E (Table S2). The fourth aFS mutant had a large deletion extending from fuzW into fuzZ.
To further explore the possible phenotypic differences among fuzY mutants, we carried out fitness assays on a selection of independent FS mutations (Table 2), including the seven sFS genotypes and three of the aFS genotypes. Small fitness differences were detected between some pairs of fuzY mutants when competed in spatially structured microcosms; however, the variance between replicates—attributable to the tendency of the competitors to diversify into a range of niche specialists over the 3-day period of the assay—was large (Figure 3A). Competition assays performed in unstructured microcosms, by way of a comparison, revealed a greater number of statistically significant fitness differences (Figure 3B). In general, mutants differing in fitness to archetypal FS generally corresponded to those with sFS or aFS phenotypes. However, fitness measured in unstructured microcosms mapped poorly to that measured in structured microcosms. Surprisingly, we observed significant differences in the ability of some fuzzy isolates to diversify (predominantly into a range of WS types) during static incubation (Figure 3C); this subset of rapidly diversifying types belongs exclusively to the aFS and sFS classes and in some instances these correspond to those with significantly different fitness as measured under shaken incubation (Figure 3B). When fitness and diversity measures were pooled by mutant class (FS vs. sFS vs. aFS), the trends were marked: for competitions in structured microcosms, aFS and sFS were equally fit and significantly fitter than FS (Figure 3D). For competitions in unstructured microcosms, aFS were significantly fitter than sFS, which were significantly fitter than FS (Figure 3E). Similarly, when the propensity of isolates to diversify was compared by mutant class, the sFS and aFS groups diversified to a significantly greater extent than the FS group (Figure 3F).
Table 2. Description of fuzY mutations within independent FS isolates included in Figure 3.
| Isolatea | Class | Nucleotide change | Amino acid change |
|---|---|---|---|
| 8 | FS | Δ4 (−1 Fsb) | M1Δ (37)c |
| 73 | FS | Δ28–33 | ΔLQ |
| 104 | sFS | 268insT (+1 Fs) | F90Δ (4) |
| 133 | sFS | C288G (Tvb) | N96K |
| 156b | sFS | G352T (Tv) | G118W |
| 145b | sFS | T376A (Tv) | Y126N |
| 108 | sFS | 432insCCTGGCGG | R144Δ (14) |
| FS | FS | T443G (Tv) | V148G |
| 152 | FS | Δ568–569 (−2 Fs) | A190Δ (93) |
| 143 | FS | G620T (Tv) | G207V |
| 56 | aFS | A638G (Tsb) | K213R |
| 163 | FS | Δ638–639 (−2 Fs) | I212Δ (71) |
| 12 | sFS | C662A (Tv) | A221E |
| 117a | FS | fuzYΔ770–1149, fuzZΔ1–220 | T256Δ (220), ΔFuzZ |
| 145a | sFS | 809ins126nt (duplication of 126 nt immediately upstream of insertion point) | Y271Δ (153) |
| 44 | FS | G857A (Ts) | G286D |
| 191 | FS | 951ins Tn pflu2158, pflu4347, pflu5832, or pflu4873 | G318Δ (10) |
| 192b | aFS | C947A (Tv) | A316E |
| 111b | aFS | G953A (Ts) | G318D |
| 142 | FS | 1059insT (+1 Fs) | R353Δ (19) |
Isolates are ordered by position of mutation (5′–3′ direction); mutant genotypes are listed in full in Table S2.
Fs, frameshift; Tv, transversion; Ts, transition.
Numbers in parentheses indicate the number of amino acid residues from the mutation site until the predicted STOP codon.
Figure 3.
Relative fitness and diversification of independent FS isolates after 3 days incubation in spatially structured or unstructured environments. Fitness was measured relative to a lacZ-marked derivative of FS (PBR742). Typical FS mutants (open bars) display the archetypal fuzzy colony morphology and accumulate at the bottom of structured microcosms. Subtle FS mutants (bars with light shading) display the archetypal fuzzy colony morphology and accumulate at the bottom of microcosms but also grow to a degree within the broth phase. Atypical FS mutants (bars with dark shading) display colony morphology intermediate between FS and SM and grow within the broth phase of structured microcosms. (A–E) Fitness measures are a ratio of Malthusian parameters and are the mean of six independent replicates; error bars show standard error. Asterisks indicate statistically significant contrasts in comparison to the archetypal FS isolate PBR367 (multiple-comparisons tests as shown: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). For competitions within spatially structured environments (A), differences in mean fitness for some pairs of mutants were statistically significant (Welch’s ANOVA, P = 0.0001), although only mutant 104 was significantly fitter than the archetypal FS (P = 0.0109, Dunnett’s multiple-comparisons test). For competitions within unstructured environments (B), differences in mean fitness for some pairs of mutants were statistically significant (Welch’s ANOVA, P = 0.0001); mutants 111b, 12, 145b, 192b, and 56 were significantly fitter than the FS archetype (P < 0.0001, = 0.034, = 0.0075, < 0.0001, < 0.0001, respectively, Dunnett’s multiple-comparisons test). (C) Population diversification was measured as the proportion of colonies whose morphology deviated from the ancestral type after 72 hr static incubation; values represent a mean of six independent replicates and error bars show standard error. Differences in diversification for some pairs of mutants were statistically significant (Welch’s ANOVA, P ≤ 0.0281), although only mutant 145a diversified to a significantly greater extent than the archetypal FS (P = 0.0025, Dunn’s multiple-comparisons test). When the data for individual mutants were pooled by FS classification, the contrasts were strongly significant [(D–F) Welch’s ANOVA: P < 0.0001]. For competitions in spatially structured environments (D), the sFS and aFS groups were of equal fitness but each was significantly more fit than the FS group (P < 0.0001, Dunnett’s multiple-comparisons test). For competitions in unstructured environments (E), each group mean was significantly different to every other group mean, with aFS being the fittest group and FS the least fit (P < 0.0001, Tukey’s multiple-comparisons test). Similarly, the sFS and aFS groups both diversified to a significantly greater extent than the FS group [(F) P < 0.0001, Dunn’s multiple comparisons test].
Taken together, the data suggest that although the majority of mutations within fuzY confer phenotypically indistinguishable fuzzy morphologies—most likely the consequence of loss of function of the enzyme—some fuzY mutations confer a more subtle phenotype. It is likely that the aFS and sFS groups express partially functional FuzY enzymes and consequently display cell surface properties intermediate to those of FS and SM. The greater propensity of aFS and sFS types to diversify in structured microcosms is likely attributable to the fact that the gene retains some functionality and that this functionality is required for expression of other niche specialist types, in particular WS.
