Abstract
In two swordtail species of the genus Xiphophorus, the onset of puberty has been shown to be modulated at the P locus by sequence polymorphism and gene copy-number variation affecting the type 4 melanocortin hormone receptor Mc4r. The system works through the interaction of two allelic types, one encoding wild type and the other dominant-negative receptors. We have analyzed the structure and evolution of the P locus in the platyfish Xiphophorus maculatus, where as many as nine alleles of P determining the onset of sexual maturity in males and females, fecundity in females, and adult size in males are located on both the X and Y chromosomes in a region linked to the master sex-determining locus. In this species, mc4r has been amplified to up to 10 copies on both the X and Y chromosomes through recent large serial duplications. Subsequently, mc4r paralogues have diverged considerably into many different subtypes. Certain copies have acquired new untranslated regions through genomic rearrangements, and transposable element insertions and other mutations have accumulated in promoter regions, possibly explaining observed deviations from the classical mc4r transcriptional pattern. In the mc4r-coding sequence, in-frame insertions and deletions as well as nonsense and missense mutations have generated a high diversity of Mc4r-predicted proteins. Most of these variants are expressed in embryos, adults, and/or tumors. Functional receptor characterization demonstrated major divergence in pharmacological behavior for Mc4r receptors encoded by different copies of platyfish mc4r, with differences in constitutive activity as well as binding and stimulation by hormones. The high degree of allelic and copy-number variation observed between individuals can explain the high level of polymorphism for sexual maturation, fecundity, and body size in the platyfish: multiple combinations of Mc4r variants with different biochemical properties might interact to modulate the melanocortin signaling that regulates the hypothalamus–pituitary–gonadal axis.
Keywords: fish, sexual maturity, melanocortin receptor, sex chromosomes, duplication
TELEOST fish of the genus Xiphophorus, which comprise two major and morphologically quite divergent groups, the platyfish and swordtails, are prominent models for the study of polymorphic traits such as sexual development, reproduction, pigmentation, and cancer (Kallman 1975, 1984; Anders and Anders 1978; Volff and Schartl 2002; Meierjohann and Schartl 2006; Schartl 2008; Schultheis et al. 2009). In the platyfish Xiphophorus maculatus and in other Xiphophorus species, many genetic loci involved in the control of these traits are located on the sex chromosomes, where they are linked to the sex-determining region (Gutbrod and Schartl 1999 and references therein).
Three types of sex chromosomes have been described in X. maculatus: X, Y, and W. Males can be XY or YY, females XX, WX, or WY (Kallman 1984). Many genetic loci have been mapped to the X and Y chromosomes of the platyfish, but information on the W chromosome is still scarce. The master sex-determining locus SD, also referred to as SEX, which determines the sexual phenotype of individuals (Kallman 1984), has not been identified so far at the molecular level. One well-studied locus linked to SD is the so-called Tumor locus (Tu), which is responsible for the formation of spontaneous melanoma in certain Xiphophorus interspecific hybrids (Gordon 1927; Kosswig 1928; Meierjohann and Schartl 2006; Schartl 2008). The critical constituent of this locus is a gene, xmrk, which encodes a mutationally activated version of the epidermal growth factor receptor (Wittbrodt et al. 1989).
Another important locus in this region on the sex chromosomes of the platyfish is the Pituitary (or Puberty) locus P, which controls the onset of sexual maturity in males and females, as well as adult size in males and fecundity in females (Kallman and Schreibman 1973; Kallman and Borkoski 1977; Schreibman et al. 1994). The P locus is highly polymorphic. As many as nine different alleles have been identified that determine different timing of puberty and adult male size (Kallman 1989). Cosegregation analyses mapped them all to both the X and Y chromosomes of the platyfish. As a consequence, X. maculatus shows a high phenotypic diversity with respect to the onset of sexual maturation, from early maturing to late-maturing animals with a continuum of intermediate phenotypes (Kallman 1989). Since males stop growing when they reach puberty, this polymorphism is also associated with great variation in adult body size: early maturing males are smaller and late-maturing animals are larger, with many intermediates in onset of puberty and, consequently, body size. In several Xiphophorus species, it has been shown that smaller vs. larger fishes display different reproductive behaviors: larger fishes defend territories visited by females and court them in a ritualized manner, while smaller fishes search for “sneak” mating opportunities by moving between territories and directly approach females (Zimmerer 1982; Ryan and Causey 1989; Ryan et al. 1990; Ryan and Rosenthal 2001). Hence, the P locus also controls reproductive behavior variation in Xiphophorus.
Due to the multiplicity of P alleles and associated phenotypes, genetic analyses are complicated to conduct in X. maculatus. In contrast, X. nigrensis and X. multilineatus, two swordtail species distantly related to X. maculatus within the genus Xiphophorus (Kang et al. 2013), present only three major P-determined male phenotypes (small/early maturing, large/late-maturing, and one intermediate phenotype). In these species, it has been shown that puberty onset, body size, and reproductive behavior are modulated by sequence polymorphism and gene copy-number variation affecting the type 4 receptor for melanocortin hormones, Mc4r (Lampert et al. 2010).
Melanocortins are neuromodulatory peptide hormones produced through post-translational processing of the precursor pro-opiomelanocortin. They include the melanocyte-stimulating hormones (α-, β-, and γ-MSH) and the adrenocorticotropic hormone ACTH. Melanocortins are expressed mainly but not exclusively in the pituitary gland and the central nervous system. These hormones are involved in diverse physiological processes including pigmentation, steroidogenesis, appetite and body weight regulation, inflammatory responses, cardiovascular system regulation, stimulation of exocrine gland secretion, and sexual function (Cone 2006). In vertebrates, melanocortin hormones mediate their effects through interaction with five types of G-protein-coupled receptors, Mc1r–Mc5r. The Mc4r receptor is involved in the control of food intake, energy balance, and body weight in mammals with some direct and indirect reported functions in reproduction (Adan et al. 2006; Van der Ploeg et al. 2002; Tao 2010; Israel et al. 2012 and references therein).
In X. nigrensis and X. multilineatus, both functional and defective mc4r copies have been identified. Defective copies have lost their capability for signal transduction because microdeletions have removed important cysteine residues from the carboxy-terminal region of the receptor. Such defective copies were detected only in males, being located on the Y chromosome (Lampert et al. 2010). Copy-number variation correlating with mc4r expression levels was observed for the defective mc4r copies in males: the highest copy-number and expression levels were found in large late-maturing males, while small early maturing males showed low copy number and low levels of expression. According to results from cell culture experiments, defective versions might compete with functional receptors to form Mc4r dimers. High expression of nonfunctional receptors would reduce the formation of functional Mc4r dimers. This would consequently inhibit melanocortin signaling, delay puberty, and allow further growth of the fish.
Through genetic analysis it was shown that mc4r copies are located on the sex chromosomes of X. nigrensis and X. multilineatus (Lampert et al. 2010). However, no genomic data are available to assess the origin, structure, and evolution of the P locus in any Xiphophorus species. In addition, characterization of the multiple P locus alleles involved in the very high polymorphism affecting body size and onset of puberty in X. maculatus requires a precise molecular characterization of the different mc4r copies involved. The genome sequence of a platyfish female (n = 28, XX) has been recently published (Schartl et al. 2013), but the structural complexity of the sex-determining region, with many recent large duplications and a high concentration of transposable elements and other repeat sequences, makes it very difficult to assemble from shotgun genomic data. In addition, this genome project, performed on female DNA, does not provide any insight into Y-linked mc4r copies in X. maculatus.
To characterize the structure of the P locus in the platyfish, a bacterial artificial chromosome (BAC) library has been established from XY platyfish males (Froschauer et al. 2002). BAC clones linked to the sex-determining region of the sex chromosomes have been isolated, assembled, and partially sequenced (Froschauer et al. 2002; Volff et al. 2003; Schultheis et al. 2006). We report here that as many as 10 copies of the mc4r gene are present on each X and Y chromosome of the platyfish. Expression and pharmacological analyses strongly support functional diversification of Mc4r receptors after duplication through mutation of both regulatory and coding regions.
Materials and Methods
Fish and cell culture
Fish were reared at the Plateau de Recherche Expérimentale de Criblage in Vivo of the SFR BioSciences Gerland-Lyon Sud (US8/UMS3444, Lyon, France) and at the fish facility of Biozentrum at the University of Würzburg (Germany). Tumor tissues were obtained from backcross hybrids between X. maculatus and X. hellerii. Xiphophorus melanoma cell line PSM (Wakamatsu 1981) and embryonic cell line A2 (Kuhn et al. 1979) were used to test mc4r expression.
For receptor assays, human embryonic kidney (HEK)-293 cells were cultivated in Dulbecco’s modified Eagle medium (DMEM) with 4.5 g/liter glucose, 10% fetal calf serum (FCS), 100 U/ml penicillin G, and 100 µg/ml streptomycin sulfate at 37°, 5% CO2. All cells were routinely passaged every 2–3 days. Culture medium for cells stably expressing Mc4r receptors was additionally supplemented with 200 μg/ml G-418.
DNA and RNA analyses
DNA extraction was performed according to standard protocols (Sambrook et al. 1989). Southern blot hybridization was done as described using PCR-amplified fragments as probes (Selz et al. 2007). Small-scale sequencing reactions were performed using the CEQ-DTCS dye terminator cycle sequencing kit and run on a CEQ 8000 DNA sequencing system (Beckmann Coulter). Sanger-based shotgun sequencing of BAC clones was done at Genoscope (Evry, France).
Total RNA was extracted using the TRIZOL Reagent (Invitrogen Life Technologies). After reverse transcription, PCR was performed using primer F2 (attcctctgctggctgctac) in combination with either primer R3 (agaagatctccttaaaggtc, no UTR specificity), primer R5 (gacatttcaggctcttcatcc, specific for the “wild-type” 3′ UTR), and primer 2-R1 (gtttgactgaaaataaaaacagg, specific for the alternative 3′ UTR*).