Genes fuzVWXYZ encode phenotypically related functions, contributing to the modification of cell surface structures
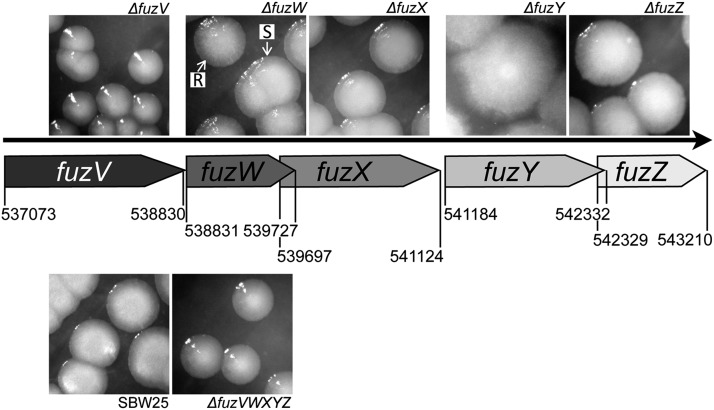
Having mapped the full spectrum of FS mutations exclusively to fuzY, we sought to determine the contribution of flanking genes. Deletion of the entire operon (fuzVWXYZ) generated a small-colony variant (Table 3, Figure 4). With the exception of fuzX, all single-gene deletion mutants gave rise to colonies with distinct phenotypes (Table 3, Figure 4); however, only deletion of fuzY generated the FS phenotype. Notable was the fuzW mutant, which reproducibly generated colonies of both “rough” (PBR746-R) and “smooth” morphotypes (PBR746-S) (Figure 4).
Table 3. Phenotypes of fuzVWXYZ single-gene knockout mutants.
| Mutant | Colony morphologya | Niche occupancyb (static microcosm) | SBW25Φ2 sensitivityc |
|---|---|---|---|
| Δ fuzV | Small, smooth | Broth phase (slow growth) | Resistant |
| Δ fuzW | Smooth (like SM) | Broth phase | Resistant (rough) |
| Rough | Microcosm floor | Sensitive (smooth) | |
| Δ fuzX | Smooth (like SM) | Broth phase | Sensitive |
| Δ fuzY | Fuzzy spreader | Microcosm floor | Resistant |
| Δ fuzZ | Large, smoothd | Broth phase | Resistant |
| Δ fuzVWXYZ | Small, smooth | Microcosm floor (slow growth) | Resistant |
Colony morphologies were scored after 48 hr growth on KB plates.
Niche occupancy was scored after 72 hr growth in 6 ml KB microcosms at 28°.
All mutants were tested for sensitivity to SBW25Φ2.
The fuzZ mutant appears as a fuzzy spreader when cultured on LBA plates.
Figure 4.
Colony morphologies of single fuz gene deletion mutants and a fuzVWXYZ deletion mutant. Deletion of the fuzVWXYZ operon produced a small-colony variant, distinct in phenotype from both FS and SM, indicating that one or more of the genes surrounding fuzY also contribute to cell surface characteristics. Each single-gene deletion (except for fuzX) produced a mutant with distinctly altered colony morphology, unlike either FS or SM (Table 3). Deletions were constructed in a SBW25 background via a two-step allelic exchange strategy as described previously (Rainey 1999; Bantinaki et al. 2007). Base numbers refer to the SBW25 genome sequence [NC_012660.1 (Silby et al. 2009)]. Images show 48-hr colonies on KB plates.
Revision of the FS niche preference
The explanatory model for the maintenance of niche specialist types arising during the course of the Pseudomonas radiation involves negative frequency-dependent interactions between SM, WS, and FS arising from the emergence of trade-offs due to diversifying selection. This model is based on data from reciprocal “invasion from rare” experiments combined with observation of the niche preference of the three dominant types: SM colonizes the broth phase, WS the AL interface, and FS the microcosm floor (Rainey and Travisano 1998). While the existence of frequency-dependent interactions among the primary types is firmly established, there is reason to reexamine the niche preference of FS. This stems from the fact that oxygen is removed from the broth phase by the founding genotype within the first 12 hr of growth (B. Ibelings and P. B. Rainey, unpublished data; Koza et al. 2010), leading to the seemingly contradictory observation that FS, evolved from the obligate aerobe SBW25, apparently grows in an environment with no oxygen. It is difficult to imagine how LPS modification might confer the property of anaerobic growth upon FS. To further investigate the connection between genotype and phenotype we revisited the question of FS niche occupancy. Structured microcosms were inoculated with FS cells and their growth was recorded by time-lapse photography (File S1). As expected, no substantial growth of FS took place in the broth phase, unlike that observed in microcosms seeded with ancestral SBW25 (File S2). Instead, the first visually discernible growth took place at the meniscus, with small rafts of cells observed to propagate both inward toward the center of the AL interface and downward in a stalactite-like fashion (File S1). As these rafts increase in size, they fall from the interface to the floor of the microcosm as snowflakes, but repeatedly reform. No substantial biofilm ever forms at the surface, but growth clearly occurs. Thus FS, like WS, specializes in colonizing the AL interface, albeit by a distinctively different strategy. Rather than forming a mat via the production of adhesive polymers (Spiers et al. 2002, 2003), FS forms transient rafts due to changes in a cell surface component.
To explore the basis of the fuzY-dependent raft-forming behavior the flocculating ability of FS cells was examined by growing cells for 3 days in structured microcosms (SM and WS were included as controls). Cells were then dispersed into suspension by vortex mixing and incubated statically for a further 24 hr, with the addition of tetracycline to prevent further growth. Time-lapse photography revealed rapid flocculation of FS cells with an almost complete clearing of the broth phase within 12 hr (File S3). In contrast, SBW25 and WS cells remained largely in suspension. From this we conclude that clumping is mediated by cell–cell attraction and is not a consequence of failure of mother and daughter cells to disperse following cell division. That flocculation requires specific interactions between FS cells is supported by the observation that the ancestral (SM) genotype does not flocculate and remains in suspension when mixed 1:1 with FS in statically incubated microcosms (data not shown).
Although FS grows poorly in the broth phase of structured microcosms, growth sometimes occurs after several days of incubation. As SBW25 is known to modify its growth environment with a resultant increase in pH over time (Table 4B), we speculated that the tendency for FS to form flocs might be pH dependent. To test the effect of pH on growth and flocculation, we recorded (again by time-lapse photography) the growth of FS across a range of pHs from 6.5 to 8.5 (File S4). At pH 8 and above, growth occurred predominantly in the broth phase. At pH 7.5, growth occurred initially at the AL interface, with rafts of cells falling to the microcosm floor, but after 24 hr the broth phase began to cloud with growth. Broth phase growth was less marked at pH 7 and did not occur at pH 6.5. The pH-dependent differences in FS growth were not due to an effect of pH on growth rate: populations were titered at 72 hr and cell density (as CFU⋅ml−1) did not differ between treatments (Table 4A). Similarly, postgrowth flocculation of dispersed cells occurred only at low pH (File S5). From this we conclude that the failure of FS to grow freely in the broth phase of structured microcosms is a consequence of flocculation following cell separation and that the particular cell surface characteristics that promote flocculation are charge mediated.