In Silico DNA sequence analysis
Multiple sequence alignments were generated using ClustalX (Thompson et al. 1997). Mc4r phylogeny was determined using MrBayes (Huelsenbeck and Ronquist 2001). Maximum likelihood (ML), maximum parsimony (MP), and neighbor joining (NJ) analyses were also performed. The ML tree was obtained with PhyML (Guindon and Gascuel 2003) using a JTT model, a discrete γ-model with four categories. The γ-shape parameter and the proportion of invariable sites were calculated by ML. Both MP and NJ trees were constructed with Phylo_Win (Galtier et al. 1996) using pairwise gap removal. Selection parameter ω (dN/dS) was estimated using CodeML from the PAML 3.15 package (Yang 1997). Recombination rate was calculated with DnaSP (Librado and Rozas 2009). Mc4r sequences from public databases were retrieved using the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) and ENSEMBL (http://www.ensembl.org/Multi/blastview) BLAST servers. X. maculatus mc4r sequences have been deposited in GenBank under accession nos. KF650640–KF650658.
Transient transfection of HEK-293 cells and selection of stable cell lines
HEK-293 cells were seeded on 10-cm culture plates to ∼50% confluence. After attachment of the cells (4–6 hr), the cells were transfected with 6 μg of DNA using Effectene according to the manufacturer’s instructions. All constructs were cloned in pcDNA3, and the empty pcDNA3 vector was used for control experiments. Medium was changed 12–16 hr later, and cells were harvested 48 hr after transfection. Positive clones were selected with 600 μg/ml of the neomycin analog G-418, and single clonal lines were isolated by limiting dilution. Cell lines were maintained at 200 μg/ml G-418. Expression of the receptor was verified by radioligand binding.
Membrane preparation
HEK-293 cells transiently transfected with Mc4r receptor constructs or stably expressing Mc4r receptors were grown adherently and cultivated in DMEM with 4.5 g/liter glucose, 10% FCS, 100 U/ml penicillin G, and 100 µg/ml streptomycin sulfate at 37°, 5% CO2, as described above. Cells stably expressing Mc4r receptor constructs were split two or three times per week at a ratio between 1:5 and 1:10. For harvesting cells, culture medium was removed, cells were washed twice with phosphate buffered saline puffer, and membranes were prepared or cells were frozen on the dishes for later preparation of membranes. Crude membrane fractions were prepared from fresh (measurement of adenylyl cyclase) or frozen cells (radioligand binding) according to two different protocols, which have been previously described (Klotz et al. 1998). The resulting membrane pellets were resuspended in 50 mM Tris–HCl buffer, pH 7.4, to give a final protein concentration of 1–2 mg/ml.
Adenylyl cyclase assay
Adenylyl cyclase activity in cell membranes was determined according to Jakobs et al. (1976). Fifty micrograms of membrane protein was added to an incubation mixture with final concentrations of 50 mM Tris–HCl, pH 7.4, 100 μM cAMP, 0.2% BSA, 10 μM GTP, 100 μM ATP, 1 mM MgCl2, 100 μM isobutylmethylxanthine, 15 mM phosphocreatine, and 300 units/ml of creatine kinase. Membranes were incubated with ∼25 kBq of [α-32P]ATP for 20 min in the incubation mixture as described (Hoffmann et al. 2004). Accumulation of [α-32P]cAMP was linear over at least 20 min under all conditions. The reaction was stopped by addition of 400 μl 125 mM zinc acetate solution and 500 μl 144 mM Na2CO3. Samples were centrifuged for 5 min at 14,000 × g. A total of 800 μl of the resulting supernatant was finally applied to aluminia WN-6 (Sigma) columns that were eluted twice with 2 ml 100 mM Tris–HCl, pH 7.4. Eluates were counted in a β-counter (Beckmann LS 1801). Data were analyzed using Origin 6.1 (OriginLab).
Radioligand binding
Radioligand-binding experiments were performed with membranes prepared as described above. Assays were done in a volume of 200 μl in 25 mM HEPES (pH 7.4) containing 2.5 mM CaCl2 and 1 mM MgCl2 in the presence of 100 μM GTP to ensure monophasic binding for agonists. For saturation binding experiments with different platyfish, we used Mc4r constructs up to 1600 pM 125I-NDP-MSH. Nonspecific binding was determined in the presence of 2 μM of unlabeled NDP-MSH. For competition binding, we used between 120 and 200 pM 125I-NDP-MSH and serial dilutions of competing unlabeled ligands: α-MSH, NDP-MSH, or ACTH. Membranes were incubated for 120 min at 30°, filtered through Whatman GF/C filters, and washed three times with ice-cold assay buffer. Samples were counted in a γ-counter (Wallac 1480 wizard 3′). KD values for 125I-NDP-MSH were calculated by nonlinear curve fitting with the program SCTFIT (De Lean et al. 1982). Ligand IC50 values were calculated using Origin 6.1 (OriginLab) and were transformed to Ki values according to Cheng and Prusoff (1973).
Results
Multiple copies of the mc4r gene in the platyfish X. maculatus
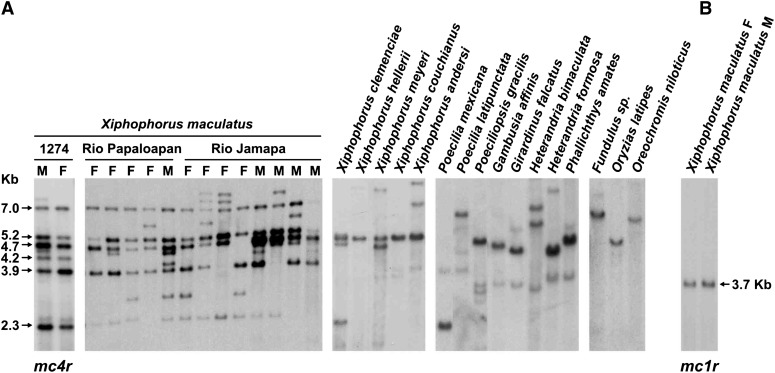
To obtain a first estimate on mc4r copy number in fish by Southern blot hybridization analysis, a molecular probe originating from one mc4r sequence linked to xmrk (Volff et al. 2003) was used against genomic DNA from different poeciliid and non-poeciliid fish species (Figure 1). Even under high-stringency conditions, several hybridizing fragments were observed in both males and females of X. maculatus as well as in some other Xiphophorus species including X. clemenciae and X. meyeri (Figure 1A). In most other poeciliid species, as well as in Fundulus, in the medaka Oryzias latipes and in the Nile tilapia Oreochromis niloticus, only one strong intensity fragment was observed (additional fainter signals were identified as being due to cross-hybridization with other mcr genes such as mc1r) (Selz et al. 2007 and data not shown). The presence of a single copy mc4r gene was confirmed in different Poecilia species and in other non-poeciliid fish through PCR and BLAST analysis of publicly available genome assemblies. Unlike mc4r, another mcr gene, mc1r, is present as a single-copy gene in the platyfish (Figure 1B) (Selz et al. 2007). Hence, in contrast to other vertebrates analyzed so far, the platyfish X. maculatus and several other Xiphophorus species possess several copies of the mc4r gene (see also Lampert et al. 2010). Variations in the number and intensity of hybridizing fragments were observed even between individuals from the same natural population, suggesting intra- and interpopulation polymorphism for type and/or number of mc4r copies (tested for two natural populations, Rio Jamapa and Rio Papaloapan; Figure 1A). Hence, copy-number variation (CNV) of the mc4r gene is apparent in X. maculatus natural populations.
Figure 1.
Southern blot hybridization analysis of mc4r in the platyfish X. maculatus and other fish species. Genomic DNA was cut with HindIII and hybridized with a mc4r (A) or mc1r (B) (Selz et al. 2007) internal fragment cloned from X. maculatus. All species belong to the family Poeciliidae except Fundulus sp. (Fundulidae), O. latipes (medaka, Adrianichthyidae), and Oreochromis niloticus (Nile tilapia, Cichlidae). The designation “1274” shows typical male and female patterns for the X. maculatus laboratory reference population WLC1274, which is derived from the Rio Jamapa natural population and has been used for the BAC genomic library (Froschauer et al. 2002). M, male; F, female.
All mc4r copies are linked to SD and Xmrk on the sex chromosomes of X. maculatus
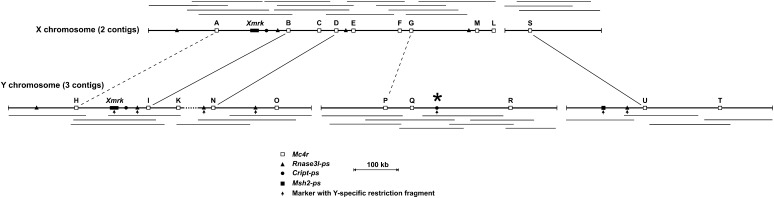
To isolate BAC clones containing the different mc4r copies present in the genome of the platyfish, a BAC library of XY males (10× genome coverage; Froschauer et al. 2002) was screened by Southern blot hybridization using a mc4r-specific molecular probe. The 42 positive BAC clones obtained were further analyzed by restriction fragment length polymorphism and Southern blot hybridization (data not shown). Some mc4r-positive BAC clones belonged to the same ∼800-kb large BAC contig containing the X-chromosomal allele of Xmrk (Froschauer et al. 2002) (Figure 2). As many as nine mc4r copies were localized in this X-linked contig. Five copies were detected in a BAC contig containing the Y-chromosomal allele of Xmrk (Froschauer et al. 2002). Five other copies were present in two other BAC contigs containing previously identified Y-chromosomal copies of pseudogenes (rnase3L-ps, also known as drosha-ps; msh2-ps; cript-ps) (Volff et al. 2003; Figure 2). Sex chromosomal localization close to xmrk was confirmed by fluorescent in situ cohybridization on chromosome preparations of BAC clones from these contigs with BAC clones from the xmrk contigs (data not shown).
Figure 2.
Localization of mc4r copies in BAC contigs from the X and Y sex chromosomes of X. maculatus. BAC clones are shown by horizontal lines. Identical mc4r copies are linked by continuous lines and very similar copies by dashed lines. The asterisk shows the position of a Y-specific pseudogene closely linked to the master sex-determining gene of X. maculatus. ps, pseudogene.
The group of BAC clones containing mc4rS could not be linked to xmrk contigs and did not present any informative Y-linked marker. However, fluorescent in situ cohybridization of a BAC clone containing mc4R-S with clones from the Xmrk contigs showed that mc4rS is also sex chromosomal and located close to the xmrk contig (data not shown). Taken together, all BAC clones obtained through genomic library screening could be assigned either to the X or to the Y chromosome. Hence, all mc4r copies of the platyfish are sex chromosomal and linked to xmrk and SD, with 10 copies on the X and 10 copies on the Y chromosome.
Sequence diversity of duplicated mc4r copies
For each mc4r copy, one BAC clone was sequenced to completion. The sequence of the open reading frame of each individual mc4r copy was determined (Figure 3 and Figure 4). All mc4r copies were found to be intronless as in all other known vertebrate mc4r genes. Substitutions and insertions/deletions (indels) were identified in mc4r-coding regions through comparison with the single-copy mc4r gene from other species. Several identical mutations were found in different copies, suggesting either mutation occurrence before gene duplication or, alternatively, events of gene conversion or recombination between duplicated copies.
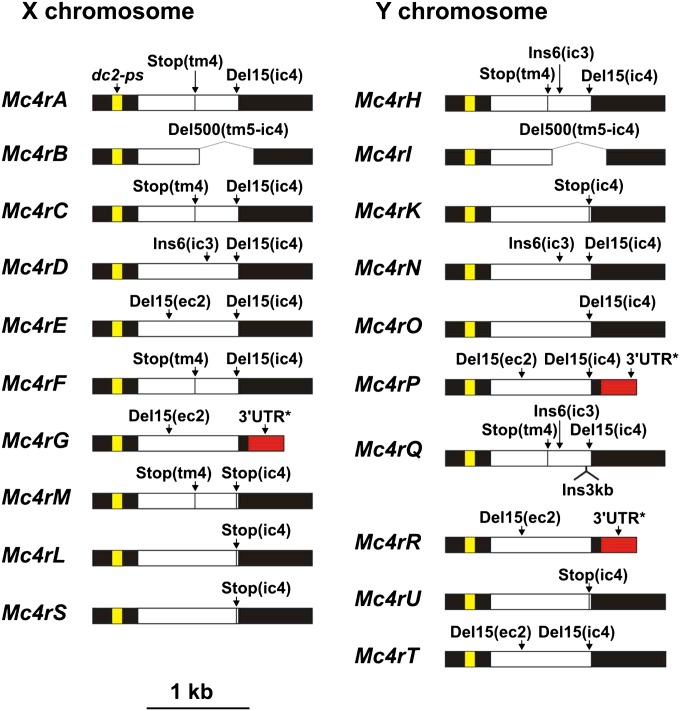
Figure 3.
Structure of X-chromosomal and Y-chromosomal mc4r copies in X. maculatus. Dc2-ps, part of the 5′ UTR derived from the dc2 gene (in yellow). Del, deletion; ec, extracellular domain; ic, intracellular domain; ins, insertion; stop, stop codon; tm, transmembrane domain. See Figure 4 for a detailed description of the effect of mutations at the amino-acid level. UTRs are in black, alternative 3′ UTR* in red, open reading frame in white.
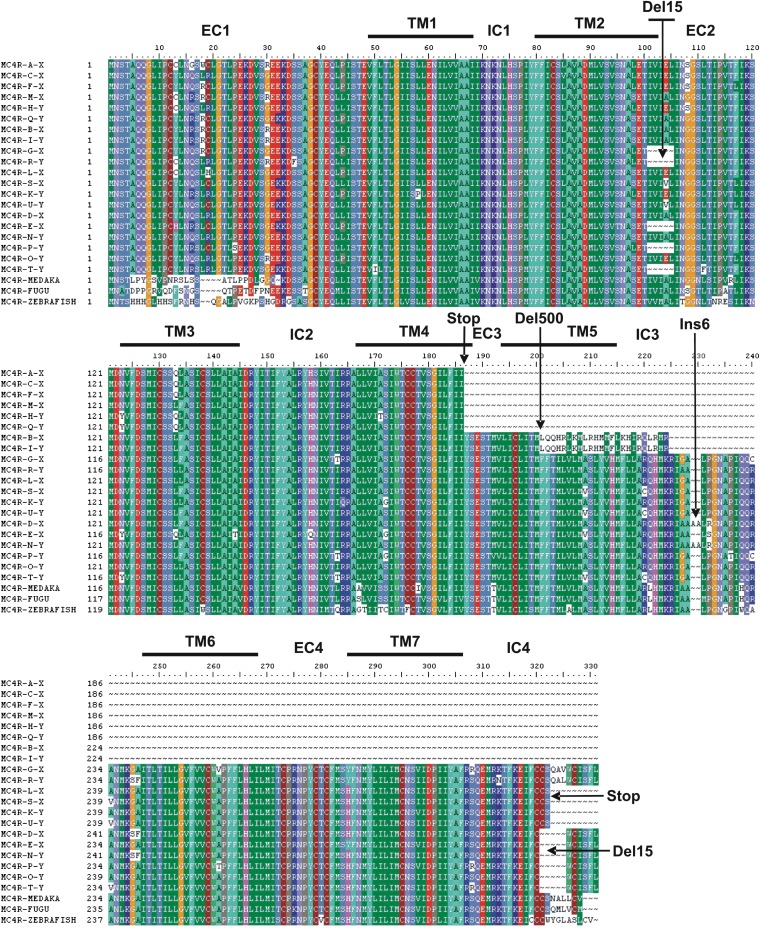
Figure 4.
Sequence alignment of Mc4r conceptual translation products. Del, deletion; EC, extracellular domain; IC, intracellular domain; Ins, insertion; stop, stop codon; TM, transmembrane domain.
Indels observed in platyfish mc4r copies can be summarized as follows (Figure 3 and Figure 4): a deletion of 15 nucleotides at the junction between transmembrane domain 2 and extracellular domain 2 (two copies on the X and three copies on the Y); a 500-nt deletion removing the coding sequence downstream from extracellular domain 3 (one copy on the X and one copy on the Y); a 6-nt insertion resulting in the addition of two alanine residues in intracellular domain 3 (one copy on the X and two copies on the Y); and a 15-nt deletion in intracellular domain 4 at the C-terminal end of the protein, removing 5 of the 11 carboxy-terminal amino-acid residues (five copies on the X and six copies on the Y). Most indels are in frame and therefore do not alter the primary sequence of the receptor downstream from the mutation. In addition, four copies on the X and two copies on the Y chromosome show a stop codon interrupting the coding region after transmembrane domain 4. Three copies on the X and two copies on the Y have a premature stop codon in the C-terminal coding region (intracellular domain 4), removing the last nine amino-acid residues of the protein. Strikingly, according to the Mc4R sequences from other fish species, including two swordtails of the genus Xiphophorus (Lampert et al. 2010), the wild-type mc4r sequence was no longer present in X. maculatus: all copies were affected by indels and/or premature stop codons at positions otherwise highly conserved in mc4r of vertebrates. No mutation in mc4r was found to be common to X. nigrensis/X. multilineatus (Lampert et al. 2010) and X. maculatus, suggesting independent mutational processes.
At the nucleotide sequence level, several mc4r copies from the X and the Y chromosome were identical (B and I; D and N; S and U) or very similar (A and H; G and P) (Figure 2). These copies might be included in discontinuous pseudoautosomal regions of the sex chromosomes, where recombination between the X and the Y chromosomes still occurs. In contrast, other copies did not possess any clear allelic counterpart on the other sex chromosome. If these copies are expressed, they might hence represent an initiation of sex-chromosome-specific variations for Mc4r functions.
The sequence of the 5′ UTR and 3′ UTR regions was determined through the sequencing of mc4r complementary DNA (cDNA) clones from a melanoma cell line library. Surprisingly, the 5′ region of all mc4r copies includes a short segment with high similarity to a gene called dc2, encoding a putative transmembrane protein reported as a component of the oligosaccharyltransferase complex in mammals (Shibatani et al. 2005). Sequence comparison with the platyfish dc2 gene indicated that 218 nt from the ∼500-nt mc4r UTR originated from dc2, this sequence being located only 38 nt upstream from the translational start codon of mc4r. Interestingly, the inserted dc2-like region corresponded to the 3′ part of the last protein-coding exon of dc2 and the 5′ part of the next noncoding exon (the last exon), but without the intervening intron. Hence, the inserted segment was derived from the reverse transcription of a spliced dc2-mRNA. The dc2-related sequences were absent in the mc4r gene of other non-Xiphophorus fish species. All platyfish mc4r copies harbored the dc2-derived sequence, indicating that the dc2 insertion occurred before mc4r serial duplication.
Two types of 3′ UTR regions were detected in platyfish mc4r copies, presenting >90% identity of their first 114 nt and diverging completely after this breakpoint. One type of 3′ UTR (∼550 nt in length) was found in most copies (17/20) while the other, 3′ UTR* (∼370 nt), was found in only three copies (G on the X chromosome and P/R on the Y).