Table 4. The effect of pH on the growth of P. fluorescens.
| A. | pH | FS: mean CFU⋅ml−1 ± SEM (P-value) | SBW25 ΔfuzY: mean CFU⋅ml−1 ± SEM (P-value) |
|---|---|---|---|
| 6.5 | 8.7 × 108 ± 1.3 × 108 (0.0733) | 1.2 × 109 ± 5.3 × 108 (0.3213) | |
| 7.0 | 5.5 × 108 ± 4.9 × 107 | 5.7 × 108 ± 1.3 × 108 | |
| 7.5 | 4.9 × 108 ± 1.4 × 108 (0.7372) | 3.8 × 108 ± 1.3 × 108 (0.3578) | |
| 8.0 | 6.5 × 108 ± 1.3 × 107 (0.1084) | 6.7 × 108 ± 7.9 × 107 (0.5265) | |
| 8.5 | 5.9 × 108 ± 1.3 × 108 (0.7403) | 8.6 × 108 ± 2.9 × 108 (0.4047) | |
| B. | pH | FS: pH prior to growth | SBW25 ΔfuzY: pH after growth |
| 6.5 | 6.55 ± 0.01 | 8.21 ± 0.09 | |
| 7.5 | 7.60 ± 0.03 | 8.61 ± 0.10 | |
| 8.5 | 8.27 ± 0.03 | 8.86 ± 0.07 |
(A) Rows give the viable titer (as CFU⋅ml−1) of FS and SBW25 ΔfuzY in 6-ml KB microcosms of differing pH following 72 hr static incubation at 28°. (B) The pH of KB was measured postautoclaving but prior to inoculation with FS. Inoculated microcosms were incubated statically at 28° for 72 hr. Spent media were sterilized by filtration through 0.22-μm filters and the pH was again measured. Titers and pH measurements are a mean of three independent replicates and variance is given as the standard error of the mean. P-values are derived from unpaired t-tests comparing the titer at each pH with that at pH 7. No statistically significant differences were found for any of the contrasts.
fuzY mutation in WS results in a compromised ability to form mats
The fuzVWXYZ locus was previously identified as a suppressor of the WS phenotype in the large spreading wrinkly spreader (LSWS; wspFA901C) and alternate wrinkly spreader (AWS; awsX Δ100–138) mutants (McDonald et al. 2009; Figure S1), suggesting a genetic link between the WS and FS phenotypes. To explore this further, fuzYT443G and ΔfuzY mutations were reconstructed in a wspF236insGACCGTC background (WS; PBR366). The resulting mutants were compromised in their ability to form substantive mats at the AL interface: WS-fuzYT443G (PBR754) and WSΔfuzY (PBR755) mutant colonies show a decreased wrinkled appearance on KB plates and produce feeble mats (fWS) that readily collapse to the microcosm floor (Figure 5B). Comparison of mat growth over time revealed striking differences between WS and WS-fuzYT443G: WS mat formation initiates with the appearance of cell aggregates at the AL interface, which in time grow together to form a thin but robust film, interspersed with denser aggregates of rope-like growth (File S6). In contrast, WS-fuzYT443G, while also initiating growth at the AL interface, forms “rafts”, many of which fall to the microcosm floor. Over time these rafts expand to form a dense mat of surface growth, this, however, being unstable with the periodic appearance of “holes” as biomass drops to the microcosm floor. No networks of rope-like structures were observed. Mat strength was measured by the glass bead assay (Rainey and Rainey 2003). Whereas 3-day-old WS mats were able to support an average of 0.24 ± 0.07 g of 2-mm glass beads (0.01 g) before collapsing, WS-ΔfuzY mats were unable to support even a single bead. Wild-type mat formation and wrinkled colony morphology were restored to WS-ΔfuzY by overexpression of fuzYwt from pSX (PBR756; Figure 5, F and G). As with SBW25, overexpression of fuzYwt in WS (PBR757) produced no discernible effect on either phenotype or growth or the ability to support beads (data not shown).
Figure 5.
fuzY mutation in LSWS compromises mat integrity. (A and B) fuzYT443G (not shown) and ΔfuzY mutations reconstructed in WS (PBR366) produced colonies with a decidedly less wrinkled appearance and, although to the eye capable of forming a thick mat at the AL interface, produced weak mats incapable of supporting 2-mm glass beads (top panels show 48-hr colonies on KB plates; bottom panels show growth after 48-hr static incubation in KB microcosms). (C–E) Phage SBW25Φ2 can plaque on WS (C) but not on either WS-fuzYT443G (PBR754) (D) or WS ΔfuzY (PBR755) (E). (F and G) fuzYT443G was complemented by overexpression of the wild-type fuzY gene in trans from the vector pSX (top panels show 48-hr colonies on KB plates; middle and bottom panels show growth after 48-hr static incubation in KB microcosms).
In contrast to FS, the genetic routes to WS are many and varied, with mutation within three distinct diguanylate cyclase-containing pathways (Wsp, Aws, and Mws) conferring cellulose overproduction and a mat-forming phenotype (McDonald et al. 2009; Figure S1). However, deletion of fuzY had the same phenotypic effect on each of three distinct WS genotypes (PBR755, PBR758, and PBR759; data not shown). Thus, the putative LPS defect caused by loss of FuzY function has consequences for the downstream effect of the WS mutations, most likely the secretion and arrangement of acetylated cellulose polymers, rather than a specific interaction with the particular genes or protein components encoded by the wsp operon (Spiers and Rainey 2005).
fuzYT443G confers resistance to SBW25Φ2
A previously noted feature of fuzzy spreader genotypes is their resistance to the lytic bacteriophage SBW25Φ2. Droplets of phage suspension produce clear zones on lawns of SBW25 but no clearing is observed on lawns of FS (Figure 6). All typical FS isolates, including the sFS subclass and SBW25ΔfuzY, are phage resistant (Table S2). Interestingly, the three broth-dwelling aFS isolates with mutations within fuzY are also phage resistant. We conclude that all (known) mutations in fuzY confer resistance to phage, regardless of any other effects of the mutations on niche preference. Single deletions of genes fuzVWX and fuzZ (but not fuzX) also conferred resistance to SBW25Φ2 (Table 3). Rough fuzW mutants (PBR746-R) are phage resistant whereas smooth fuzW mutants (PBR746-S) are phage sensitive.
Figure 6.
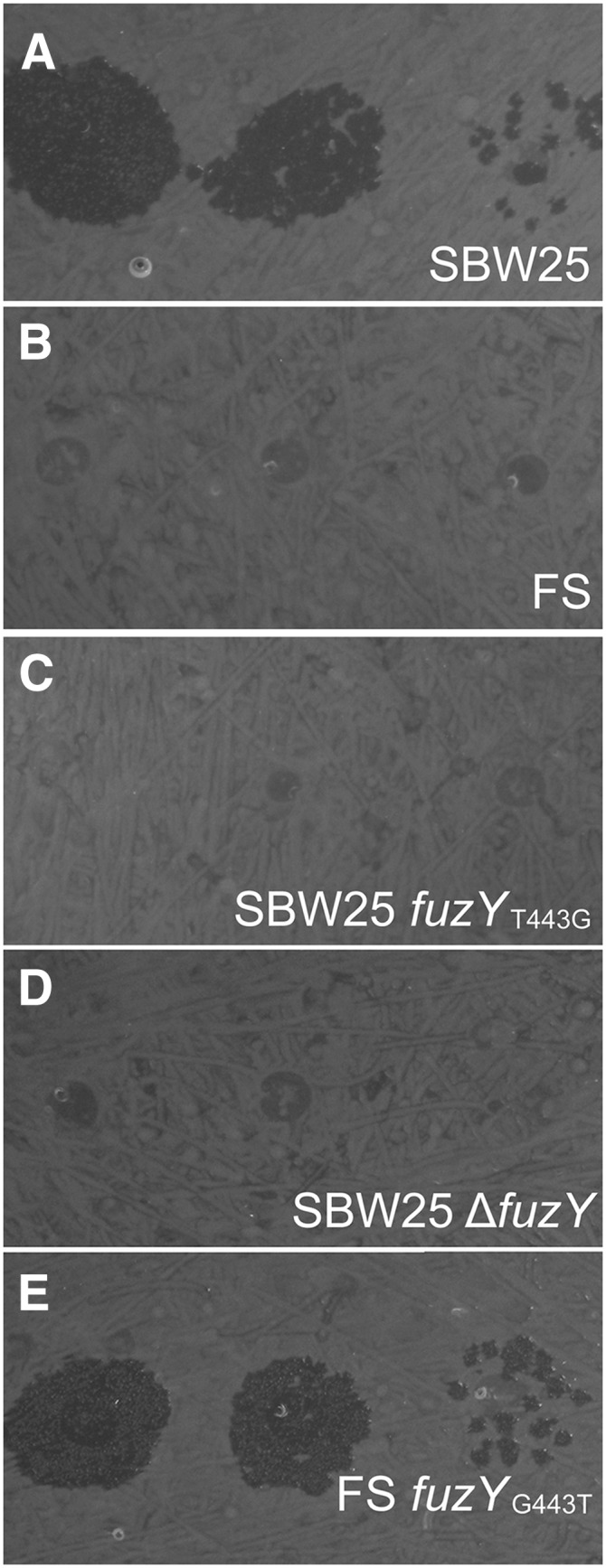
fuzYT443G confers resistance to SBW25Φ2. (A–E) Phage SBW25Φ2 forms plaques on P. fluorescens SBW25 (A) and the reconstructed wild-type FS-fuzYG443T (E) but is unable to form plaques on fuzY mutants (B–D).
Location of the FS causal mutation to a gene product predicted to modify LPS allows us to speculate upon the Φ2 receptor: LPS serves as a receptor for a broad range of bacteriophage in many bacterial species (for examples see Michel et al. 2010; Filippov et al. 2011; Garbe et al. 2011; Shin et al. 2012). It appears likely that phage resistance in FS is conferred by alterations in LPS structure. SBW25Φ2 proteins involved in phage attachment and entry are most closely related to P. aeruginosa-specific ΦKMV-like phages of the T7 supergroup, including ΦLKA1 (BLASTp, data not shown). Notably, ΦLKA1 infection requires expression of algC (Ceyssens et al. 2011)—a gene encoding phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core (Coyne et al. 1994).
An evolutionary arms race at the FS locus
The fact that fuzY mutations confer resistance to bacteriophage SBW25Φ2 but have deleterious pleiotropic consequences for subsequent evolution of WS—the superior competitor at the AL interface (Rainey and Travisano 1998)—raises the possibility that a trade-off between phage resistance and FS vs. WS strategies to colonize the AL interface might underlie—at least in part—the previously reported arms race between P. fluorescens SBW25 populations and SBW25Φ2 (Buckling and Rainey 2002a). Such a possibility can be envisaged as follows: in the absence of phage the ancestral SM genotype rapidly diversifies into numerous niche specialist types, including WS and FS. Presence of phage selects for phage-resistant genotypes, in particular, FS morphotypes, while at the same time selecting against phage-sensitive genotypes such as WS. As FS types increase in frequency, the phage titer—under a serial transfer regime—declines. Once at low density, opportunity exists for the competitively superior WS types to reemerge—either from preexisting sensitive types or, de novo, by mutation, from the dominant FS population. However, for WS to evolve from FS, a mutation that reverts (or compensates for) the fuzY mutation is required. Given that FS and WS share dependence on a functional fuz operon, it is possible that the bacterial response to the presence of phage may involve continual cycling between phage-sensitive WS and phage-resistant FS, effected by mutational changes at the fuzY locus.
We explored this possibility by asking whether WS genotypes can evolve from FS and, if they do, whether the FS-derived WS are phage sensitive. Fifty-six independent FS mutants representative of the range of fuzY mutational types were propagated for 3-day periods by serial dilution in structured microcosms until the emergence of colony morphologies resembling WS types (Table S3). All (except for 3) FS genotypes gave rise to WS types within four serial transfers (with WS emergence most commonly requiring just one or two successive transfers to detect). However, the majority of the derived types produced feeble mats (fWS), indicative of the fact that the fuzY mutation had neither been reversed nor compensated for by a change restoring functionality to the fuz locus. Consistent with this thesis, most weak fWS types remained phage resistant (Table S3). However, strong mat-forming WS derivatives (typical of regular WS mats) did arise from a number of FS isolates and repeatedly arose from 2 FS isolates in particular: FS108 (fuzY432insCCTGGCGG) and FS156a (fuzYC1096T) (Table S2). These derived WS types were phage sensitive. A more extensive collection of FS-derived WS genotypes was selected from 12 replicate populations of each of FS108 and FS156a after 5 days growth in structured microcosms (data not shown). Sequencing of fuzY from 22 phage-sensitive FS108-derived WS isolates revealed a reversion to the wild-type sequence (by retraction of an 8-bp duplication of adjacent sequence at base pair 432) in every case. Sequencing of fuzY from 17 phage-sensitive FS156a-derived WS isolates revealed a reversion of C1096T to the wild-type T1096C in 7 cases (restoring a Gln codon from an amber stop) and an additional SNP (A1097G) in 10 cases (creating a Trp codon from an amber stop). Taken together, these data demonstrate that while fWS are more likely to evolve from FS (probably by a single mutation), true WS do emerge and are most likely the consequence of two mutations: the first one reverting the fuzY gene to wild type (and thus generating the SM morph), followed by a second mutation—in the wsp-, aws-, or mws-determined pathways (McDonald et al. 2009)—to give WS. The extent to which this is possible is likely to depend on the nature of the FS-causing fuzY mutation. Certain fuzY mutations, by virtue of their genetic context (for example, those involving expansion or contraction of repeated sequence) are more likely to revert than others. When reversion of fuzY occurs, phage-sensitive WS are inevitable and are likely competitively superior to phage-resistant fWS: the relative fitness of fWS (WS-fuzYT443G) in 3-day structured microcosms was just 0.80 ± 0.06 whereas the relative fitness of WS was 0.97 ± 0.05 (t-test: P < 0.0001, n = 8). The strong mat-forming WS is substantially fitter than weak mat-forming WS-fuzYT443G and the major component of the differential fitness is the ability to exploit the AL interface when competed in a structured environment. Thus, given sufficient generational time, it is predicted that phage-sensitive WS types will derive from FS progenitors in spatially structured environments once the dominance of FS genotypes has driven SBW25Φ2 to near extinction.
An on-going arms race requires that phage-sensitive WS can generate phage-resistant FS by mutation. From knowledge of the underlying genetics this is highly probable, but is likely to require two mutations: one to revert the WS to SM and a second to generate FS. From 12 replicate populations of each of three distinct FS-derived WS genotypes propagated for two successive 3-day periods and three successive 7-day periods by serial dilution in structured microcosms, 23 produced FS at detectable frequencies, with earlier emergence of “step one” SM and/or fWS types as predicted (Table S4). All of the derived FS types were phage resistant. Thus, the plausibility of a continuing arms race with changes wrought at the fuzY locus is demonstrated.