Amplification of mc4r by large serial sex chromosomal duplications
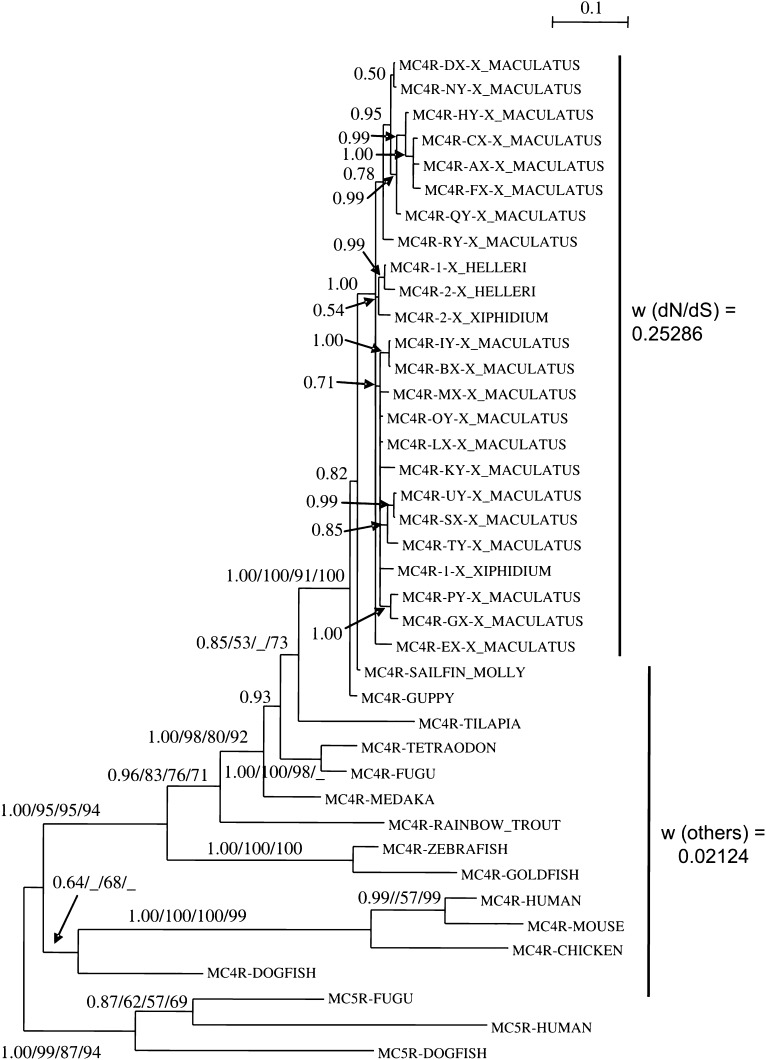
Molecular phylogeny based on the mc4r-coding sequence indicated that duplication events were relatively recent after divergence of the genus Xiphophorus and the related genus Poecilia (Figure 5). While several phylogenetic relationships between X. maculatus copies received strong statistical support, most of them could not be established unambiguously. In addition to the absence of an informative phylogenetic signal, one possible explanation might be gene conversion or recombination between copies obscuring the duplication history of mc4r genes. Accordingly, multiple recombination events were predicted between copies using DNAsp (Librado and Rozas 2009; data not shown). Using the program codeml from the PAML 3.15 package (Yang 1997), the natural selection parameter ω, corresponding to the ratio of nonsynonymous (dN) vs. synonymous (dS), was calculated. This indicated a relaxation of purifying selection probably reflecting a reduction of functional constraints (ω = 0.25286) compared to the value globally observed for mc4r in other species (ω = 0.03766).
Figure 5.
Molecular phylogeny of Mc4r sequences. Phylogeny was calculated on a 578-nucleotide sequence alignment using MrBayes (Huelsenbeck and Ronquist 2001). First or unique value gives the posterior probability value. For a given branch, when multiple values are available, a second value was obtained using maximum likelihood, a third value using maximum parsimony, and a final value using neighbor joining (see Materials and Methods). Selection parameter ω was calculated as the nonsynonymous substitution rate/synonymous substitution rate (dN/dS) ratio using CodeML from the PAML 3.15 package (Yang 1997).
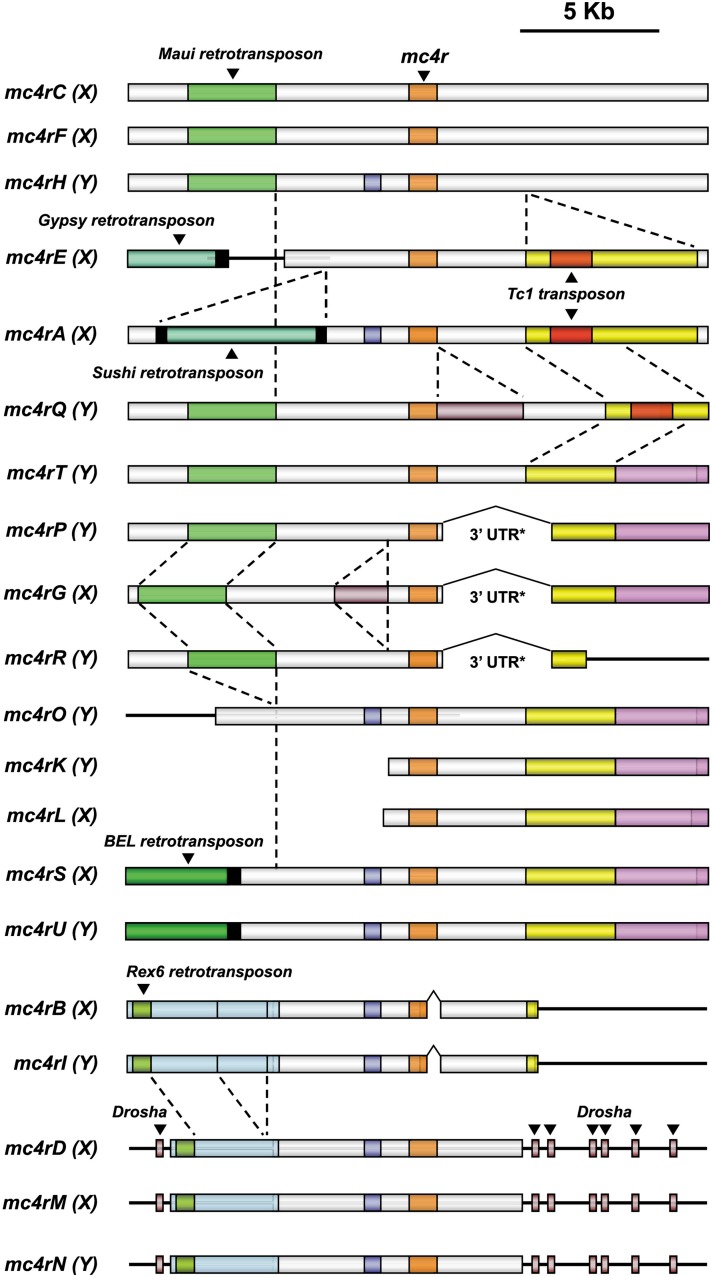
Regions containing mc4r copies were compared over a sequence window including 10 kb upstream and 10 kb downstream from the open reading frame (Figure 6). In some cases, global nucleotide identities of >90% were observed between sequences on the same chromosome over the complete length of the region analyzed, with the exception of indels ranging from a few to several hundreds of nucleotides. Hence, certain events of mc4r duplication involved genomic regions of at least 20 kb in length.
Figure 6.
Comparative structure analysis of mc4r-containing regions reveals large duplications on the sex chromosomes of the platyfish. Orange, mc4r; green, retrotransposons, with long terminal repeats in black; red, Tc1 DNA transposon. Boxes in white, blue, yellow, brown, and pink show regions present in some copies but absent from others. Drosha exons are indicated by arrowheads. Dashed lines indicate deletions.
Many indels were observed in duplicated regions; for example, a 3-kb sequence in mc4rQ was inserted directly 3′ to the open reading frame. Shorter indels were also detected but are not shown in Figure 6. Some indels correspond to transposable elements, including DNA transposons and retrotransposons [long terminal repeat (LTR) and non-LTR retrotransposons]. Certain insertions arose before duplication. For example, the Maui non-LTR retrotransposon is located at the same position in 8 of 20 copies. Frequently, some indels and other mutations are found 5′ close to the mc4r genes, suggesting that they might have an effect on their transcriptional context. One 3-kb deletion removed the 3′ part of the most common 3′ UTR as well as several kilobases of neighboring DNA (in copies P, G, and R). Interestingly, this deletion generated a new 3′ UTR region (3′ UTR*; see above). Three copies (D, M, and N) are flanked by exons of the same (pseudo)gene (rna3L, aka drosha) on both sides, indicating that the duplicated mc4r was inserted into this gene.
Expression analysis of mc4r copies
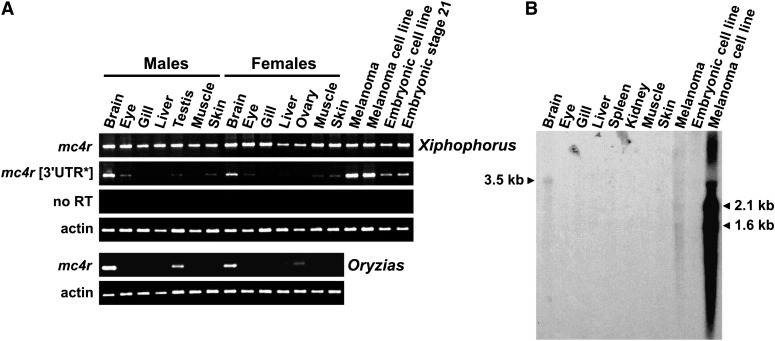
Expression of mc4r copies was analyzed by RT-PCR and Northern blot hybridization (Figure 7). RT-PCR products were sequenced to determine which type of copy was expressed in different organs and tissues. Primers specific for each type of 3′ UTR were also used in RT-PCR experiments. Results were compared with those obtained for the single-copy gene of the medaka.