Discussion
How mutational processes create the molecular variation upon which natural selection acts is a question readily addressed by experimental evolution (Beaumont et al. 2009; Blount et al. 2012). Ultimately, we seek to complete a comprehensive account of the common evolutionary trajectories during P. fluorescens SBW25 adaptive radiation and understand the underlying genetic architecture, mutational processes and targets, and the emergent ecological interactions that drive adaptive evolution. Here, we have described the genetic and ecological bases of FS fitness. Moreover, our genetic analyses have led to a reassessment of the niche preference of FS and the ecological processes underpinning the Pseudomonas radiation.
The FS mutational spectrum
Previous work has cataloged the underlying mutational bases of WS genotypes and identified the number of mutational routes to the WS phenotype (Spiers et al. 2002; Goymer et al. 2006; Bantinaki et al. 2007; McDonald et al. 2009). While a multitude of pathways lead to WS, the genetic underpinnings of FS are much simpler. Nevertheless, our catalog of FS mutants provides useful information about the mutational processes that generate the diversity upon which natural selection acts. The spectrum of FS causal mutations is highly diverse—encompassing SNPs causing both missense and nonsense mutations, small indels causing frameshifts, and both small and large deletions—with mutations spaced throughout the length of the gene. A number of studies, both experimental and comparative, have reported mutational spectra across genes and genomes in bacterial lineages experiencing hard or soft selection (Miller 1985; Leong et al. 1986; Mo et al. 1991; Garibyan et al. 2003; and reviewed in Ochman 2003; Wolff et al. 2004). More recently, experimental evolution studies, long-term experiments in particular, have provided the material for cataloging mutational spectra—both neutral and those subject to positive selection—across whole genomes (Wielgoss et al. 2011; Lee et al. 2012). These studies reveal commonalities (such as a general twofold bias for transitions over transversions, in particular the G:C > A:T substitution) but also idiosyncrasies. Indeed it can be difficult to compare mutational spectra between species, and even between genes within the same species, since mutation rates and classes are affected by gene-specific parameters such as distance of the gene from the replication origin, the direction of transcription relative to the passage of a replication fork, and the level of transcription (reviewed by Ochman 2003; Juurik et al. 2012).
Various studies have reported mutational spectra for single genes encoding proteins constrained by functions essential for viability (Garibyan et al. 2003; Wolff et al. 2004), making it difficult to draw conclusions about the relative occurrence of SNPs and indels and the six classes of substitution. Although it may be difficult, perhaps impossible, to infer how the mutations sampled in this study might compare with the full spectrum of fuzY mutations occurring through time in each evolving population, the mutations we sampled (at least those of the typical FS class) did not differ markedly in their relative fitness, indicating a phenotypic equivalence. Knowing that all confer a similar phenotype, it is possible to argue that the spectrum of sampled mutations is likely to be largely representative of the broader pool of those occurring in fuzY sequence space. Considering unique SNPs only, the transition:transversion (Ts:Tv) ratio in our study was 1:1.3. Considering the transitions, the distribution of AT > GC and GC > AT substitutions (after correcting for GC content) did not differ from that expected by chance (χ2 = 1.517, P = 0.2180, d.f. = 1, chi-square test). Similarly, the relative frequencies of the four possible transversions did not differ from that expected (χ2 = 2.000, P = 0.5724, d.f. = 3, chi-square test).
Curious is the finding that one-quarter of all the independently isolated fuzzy spreaders were caused by an identical SNP—the same T443G transversion carried by the resequenced archetypal isolate—causing valine to be substituted for glycine at position 148 (V148G). Fitness assays performed under conditions similar to those in which our independent FS isolates evolved show that this mutation does not confer a fitness benefit substantially greater than that of any other FS mutation; indeed, mathematical models conclude that the differential fitness necessary to account for the preferential selection of T443 is beyond biological reality (E. Libby, unpublished results). The more frequent occurrence of this mutation suggests that it has an increased probability of being generated, likely as a consequence of the local DNA sequence or secondary structure. A comparison of the DNA sequence surrounding the T443G hotspot with nearby sequences failed to identify any imperfect palindromic matches likely to template a T > G substitution at position 443; thus replication template switching within imperfect repeat (quasi-palindromic) sequences during passage of a replication fork is not obviously behind the generation of T443G (De Boer and Ripley 1984; Mo et al. 1991; Demarini et al. 1998; Viswanathan et al. 2000; Lovett 2004).
Ecological interactions underpinning the Pseudomonas radiation
How genetic diversity translates into phenotypic diversity and how phenotypic diversity in turn drives the interactions between species and their environment is of considerable interest. The interplay of predominant emergent types during P. fluorescens SBW25 radiations has been extensively described (Rainey and Travisano 1998; Rainey and Rainey 2003; Rainey 2005). However, closer inspection of FS growth within spatially structured microcosms—motivated initially by the overlap of the genetic architecture underpinning both FS and WS—has led to a significant revision of our understanding of the ecology of this particular morphotype: previously designated a bottom dweller, FS innocula observed over time initially begin growth at the AL interface, with flocs of cells attempting to spread as a biofilm but ultimately falling to the microcosm floor. This pattern of growth indicates that FS competes with WS for occupancy of the AL interface, which fits with the fact that oxygen is the primary limiting resource driving the Pseudomonas radiation. FS performs poorly in competition with WS (Rainey and Travisano 1998; Rainey 2005); however, it repeatedly arises and increases in frequency whenever spatially structured microcosms are seeded with the ancestral type. Moreover, that aFS and sFS types repeatedly arise in parallel adaptive radiations implies an ecological complexity within the simple structured broth microcosm that remains to be fully described. Our working model is as follows: growth of the ancestral SM genotype rapidly depletes the environment of oxygen, which establishes conditions that favor evolution of mat-forming WS types. WS mats dominate for several days before collapsing (Rainey and Rainey 2003). Mat collapse opens a niche for FS types, which exploit this opportunity via the formation of transient rafts that exist for sufficient time to enable FS cells to access oxygen that is otherwise absent from the microcosm. Interestingly, SM types, being unable to adhere to FS types, cannot hitchhike with FS and are thus unable to take advantage of FS rafts in the same way that they can take advantage of the WS mats. This accounts for the previously noted inhibitory effect of FS on SM (Rainey 2005): growth of FS takes oxygen from the uppermost layer of broth, thus depriving SM of this limiting resource. SM growth is subsequently compromised (Rainey 2005). Eventually WS types reinvade and perpetuate the cycle. Diversity is maintained by time-lagged frequency-dependent selection fueled by the fallibility of the surface-colonizing strategies of both WS and FS.
The effect of fuzY mutation on WS phenotype
The genetic link between FS and WS—the two key emergent types whose negative frequency-dependent interactions drive the dynamics within diversifying structured microcosms—is an unanticipated outcome of these studies. The fuzVWXYZ locus was previously identified as a suppressor of the LSWS (wspFA901C) and AWS (awsX Δ100–138)-based WS morphotypes (McDonald et al. 2009), an association that was reconfirmed here. WS-fuzYT443G and WSΔfuzY mutant colonies showed a decreased wrinkled appearance on KB and produced mats that collapsed easily to the microcosm floor.
The role of LPS as a major component of WS biofilm strength and integrity, as a consequence of interactions between LPS and the cellulose matrix of the biofilm, was revealed in an earlier study (Spiers and Rainey 2005) and is well supported (Lau et al. 2009; Nielsen et al. 2011; Nilsson et al. 2011). Alteration of the bacterial cell surface, with consequent changes to cell hydrophobicity, adhesive properties, and motility, likely perturbs the cell–cell interactions that maintain biofilm structure. The relationship between LPS defects and the adhesive, cohesive, and viscoelastic properties of biofilms and how these properties correlate with biofilm structure has been extensively characterized in P. aeruginosa PAO1 (Lau et al. 2009). Essentially, more dramatic LPS defects (such as full truncation of O antigen) result in stronger adhesion of cells to glass and other abiotic surfaces and stronger cell–cell interactions (evident by flocculation). Furthermore, mutants lacking O antigen produce biofilms with weaker viscoelastic properties. Recent studies with P. putida KT2440 EPS mutants support a model in which LPS mediates the necessary cell–cell interactions that maintain biofilm matrix stability following attachment (Nilsson et al. 2011), with EPS in a stabilizing role and cellulose polymers providing additional structural support (Nielsen et al. 2011; Nilsson et al. 2011).
An evolutionary arms race due to trade-offs in niche specialization
Phages are important determinants of microbial evolution and population structure (reviewed in Stern and Sorek 2010; Koonin and Wolf 2012). Their success in the face of asymmetry in the coevolutionary potential of phages, relative to that of their more complex bacterial hosts, raises questions as to their persistence (Lenski 1984). Various explanations have been put forward: one is that phages and their hosts participate in an on-going arms race mediated by endless cycles of bacterial mutations to phage resistance and phage counterdefenses (Rodin and Ratner 1983a,b). Evidence consistent with this comes from experiments with SBW25 and SBW25Φ2 (Buckling and Rainey 2002a,b; Brockhurst et al. 2004, 2006; Benmayor et al. 2008; Paterson et al. 2010; Gomez and Buckling 2011) and with other bacterial systems (for examples see Kashiwagi and Yomo 2011; Dennehy 2012; Levin et al. 2013; Seed et al. 2013). An alternative explanation posits that phages may persist if there exists a mixture of sensitive and resistant hosts where the resistant types are less fit in competition for resources than the sensitive types (Levin et al. 1977). Such a scenario stands to allow phage persistence if resistant bacteria revert to sensitivity and in so doing restore some capacity that is lost by the resistance-generating mutation (Lenski 1984). Such mutations would be likely once phages are rare, thus allowing phage reinvasion. This scenario appears to be relevant to our findings—at least in part—thus providing potential for persistent coevolution. Phage-sensitive WS are superior colonizers of the air–liquid interface; however, in the presence of phages their success is compromised: FS instead dominate. However, as phage titers decline (with declining availability of WS hosts), opportunity exists for mutants that revert to phage-sensitive, competitively superior, WS types. We show that this requires two mutations; nonetheless, such a possibility clearly exists.
The necessity of phage resistance in an environment where phages are abundant entails a trade-off against the optimal strategy for bacterial colonization: in the case of our simple experimental system, phage resistance is traded off against the ability to generate stable biofilms, enabling optimal access to O2 for growth; in native environments, phage resistance is predicted to trade off against strategies for the establishment of infection and colonization of host organisms—strategies typically mediated by bacterial surface structures. Indeed, O antigen is essential for the establishment of infection (or protection against host defenses) in a diverse array of plant and animal pathogens (Cava et al. 1989; Dekkers et al. 1998; Yang et al. 2000; Kannenberg and Carlson 2001; Poon et al. 2008; Kesawat et al. 2009); moreover, the O antigen serves as a receptor for phages (Michel et al. 2010; Filippov et al. 2011; Garbe et al. 2011; Shin et al. 2012). Thus it would appear that O antigens are subject to two conflicting selective pressures: pressure to change to avoid phage predation and pressure to be maintained to enable colonization of host species during infection. Such a trade-off may be widely relevant to persistence of phage and their bacterial hosts.
Supplementary Material
Acknowledgments
We thank members of the Rainey group for discussion and comments on the manuscript and Eric Libby for mathematical modeling. pSX was a gift from David Ackerley. Financial support was provided by the Foundation for Research, Science, and Technology; the Allan Wilson Centre; and Massey University. P.B.R. is a James Cook Research Fellow and gratefully acknowledges the Royal Society of New Zealand.
Footnotes
Communicating editor: J. Lawrence
Literature Cited
- Bantinaki E., Kassen R., Knight C. G., Robinson Z., Spiers A. J., et al. , 2007. Adaptive divergence in experimental populations of Pseudomonas fluorescens. III. Mutational origins of Wrinkly Spreader diversity. Genetics 176: 441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont H. J. E., Gallie J., Kost C., Ferguson G. C., Rainey P. B., 2009. Experimental evolution of bet hedging. Nature 462: 90–93. [DOI] [PubMed] [Google Scholar]
- Benmayor R., Buckling A., Bonsall M. B., Brockhurst M. A., Hodgson D. J., 2008. The interactive effects of parasites, disturbance, and productivity on experimental adaptive radiations. Evolution 62: 467–477. [DOI] [PubMed] [Google Scholar]
- Blount Z. D., Barrick J. E., Davidson C. J., Lenski R. E., 2012. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braendle C., Baer C. F., Felix M.-A., 2010. Bias and evolution of the mutationally accessible phenotypic space in a developmental system. PLoS Genet. 6: e1000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst M. A., Rainey P. B., Buckling A., 2004. The effect of spatial heterogeneity and parasites on the evolution of host diversity. Proc. Biol. Sci. 271: 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst M. A., Buckling A., Rainey P. B., 2006. Spatial heterogeneity and the stability of host-parasite coexistence. J. Evol. Biol. 19: 374–379. [DOI] [PubMed] [Google Scholar]
- Buckling A., Rainey P. B., 2002a Antagonistic coevolution between a bacterium and a bacteriophage. Proc. Biol. Sci. 269: 931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A., Rainey P. B., 2002b The role of parasites in sympatric and allopatric host diversification. Nature 420: 496–499. [DOI] [PubMed] [Google Scholar]
- Buckling A., Maclean R. C., Brockhurst M. A., Colegrave N., 2009. The Beagle in a bottle. Nature 457: 824–829. [DOI] [PubMed] [Google Scholar]
- Cava J. R., Elias P. M., Turowski D. A., Noel K. D., 1989. Rhizobium leguminosarum CFN42 genetic regions encoding lipopolysaccharide structures essential for complete nodule development on bean plants. J. Bacteriol. 171: 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceyssens P.-J., Glonti T., Kropinski A. M., Lavigne R., Chanishvili N., et al. , 2011. Phenotypic and genotypic variations within a single bacteriophage species. Virol. J. 8: 134–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.-H., Gaynor J. B., White K. G., Lopez C., Bosio C. M., et al. , 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2: 443–448. [DOI] [PubMed] [Google Scholar]