Figure 7.
Mc4r expression analysis. (A) RT-PCR analysis in Xiphophorus using a reverse primer matching the “classical” 3′ UTR (“mc4r”) or the alternative 3′ UTR [“mc4r (3′ UTR*”)]. Expression of the single copy mc4r of the medaka O. latipes was also analyzed. For calibration, the cDNA amount used in the PCR reaction was monitored with actin PCR. “no RT” indicates treatment without reverse transcriptase, indicating absence of contaminating genomic DNA. (B) Northern blot analysis in Xiphophorus using an internal mc4r fragment from X. maculatus as a probe.
In medaka adult tissues, expression of mc4r was predominantly observed in brain, testis, and ovary. This is consistent with expression patterns found in other teleosts (Kobayashi et al. 2008). In contrast, mc4r copies were ubiquitously expressed in adult platyfish (Figure 7), indicating that at least some copies have a modified transcriptional pattern. Expression was also detected in embryos and an embryo-derived cell line. Copies with an alternative 3′ UTR region presented a more restricted expression pattern more similar to that observed for the single gene of the medaka. Notably, a very strong mc4r expression was observed in a melanoma cell line.
Sequencing of 171 cloned RT-PCR products obtained from different organs and tissues from X. maculatus males and females as well as from embryos and a melanoma cell line revealed detectable expression for most copies. No characteristic cDNA sequence could be identified only for copies P (Y) and E (X). Copies I (Y) and B (X) were not analyzed since the region matching the reverse PCR primer used for amplification is deleted in these copies. Expression was found for all structural types of copies (with different 3′ UTRs, indels, or stop codons). Expression of several copies was detected in all tissues and organs tested (6 different copies among 14 cDNA sequences in eyes, 5/13 in brain, 5/15 in skin, 5/14 in gills, 3/8 in liver, 4/11 in muscle, 6/9 in testis, and 4/7 in ovary). At least 5 different copies were identified in melanoma, with strong expression of copy G. Copies D/N (with two additional alanine residues in intracellular domain 3) are the most widely expressed, with expression in all nonmelanoma tissues (41/96 sequenced cDNA molecules). The proportion of expression of copies with a stop codon interrupting the coding region after transmembrane domain 4 is variable (from 1/13 in brain to 6/7 in ovary).
Functional analysis of mc4r copies
Because all mc4r copies appeared to still encode complete or partial receptors, the question about their functionality arose. We chose representatives of major paralogue classes to test for their ability to elicit a hormone response by generation of cAMP and for ligand binding.
Of the 20 different mc4r copies that were cloned from X. maculatus (Figure 4), we selected 11 for further pharmacological evaluation in cell culture experiments. These 11 copies are representatives for each possible class of observed changes in the coding sequence.
The initial functional screening was done by transient transfection of HEK-293 cells. Membranes were prepared from transfected cells and were tested for receptor function by adenylylcyclase stimulation. Vector-transfected cells served as negative controls, while cells transfected with human mc1r, Tetraodon mc1r, and Tetraodon mc4r served as positive controls (Figure 8).
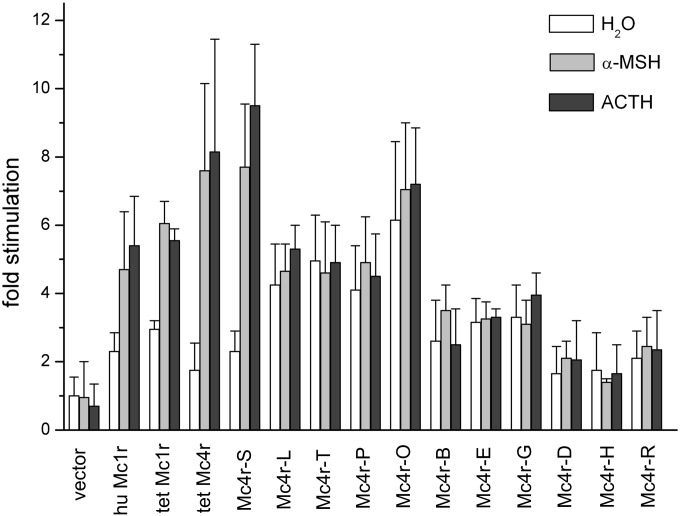
Figure 8.
Agonist-stimulated adenylyl cyclase activity in membranes isolated form HEK-293 cells transiently expressing different platyfish Mc4r receptors and wild-type melanocortin control receptors from other species. Adenylyl cyclase stimulation was calculated as fold stimulation of basal activity in untransfected control cells. A total of 100 μg membranes were stimulated with H2O, 100 μM α-MSH, or 10 μM ACTH. Open bars represent data for H2O, light-gray bars represent data for stimulation with 100 μM α-MSH, and dark-gray bars represent data for 10 μM ACTH. Data are representative of four or five experiments done in duplicate.
Membranes were stimulated with buffer, α-MSH, or ACTH, respectively. HEK-293 cells do not express endogenous functional Mcr receptors since no increase in cAMP production was observed upon stimulation using α-MSH or ACTH. In contrast, membranes prepared from cells transfected with cDNAs encoding the human Mc1r, Tetraodon Mc1r, or Tetraodon Mc4r clearly exhibited an increase in cAMP production upon agonist stimulation. These controls verified that HEK293 can be used to detect functional Mcr receptors.
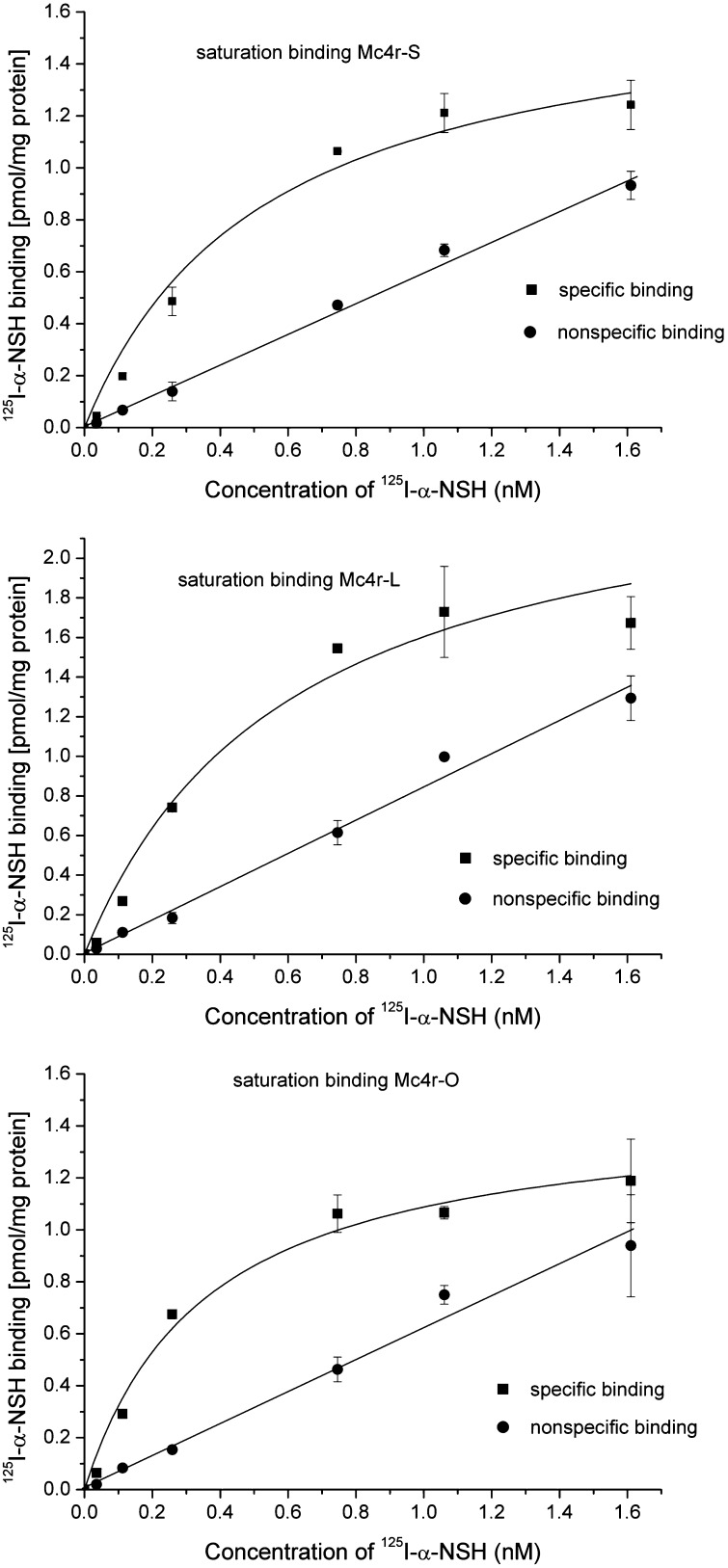
For the platyfish Mc4r tested, of the 11 copies selected for testing, only the copy termed “Mc4r-S” exhibited a significant increase in cAMP production upon stimulation with α-MSH or ACTH. From the remaining clones, Mc4r-L, T, P, and O showed a higher level of basal cAMP production when compared to clone S. Since P, T, and O form a similarity cluster with respect to their changes in amino-acid sequence, we selected S, O, and L for further analysis.