- Colosimo P. F., Hosemann K. E., Balabhadra S., Villarreal G. J., Dickson M., et al. , 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Conte G. L., Arnegard M. E., Peichel C. L., Schluter D., 2012. The probability of genetic parallelism and convergence in natural populations. Proc. Biol. Sci. 279: 5039–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne M. J., Jr, Russell K. S., Coyle C. L., Goldberg J. B., 1994. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176: 3500–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. R., 1859. The Origin of Species. John Murray, London. [Google Scholar]
- de Boer J. G., Ripley L. S., 1984. Demonstration of the production of frameshift and base-substitution mutations by quasipalindromic DNA sequences. Proc. Natl. Acad. Sci. USA 81: 5528–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers L. C., van der Bij A. J., Mulders I. H., Phoelich C. C., Wentwoord R. A., et al. , 1998. Role of the O-antigen of lipopolysaccharide, and possible roles of growth rate and of NADH:ubiquinone oxidoreductase (nuo) in competitive tomato root-tip colonization by Pseudomonas fluorescens WCS365. Mol. Plant Microbe Interact. 11: 763–771. [DOI] [PubMed] [Google Scholar]
- DeMarini D. M., Shelton M. L., Abu-Shakra A., Szakmary A., Levine J. G., 1998. Spectra of spontaneous frameshift mutations at the hisD3052 allele of Salmonella typhimurium in four DNA repair backgrounds. Genetics 149: 17–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy J. J., 2012. What can phages tell us about host-pathogen coevolution? Int. J. Evol. Biol. 2012: 396165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1951. Genetics and the Origin of Species. Columbia University Press, New York. [Google Scholar]
- Figurski D., Helinski D., 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76: 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippov A. A., Sergueev K. V., He Y., Huang X. Z., Gnade B. T., et al. , 2011. Bacteriophage-resistant mutants in Yersinia pestis: identification of phage receptors and attenuation for mice. PLoS ONE 6: e25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe J., Bunk B., Rohde M., Schobert M., 2011. Sequencing and characterization of Pseudomonas aeruginosa phage JG004. BMC Microbiol. 11: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibyan L., T. Huang, M. Kim, E. Wolff, A. Nguyen et al, 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2: 593–608. [DOI] [PubMed] [Google Scholar]
- Gomez P., Buckling A., 2011. Bacteria-phage antagonistic coevolution in soil. Science 332: 106–109. [DOI] [PubMed] [Google Scholar]
- Goymer P., Kahn S. G., Malone J. G., Gehrig S. M., Spiers A. J., et al. , 2006. Adaptive divergence in experimental populations of Pseudomonas fluorescens. II. Role of the GGDEF regulator WspR in evolution and development of the Wrinkly Spreader phenotype. Genetics 173: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindré T., Knibbe C., Beslon G., Schneider D., 2012. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat. Rev. Microbiol. 10: 352–365. [DOI] [PubMed] [Google Scholar]
- Horton R., Hunt H., Ho S., Pullen J., Pease L., 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68. [DOI] [PubMed] [Google Scholar]
- Juurik T., Ilves H., Teras R., Ilmjärv T., Tavita K., et al. , 2012. Mutation frequency and spectrum of mutations vary at different chromosomal positions of Pseudomonas putida. PLoS ONE 7: e48511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannenberg E. L., Carlson R. W., 2001. Lipid A and O-chain modifications cause Rhizobium lipopolysaccharides to become hydrophobic during bacteroid development. Mol. Microbiol. 39: 379–391. [DOI] [PubMed] [Google Scholar]
- Kashiwagi A., Yomo T., 2011. Ongoing phenotypic and genomic changes in experimental coevolution of RNA bacteriophage Qβ and Escherichia coli. PLoS Genet. 7: e1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R., 2009. Toward a general theory of adaptive radiation: insights from microbial experimental evolution. Ann. N. Y. Acad. Sci. 1168: 3–22. [DOI] [PubMed] [Google Scholar]
- Kesawat M. S., Das B. K., Bhaganagare G. R., Sharma V., and Manorama, 2009. Isolation and characterization of lipopolysaccharides from different rhizobial isolates. J. Crop Sci. Biotechnol. 12: 109–113. [Google Scholar]
- King E. O., Ward M. K., Raney D. E., 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44: 301–307. [PubMed] [Google Scholar]
- Kitten T., Kinscherf T. G., McEvoy J. L., Willis D. K., 1998. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 28: 917–929. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Wolf Y. I., 2012. Evolution of microbes and viruses: A paradigm shift in evolutionary biology? Front. Cell. Infect. Microbiol. 2: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koza A., Moshynets O., Otten W., Spiers A. J., 2010. Environmental modification and niche construction: developing O2 gradients drive the evolution of the Wrinkly Spreader. ISME J. 5: 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack D., 1947. Darwin’s Finches. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Lau P. C. Y., Lindhout T., Beveridge T. J., Dutcher J. R., Lam J. S., 2009. Differential lipopolysaccharide core capping leads to quantitative and correlated modifications of mechanical and structural properties in Pseudomonas aeruginosa biofilms. J. Bacteriol. 191: 6618–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Popodi E., Tang H., Foster P. L., 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 109: E2774–E2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R. E., 1984. Coevolution of bacteria and phage: Are there endless cycles of bacterial defenses and phage counterdefenses? J. Theor. Biol. 108: 319–325. [DOI] [PubMed] [Google Scholar]
- Lenski R., Rose M., Simpson S., Tadler S., 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138: 1315–1341. [Google Scholar]
- Lenski R. E., Mongold J. A., Sniegowski P. D., Travisano M., Vasi F., et al. , 1998. Evolution of competitive fitness in experimental populations of E. coli: What makes one genotype a better competitor than another? Anton. Leeuw. 73: 35–47. [DOI] [PubMed] [Google Scholar]
- Leong P. M., Hsia H. C., Miller J. H., 1986. Analysis of spontaneous base substitutions generated in mismatch-repair-deficient strains of Escherichia coli. J. Bacteriol. 168: 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B. R., Stewart F. M., Chao L., 1977. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am. Nat. 111: 3–24. [Google Scholar]
- Levin B. R., Moineau S., Bushman M., Barrangou R., 2013. The population and evolutionary dynamics of phage and bacteria with CRISPR-mediated immunity. PLoS Genet. 9: e1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A., 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4(3): 203–221. [DOI] [PubMed] [Google Scholar]
- Losos J. B., 2010. Adaptive radiation, ecological opportunity, and evolutionary determinism. Am. Nat. 175: 623–639. [DOI] [PubMed] [Google Scholar]
- Lovett S. T., 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 52: 1243–1253. [DOI] [PubMed] [Google Scholar]
- MacLean R. C., 2005. Adaptive radiation in microbial microcosms. J. Evol. Biol. 18: 1376–1386. [DOI] [PubMed] [Google Scholar]
- McDonald M. J., Gehrig S. M., Meintjes P. L., Zhang X. X., Rainey P. B., 2009. Adaptive divergence in experimental populations of Pseudomonas fluorescens. IV. Genetic constraints guide evolutionary trajectories in a parallel adaptive radiation. Genetics 183: 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A., Clermont O., Denamur E., Tenaillon O., 2010. Bacteriophage PhiX174’s ecological niche and the flexibility of its Escherichia coli lipopolysaccharide receptor. Appl. Environ. Microbiol. 76: 7310–7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., 1985. Mutagenic specificity of ultraviolet light. J. Mol. Biol. 182: 45–65. [DOI] [PubMed] [Google Scholar]
- Mo J. Y., Maki H. H., Sekiguchi M. M., 1991. Mutational specificity of the dnaE173 mutator associated with a defect in the catalytic subunit of DNA polymerase III of Escherichia coli. J. Mol. Biol. 222: 925–936. [DOI] [PubMed] [Google Scholar]
- Mortlock R. P., 1982. Metabolic acquisitions through laboratory selection. Annu. Rev. Microbiol. 36: 259–284. [DOI] [PubMed] [Google Scholar]
- Nielsen L., Li X., Halverson L. J., 2011. Cell-cell and cell-surface interactions mediated by cellulose and a novel exopolysaccharide contribute to Pseudomonas putida biofilm formation and fitness under water-limiting conditions. Environ. Microbiol. 13: 1342–1356. [DOI] [PubMed] [Google Scholar]
- Nilsson M., Chiang W. C., Fazli M., Gjermansen M., Givskov M., et al. , 2011. Influence of putative exopolysaccharide genes on Pseudomonas putida KT2440 biofilm stability. Environ. Microbiol. 13: 1357–1369. [DOI] [PubMed] [Google Scholar]
- Ochman H., 2003. Neutral mutations and neutral substitutions in bacterial genomes. Mol. Biol. Evol. 20: 2091–2096. [DOI] [PubMed] [Google Scholar]
- Owen J. G., Ackerley D. F., 2011. Characterization of pyoverdine and achromobactin in Pseudomonas syringae pv. phaseolicola 1448a. BMC Microbiol. 11: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson S., Vogwill T., Buckling A., Benmayor R., Spiers A. J., et al. , 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464: 275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole A. M., Phillips M. J., Penny D., 2003. Prokaryote and eukaryote evolvability. Biosystems 69: 163–185. [DOI] [PubMed] [Google Scholar]
- Poon K. K. H., Westman E. L., Vinogradov E., Jin S., Lam J. S., 2008. Functional characterization of MigA and WapR: putative rhamnosyltransferases involved in outer core oligosaccharide biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 190: 1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey P. B., 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1: 243–257. [DOI] [PubMed] [Google Scholar]
- Rainey, P. B., 2005 Bacteria evolve and function within communities: observations from experimental Pseudomonas populations, pp. 83–102 in The Influence of Cooperative Bacteria on Animal Host Biology, edited by M. McFail-Ngai, B. Henderson, and N. Ruby. Cambridge University Press, Cambridge. [Google Scholar]
- Rainey P. B., Bailey M. J., 1996. Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome. Mol. Microbiol. 19: 521–533. [DOI] [PubMed] [Google Scholar]
- Rainey P. B., Rainey K., 2003. Evolution of cooperation and conflict in experimental bacterial populations. Nature 425: 72–74. [DOI] [PubMed] [Google Scholar]
- Rainey P. B., Travisano M., 1998. Adaptive radiation in a heterogeneous environment. Nature 394: 69–72. [DOI] [PubMed] [Google Scholar]
- Rainey P. B., Buckling A., Kassen R., Travisano M., 2000. The emergence and maintenance of diversity: insights from experimental bacterial populations. Trends Ecol. Evol. 15: 243–247. [DOI] [PubMed] [Google Scholar]
- Rodin S. N., Ratner V. A., 1983a Some theoretical aspects of protein coevolution in the ecosystem “phage-bacteria” I. The problem. J. Theor. Biol. 100: 185–195. [Google Scholar]
- Rodin S. N., Ratner V. A., 1983b Some theoretical aspects of protein coevolution in the ecosystem “phage-bacteria” II. The deterministic model of microevolution. J. Theor. Biol. 100: 197–210. [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., et al. , 2000. Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schluter D., 2000. The Ecology of Adaptive Radiation. Oxford University Press, Oxford. [Google Scholar]
- Schluter D., 2009. Evidence for ecological speciation and its alternative. Science 323: 737–741. [DOI] [PubMed] [Google Scholar]
- Seed K. D., Lazinski D. W., Calderwood S. B., Camilli A., 2013. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494: 489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Lee J. H., Kim H., Choi Y., Heu S., et al. , 2012. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. PLoS ONE 7: e43392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby M. W., Cerdeño-Tárraga A. M., Vernikos G. S., Giddens S. R., Jackson R. W., et al. , 2009. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 10: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G. G., 1953. The Major Features of Evolution. Columbia University Press, New York. [Google Scholar]
- Spiers A. J., Rainey P. B., 2005. The Pseudomonas fluorescens SBW25 Wrinkly Spreader biofilm requires attachment factor, cellulose fibre and LPS interactions to maintain strength and integrity. Microbiology 151: 2829–2839. [DOI] [PubMed] [Google Scholar]
- Spiers A. J., Kahn S. G., Bohannon J., Travisano M., Rainey P. B., 2002. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of Wrinkly Spreader fitness. Genetics 161: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers A. J., Bohannon J., Gehrig S. M., Rainey P. B., 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 Wrinkly Spreader requires an acetylated form of cellulose. Mol. Microbiol. 50: 15–27. [DOI] [PubMed] [Google Scholar]
- Stein L. D., Mungall C., Shu S., Caudy M., Mangone M., et al. , 2002. The generic genome browser: a building block for a model organism system database. Genome Res. 12: 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A., Sorek R., 2010. The phage-host arms race: shaping the evolution of microbes. Bioessays 33: 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M., Lacirignola J. J., Hurley R. L., Lovett S. T., 2000. A novel mutational hotspot in a natural quasipalindrome in Escherichia coli. J. Mol. Biol. 302: 553–564. [DOI] [PubMed] [Google Scholar]
- Wagner G. P., Zhang J., 2011. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 12: 204–213. [DOI] [PubMed] [Google Scholar]
- Wielgoss, S., J. E. Barrick, O. Tenaillon, S. Cruveiller, B. Chane-Woon-Ming et al., 2011. Mutation rate inferred from synonymous substitutions in a long-term evolution experiment with Escherichia coli. G3 1: 183–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor G. L., Van Rossum T., Lo R., Khaira B., Whiteside M. D., et al. , 2009. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 37: D483–D488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff E., Kim M., Hu K., Yang H., Miller J. H., 2004. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J. Bacteriol. 186: 2900–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. J., Matewish M., Loubens I., Storey D. G., Lam J. S., et al. , 2000. migA, a quorum-responsive gene of Pseudomonas aeruginosa, is highly expressed in the cystic fibrosis lung environment and modifies low-molecular-mass lipopolysaccharide. Microbiology 146: 2509–2519. [DOI] [PubMed] [Google Scholar]
- Zhang X.-X., Rainey P. B., 2007. Construction and validation of a neutrally-marked strain of Pseudomonas fluorescens SBW25. J. Microbiol. Methods 71: 78–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.