HEK-293 cells were stably transfected with each corresponding construct. Since the cauMcr4 receptor from the goldfish Carassius auratus has been pharmacologically well characterized (Cerda-Reverter et al. 2003), a control cell line was generated using this receptor. Membranes were prepared and saturation binding experiments performed using 125I-NDP-MSH as radioligand. Control HEK-293 did not exhibit any specific binding of 125I-NDP-MSH (data not shown), while each clone tested exhibited specific saturable binding of 125I-NDP-MSH (Figure 9). Next, competition binding experiments were performed to evaluate the ligand-binding properties of the selected clones (Table 1). The ligand-binding data show that all Mc4r copies tested have comparable binding affinities for NDP-MSH and α-MSH, while the affinity for ACTH varied more than fivefold between clone O and S. Differential binding affinity was also observed when the cauMc4r receptor was tested. The binding affinities for NDP-MSH and α-MSH are comparable between these species, while the cauMcr4 receptor has a 20- to 100-fold higher affinity for ACTH compared to platyfish.
Figure 9.
125I-α-NDP-MSH radioligand saturation binding to different platyfish Mc4 receptor constructs expressed in HEK-293 cells. The results of representative saturation binding experiments with the radioligand 125I-α-NDP-MSH are shown as specific (square) and nonspecific binding (circle). (Top) Saturation binding at the platyfish Mc4r-S. (Middle) Saturation binding at platyfish Mc4r-L. (Bottom) Saturation binding at platyfish Mc4r-O. The results of the saturation binding experiments are summarized in Table 1.
Table 1. Binding affinities from competition experiments for agonists at different Mc4r receptor subtypes.
| ACTH | NDP-MSH | α-MSH | ||||
|---|---|---|---|---|---|---|
| Receptor construct | Ki (nM) | 95% confidence limit | Ki (nM) | 95% confidence limit | Ki (nM) | 95% confidence limit |
| Cau Mc4r | 3.72 | 2.54–5.44 | 0.47 | 0.37–0.61 | 260.1 | 155.7–434.4 |
| Cau Mc4r (Lit) | ND | 1.43 | 988 | |||
| Mc4r-S | 466 | 214–1018 | 2.00 | 1.63–2.45 | 4083 | 2548–6543 |
| Mc4r-L | 116.6 | 69.4–196 | 4.09 | 2.20–7.58 | 5253 | 2595–10,633 |
| Mc4r-O | 76.6 | 52.1–112.7 | 2.44 | 1.18–5.04 | 2590 | 2041–3286 |
A total of 120–200 pM 125I-NDP-MSH was used as radioligand. Experiments were done in the presence of 100 μM GTP. Ki values were calculated from IC50 values using the Cheng–Prussow equation and represent geometric mean values of at least three different experiments done in duplicate. “Lit” indicates data from the literature (Cerda-Reverter et al. 2003).
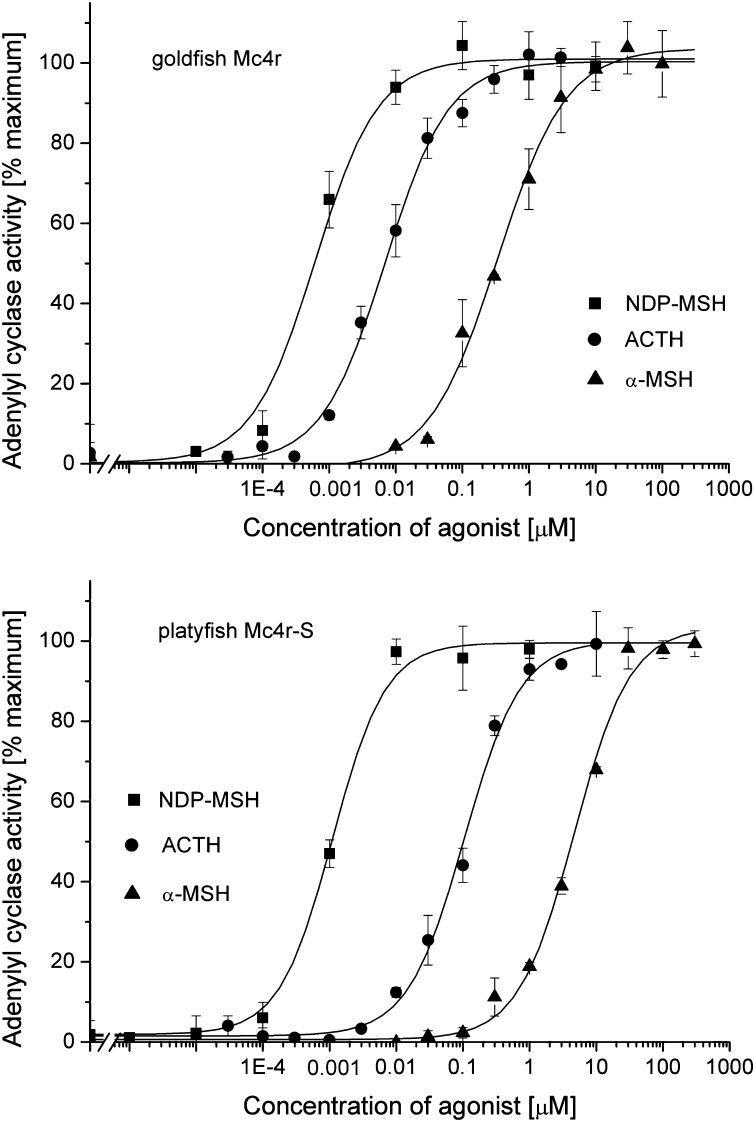
Since clone S was the only receptor copy that clearly exhibited robust agonist-dependent adenylylcyclase stimulation, we compared this receptor copy to cauMc4r for agonist-dependent adenylylcyclase stimulation parameters. Concentration response curves for ligand-dependent cAMP production of cauMc4r in comparison to platyfish S are highly similar (Figure 10). For both receptors, the agonist rank order remained constant, with NDP-MSH being more potent than ACTH followed by α-MSH. The functional data observed are in good agreement with the binding data (Table 1) and show that receptor copy S is probably the (only) physiologically functional platyfish Mc4r receptor.
Figure 10.
Concentration response curves for agonist-stimulated adenylyl cyclase activity in membranes isolated from HEK-293 cells stably expressing goldfish Mc4r or platyfish Mc4r-S. Adenylyl cyclase stimulation was calculated as a percentage of the stimulation of maximal agonist-induced stimulation. For all ligands, the maximal stimulation was similar. (Top) Data for goldfish Mc4r. (Bottom) Data for platyfish Mc4r-S. Solid squares represent data for stimulation with NDP-MSH, solid circles represent data for stimulation with ACTH, and solid triangles represent data for stimulation with α-MSH. Data are representative of four to seven experiments done in duplicate.
Discussion
Mc4r is well conserved with respect to primary structure in vertebrates (Schiöth et al. 2003, 2005; Haitina et al. 2004, 2007a,b; Metz et al. 2006; Takahashi and Kawauchi 2006). In mammals, Mc4r is involved in the control of food intake, energy balance, and body weight (for review, Adan et al. 2006; Tao 2010). Over 150 distinct mutations in this gene have been associated with hereditary obesity in humans, and disruption of mc4r in mice leads to an obese phenotype, too (Stäubert et al. 2007 and references therein; for review: Adan et al. 2006; Tao 2010). In addition, roles have been reported for Mc4r in the regulation of skin pigmentation, in cardiovascular function, and in reproduction and sexual function (Van der Ploeg et al. 2002; Spencer and Schallreuter 2009; Israel et al. 2012). In adult mammals, mc4r is predominantly expressed in different regions of the brain including the hypothalamus, septum, hippocampus, spinal cord, striatum, thalamus, brainstem, and cortex.
In teleost fish, the Mc4r receptor has been analyzed in different species (Ringholm et al. 2002; Cerdá-Reverter et al. 2003; Haitina et al. 2004; Klovins et al. 2004; Kobayashi et al. 2008; Sánchez et al. 2009; Lampert et al. 2010; Wan et al. 2012). From the pharmacological point of view, teleost Mc4r receptors have a much higher affinity for ACTH than mammalian receptors (Haitina et al. 2005). Several convergent observations support an important role of the melanocortin hormone system in the regulation of food intake and energy homeostasis in fish, probably mediated by the Mc4r receptor (Cerdá-Reverter et al. 2003; Song and Cone 2007; Sánchez et al. 2009; Schjolden et al. 2009; Wan et al. 2012; Zhang et al. 2012; Sebag et al. 2013).
Interestingly, analysis of mc4r sequence polymorphism and copy-number variation in two swordtail species, X. nigrensis and X. multilineatus, has uncovered a new Mc4r-mediated physiological link between energy balance and reproduction. In these species, different versions of mc4r were found to modulate not only male adult size but also the onset of sexual maturity and mating behavior (Lampert et al. 2010). Large late-maturing fishes, which perform courtship mating and are preferred by females, display high copy number and high levels of expression of defective mc4r copies located on the Y chromosome. These copies encode receptors mutated in their C-terminal domain that are unable to transduce melanocortin-mediated signals. In contrast, small early maturing males that reproduce through “sneak” mating have a lower copy number and lower expression levels of these defective receptors. An intermediate male phenotype for body size, puberty onset, reproductive behavior, and defective mc4r expression is also observed in both swordtail species. Hence, mc4r receptor genes have been proposed to be major constituents of the sex chromosomal P locus, which modulates body size, sexual development, and reproductive behavior in Xiphophorus (Kallman and Schreibman 1973; Kallman and Borkoski 1977; Schreibman et al. 1994). However, no genomic data were available to analyze the structure, origin, and evolution of the mc4r-containing P locus in any species.
We report here the structure of the mc4r-containing loci on the X and Y chromosomes of the platyfish X. maculatus. In this species, which is relatively divergent from the two swordtails studied so far, the P locus controls the onset of sexual maturity not only in males but also in females and, in addition to adult size in males, also fecundity in females (Kallman and Borkoski 1977). In addition, the degree of phenotypic and genetic polymorphism is much higher than in X. nigrensis and X. multilineatus: as many as nine P alleles have been identified on both the X and Y chromosomes in association with a high diversity of phenotypes (Kallman 1989). In this study, we have identified 10 mc4r copies, which have been produced through large serial duplications on both the X and Y chromosomes of the platyfish.
Expression analysis suggested diversification of expression patterns for at least some copies after duplication. As observed in other fish, mc4r expression was found in brain, particularly for copies with the alternative 3′ UTR. However, prominent mc4r expression was also detected in all other tissues and organs tested, with a surprisingly high expression in melanoma and a melanoma cell line. Several copies, for example, D/N, are expressed almost ubiquitously. Such deviations from classical expression patterns observed in other fish and vertebrates suggest that mc4r regulatory sequences have been modified in at least several of the duplicated copies in X. maculatus. Certain rearrangements observed in this study might have effects on gene expression, for example, the different indels and other mutations found 5′ from the mc4r genes. In some copies (P, G, and R), a 3′ deletion has formed a new 3′ UTR region. As this structure usually contains elements that regulate messenger RNA stability, the new 3′ UTR may have consequences for gene expression levels or cell-type specificity. We do not know at the moment if such new expression patterns are associated with new functions for Mc4r in the platyfish.
In addition, mc4r copies have also diverged after duplication through different types of mutations in their coding sequence. Functional analysis of some of these copies revealed that they have different pharmacological properties, and almost all copies are expressed in at least one tissue type. Hence, sequence divergence in coding sequences might be relevant in vivo for the modulation of Mc4r-mediated signaling. From the sequence, no Mc4r was identified that contained all conserved amino acid positions usually shared between the known fish Mc4Rs and other vertebrates (Schiöth et al. 2003). At the pharmacological level, only one platyfish Mc4r version, Mc4r-S/U, was shown to behave as a classical Mc4r receptor. Therefore, Mc4r-S/U might correspond to the wild-type physiologically functional Mc4r receptor of X. maculatus. The mc4rS/U gene is located in a pseudoautosomal region of the sex chromosomes, where it might be less prone to mutations associated with the process of sex chromosome differentiation and degeneration. However, compared to the putative ancestral receptor before duplication, the Mc4r-S/U receptor open reading frame is interrupted by a premature stop codon having removed the nine C-terminal residues of the protein. This deletion occurred only one amino acid after a dicysteine motif highly conserved in vertebrate Mc4r proteins. This motif, likely to be essential for receptor activation, is probably modified by palmitoylation, anchoring the receptor to the membrane and forming a fourth intracellular loop (Tao 2010). In fish, deletion of this dicysteine motif leads to inability to transduce the signal (Lampert et al. 2010). However, human receptors showing truncations after the C-C motif similar to Mc4r-S/U are fully functional (Ho and MacKenzie 1999; Yang et al. 2005). Hence, even with this short C-terminal truncation, Mc4r-S/U appears to function as the classical Mc4r receptor.
Due to premature stop codons or 3′ large deletions, several mc4r genes such as copies A, C, F, M, H, Q, B, and I encode conceptual translation products with C-terminal truncation after the fourth transmembrane domain. No significant activity was detected for the tested representative receptors Mc4r-H and Mc4r-B. Human Mc4r receptor variants harboring truncations after the first five transmembrane domains are neither able to stimulate cAMP production nor able to bind agonists and do not localize correctly to the membrane (Ho and MacKenzie 1999). Hence, platyfish truncated receptors are most likely to be inactive. Transcripts were found for at least four of these receptors, particularly in testis and ovary. The fact that cDNAs for Mc4r-H and -B gave rise to protein products in HEK-293 cells may indicate that those transcripts are also translated in the platyfish. Consequences of this expression—for example, dominant negative effects through binding to a wild-type receptor that interfere with transport to the membrane—remain to be analyzed. While such an effect was not detected for truncated human Mc4r variants, despite truncation, biological activities have been reported for other types of G-protein-coupled receptors (Ho and MacKenzie 1999 and references therein).
In addition to Mc4r-S/U, some other receptors are full or almost full length and without important truncation at their C-terminal end. However, none of them could be stimulated by α-MSH or ACTH to produce cAMP, indicating mutational loss of the capacity to transmit agonist-induced signaling to the cell. This might be due to mutations occurring in the third intracellular loop. This region, which appears particularly variable between platyfish Mc4r proteins, has been shown to be involved in receptor signaling but not in ligand binding in human Mc4r. Indeed, some of these receptors are still able to bind agonists, as demonstrated for Mc4r-L and Mc4r-O. Accordingly, amino-acid residues shown to be involved in ligand binding in mammalian Mc4r are conserved in these receptors (for review, see Tao 2010). Since expression of Mc4r-L and Mc4r-O was detected in various organs, these receptors might correspond to dominant negative versions of Mc4r that reduce the formation of functional Mc4r dimers or sequester the MSH ligand, leading to reduction of agonist-mediated signaling. Accordingly, coexpression of swordtail functional and nonfunctional Mc4R alleles in cell culture experiments decreased signal transduction (Lampert et al. 2010). Interestingly, at least for receptors L, T, P, and O, the level of cAMP production without agonist stimulation is largely higher than for vector control or heavily truncated vectors such as Mc4r-H. Hence, these receptors might possess a constitutive activity to stimulate G proteins in the absence of agonists.
In contrast to the situation observed in X. nigrensis and X. multilineatus, where mutated mc4r genes were found only on the Y chromosome, genes with any kind of mutations were found on both the X and Y chromosomes in the platyfish. This is consistent with the fact that the P locus in the platyfish controls not only the onset of sexual maturity and adult body size in males, but also puberty and fecundity in females (Kallman and Borkoski 1977). In addition, many more mc4r copies were found in X. maculatus, reflecting the complexity and diversity of phenotypes observed in this species. In both X. nigrensis and X. multilineatus, in addition to a wild-type mc4r “A allele,” only two mc4r “B alleles” with microdeletions at their carboxyl terminus have been identified. Both B alleles have a 6-nt deletion particularly removing two adjacent cysteine residues. One B allele harbors an additional 4-nt deletion leading to a frameshift, which replaced the six last residues of the receptor by 13 new amino acids (Lampert et al. 2010). Comparison with mutations affecting the C-terminal region in predicted platyfish Mc4r sequences, in which at least one of the two adjacent cysteines was always present, strongly suggested that these mutations have occurred independently in the X. nigrensis/X. multilineatus lineage, on the one hand, and in X. maculatus, on the other hand. It has been proposed in swordtails that nonsignal-transducing versions of Mc4r might reduce melanocortin signaling and delay the onset of puberty (Lampert et al. 2010). A similar mechanism, involving different combinations of the different Mc4r variants characterized in this work, might underlie polymorphism at the onset of sexual maturity in both males and females, adult size in males, and fecundity in females in X. maculatus. This might imply convergent evolution at the molecular level, where the same biochemical pathway is used to bring about a physiological signal leading to the onset of puberty, but through different changes in Mc4R.
Taken together, the data suggest that the sex-determining region of X. maculatus is a hotspot for large segmental duplications and show how sex chromosomal gene families can be formed. Y-specific duplications have been reported in other species, including threespine sticklebacks and humans (Peichel et al. 2004; Jobling 2008). However, in these cases, gene duplications were specific for one type of sex chromosome, the nonrecombining Y chromosome. In the platyfish, duplicated copies have been found on both the X and the Y chromosomes. Some copies are identical or extremely similar since they are located in pseudoautosomal regions, where X/Y recombination still occurs. Other copies are found in regions differing between the X and the Y chromosomes and may lead to the birth of new X- and Y-linked variations and even to new sex-specific Mc4r functions, with pseudoautosomal copies still working as classical receptors. Much more work is necessary to better characterize the physiological roles of each of these Mc4r variants and the in vivo consequences of their complex interactions.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SCHA408/7-1 and 7-2 to M.S. and J.-N.V.), the Biofuture program of the German Bundesministerium für Bildung und Forschung, the Association pour la Recherche contre le Cancer, the Centre National de la Recherche Scientifique (CNRS), the Ecole Nationale Supérieure de Lyon, the Fondation pour la Recherche Médicale, and the Agence Nationale de la Recherche (to J.-N.V.). C.S. and A.B. are recipients of fellowships from the CNRS and the French Ministère de l’Enseignement Supérieur et de la Recherche, respectively.
Footnotes
Communicating editor: B. A. Payseur
Literature Cited
- Adan R. A., Tiesjema B., Hillebrand J. J., La Fleur S. E., Kas M. J., et al. , 2006. The MC4 receptor and control of appetite. Br. J. Pharmacol. 149: 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders A., Anders F., 1978. Etiology of cancer as studied in the platyfish-swordtail system. Biochim. Biophys. Acta 516: 61–65. [DOI] [PubMed] [Google Scholar]
- Cerdá-Reverter J. M., Ringholm A., Schiöth H. B., Peter R. E., 2003. Molecular cloning, pharmacological characterization, and brain mapping of the melanocortin 4 receptor in the goldfish: involvement in the control of food intake. Endocrinology 144: 2336–2349. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Prusoff W. H., 1973. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22: 3099–3108. [DOI] [PubMed] [Google Scholar]
- Cone R. D., 2006. Studies on the physiological functions of the melanocortin system. Endocr. Rev. 27: 736–749. [DOI] [PubMed] [Google Scholar]
- De Lean A., Hancock A. A., Lefkowitz R. J., 1982. Validation and statistical analysis of a computer modeling method for quantitative analysis of radioligand binding data for mixtures of pharmacological receptor subtypes. Mol. Pharmacol. 21: 5–16. [PubMed] [Google Scholar]
- Froschauer A., Körting C., Katagiri T., Aoki T., Asakawa S., et al. , 2002. Construction and initial analysis of bacterial artificial chromosome (BAC) contigs from the sex-determining region of the platyfish Xiphophorus maculatus. Gene 295: 247–254. [DOI] [PubMed] [Google Scholar]
- Galtier N., Gouy M., Gautier C., 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12: 543–548. [DOI] [PubMed] [Google Scholar]
- Gordon M., 1927. The genetics of viviparous top-minnow Platypoecilus: the inheritance of two kinds of melanophores. Genetics 12: 253–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Gutbrod H., Schartl M., 1999. Intragenic sex-chromosomal crossovers of Xmrk oncogene alleles affect pigment pattern formation and the severity of melanoma in Xiphophorus. Genetics 151: 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitina T., Klovins J., Andersson J., Fredriksson R., Lagerström M. C., et al. , 2004. Cloning, tissue distribution, pharmacology and three-dimensional modelling of melanocortin receptors 4 and 5 in rainbow trout suggest close evolutionary relationship of these subtypes. Biochem. J. 380: 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitina T., Klovins J., Schiöth H. B., 2005. Pharmacological characterization of melanocortin receptors in fish suggests an important role for ACTH. Ann. N. Y. Acad. Sci. 1040: 337–339. [DOI] [PubMed] [Google Scholar]
- Haitina T., Klovins J., Takahashi A., Löwgren M., Ringholm A., et al. , 2007a Functional characterization of two melanocortin (MC) receptors in lamprey showing orthology to the MC1 and MC4 receptor subtypes. BMC Evol. Biol. 7: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitina T., Takahashi A., Holmén L., Enberg J., Schiöth H. B., 2007b Further evidence for ancient role of ACTH peptides at melanocortin (MC) receptors: pharmacology of dogfish and lamprey peptides at dogfish MC receptors. Peptides 28: 798–805. [DOI] [PubMed] [Google Scholar]
- Ho G., MacKenzie R. G., 1999. Functional characterization of mutations in melanocortin-4 receptor associated with human obesity. J. Biol. Chem. 274: 35816–35822. [DOI] [PubMed] [Google Scholar]
- Hoffmann C., Leitz M. R., Oberdorf-Maass S., Lohse M. J., Klotz K. N., 2004. Comparative pharmacology of human beta-adrenergic receptor subtypes: characterization of stably transfected receptors in CHO cells. Naunyn Schmiedebergs Arch. Pharmacol. 369: 151–159. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F., 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Israel D. D., Sheffer-Babila S., de Luca C., Jo Y. H., Liu S. M., et al. , 2012. Effects of leptin and melanocortin signaling interactions on pubertal development and reproduction. Endocrinology 153: 2408–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs K. H., Saur W., Schultz G., 1976. Reduction of adenylate cyclase activity in lysates of human platelets by the alpha-adrenergic component of epinephrine. J. Cyclic Nucleotide Res. 2: 381–392. [PubMed] [Google Scholar]
- Jobling M. A., 2008. Copy number variation on the human Y chromosome. Cytogenet. Genome Res. 123: 253–262. [DOI] [PubMed] [Google Scholar]
- Kallman K. D., 1975. The platyfish Xiphophorus maculatus, pp. 81–132 in Handbook of Genetics, Vol. 4, edited by King R. C. Plenum Press, New York. [Google Scholar]
- Kallman K. D., 1984. A new look at sex determination in poeciliid fishes, pp. 95–171 in Evolutionary Genetics of Fishes, edited by Turner B. J. Plenum Publishing, New York. [Google Scholar]
- Kallman K. D., 1989. Genetic control of size at maturation in Xiphophorus, pp. 163–184 in Ecology and Evolution of Livebearing Fishes (Poeciliidae), edited by Meffe G. K., Snelson F. F. J. Prentice-Hall, Englewood Cliffs, NJ. [Google Scholar]
- Kallman K. D., Borkoski V., 1977. A sex-linked gene controlling the onset of sexual maturity in female and male platyfish (Xiphophorus maculatus), fecundity in females and adult size in males. Genetics 89: 79–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallman K. D., Schreibman M. P., 1973. A sex-linked gene controlling gonadotrope differentiation and its significance in determining the age of sexual maturation and the size of the platyfish, Xiphophorus maculatus. Gen. Comp. Endocrinol. 21: 287–304. [DOI] [PubMed] [Google Scholar]
- Kang J. H., Schartl M., Walter R. B., Meyer A., 2013. Comprehensive phylogenetic analysis of all species of swordtails and platies (Pisces: genus Xiphophorus) uncovers a hybrid origin of a swordtail fish, Xiphophorus monticolus, and demonstrates that the sexually selected sword originated in the ancestral lineage of the genus, but was lost again secondarily. BMC Evol. Biol. 13: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz K. N., Hessling J., Hegler J., Owman C., Kull B., et al. , 1998. Comparative pharmacology of human adenosine receptor subtypes: characterization of stably transfected receptors in CHO cells. Naunyn Schmiedebergs Arch. Pharmacol. 357: 1–9. [DOI] [PubMed] [Google Scholar]
- Klovins J., Haitina T., Fridmanis D., Kilianova Z., Kapa I., et al. , 2004. The melanocortin system in Fugu: determination of POMC/AGRP/MCR gene repertoire and synteny, as well as pharmacology and anatomical distribution of the MCRs. Mol. Biol. Evol. 21: 563–579. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Tsuchiya K., Yamanome T., Schiöth H. B., Kawauchi H., et al. , 2008. Food deprivation increases the expression of melanocortin-4 receptor in the liver of barfin flounder, Verasper moseri. Gen. Comp. Endocrinol. 155: 280–287. [DOI] [PubMed] [Google Scholar]
- Kosswig C., 1928. About crosses between the teleosts Xiphophorus hellerii and Platypoecilus maculatus. Z. Indukt. Abstamm. Vererbungsl. 47: 150–158. [Google Scholar]
- Kuhn C., Vielkind U., Anders F., 1979. Cell cultures derived from embryos and melanoma of poeciliid fish. In Vitro 15: 537–544. [DOI] [PubMed] [Google Scholar]
- Lampert K. P., Schmidt C., Fischer P., Volff J.-N., Hoffmann C., et al. , 2010. Determination of onset of sexual maturation and mating behavior by melanocortin receptor 4 polymorphisms. Curr. Biol. 20: 1729–1734. [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Meierjohann S., Schartl M., 2006. From Mendelian to molecular genetics: the Xiphophorus melanoma model. Trends Genet. 22: 654–661. [DOI] [PubMed] [Google Scholar]
- Metz J. R., Peters J. J., Flik G., 2006. Molecular biology and physiology of the melanocortin system in fish: a review. Gen. Comp. Endocrinol. 148: 150–162. [DOI] [PubMed] [Google Scholar]
- Peichel C. L., Ross J. A., Matson C. K., Dickson M., Grimwood J., et al. , 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Ringholm A., Fredriksson R., Poliakova N., Yan Y. L., Postlethwait J. H., et al. , 2002. One melanocortin 4 and two melanocortin 5 receptors from zebrafish show remarkable conservation in structure and pharmacology. J. Neurochem. 82: 6–18. [DOI] [PubMed] [Google Scholar]
- Ryan M. J., Causey B. A., 1989. ‘‘Alternative’’ mating behavior in the swordtail Xiphophorus nigrensis and Xiphophorus pygmaeus (Pisces: Poeciliidae). Behav. Ecol. Sociobiol. 24: 341–348. [Google Scholar]
- Ryan M. J., Rosenthal G. G., 2001. Variation and selection in swordtails, pp. 133–148 in Model Systems in Behavioral Ecology, edited by Dugatkin L. A. Princeton University Press, Princeton, NJ. [Google Scholar]
- Ryan M. J., Hews D. K., Wagner W. E. J., 1990. Sexual selection on alleles that determine body size in the swordtail Xiphophorus nigrensis. Behav. Ecol. Sociobiol. 26: 231–237. [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual, Ed. 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sánchez E., Rubio V. C., Thompson D., Metz J., Flik G., et al. , 2009. Phosphodiesterase inhibitor-dependent inverse agonism of agouti-related protein on melanocortin 4 receptor in sea bass (Dicentrarchus labrax). Am. J. Physiol. Regul. Integr. Comp. Physiol. 296: R1293–R1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M., 2008. Evolution of Xmrk: An oncogene, but also a speciation gene? Bioessays 30: 822–832. [DOI] [PubMed] [Google Scholar]
- Schartl M., Walter R. B., Shen Y., Garcia T., Catchen J., et al. , 2013. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat. Genet. 45: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiöth H. B., Raudsepp T., Ringholm A., Fredriksson R., Takeuchi S., et al. , 2003. Remarkable synteny conservation of melanocortin receptors in chicken, human, and other vertebrates. Genomics 81: 504–549. [DOI] [PubMed] [Google Scholar]
- Schiöth H. B., Haitina T., Ling M. K., Ringholm A., Fredriksson R., et al. , 2005. Evolutionary conservation of the structural, pharmacological, and genomic characteristics of the melanocortin receptor subtypes. Peptides 26: 1886–1900. [DOI] [PubMed] [Google Scholar]
- Schjolden J., Schiöth H. B., Larhammar D., Winberg S., Larson E. T., 2009. Melanocortin peptides affect the motivation to feed in rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 160: 134–138. [DOI] [PubMed] [Google Scholar]
- Schreibman M. P., Schartl M., Kallman K. D., Magluilo-Cepriano L., 1994. Molecular approaches to study the genetic regulation of the fish reproductive system, pp. 343–351 in Perspectives in Comparative Endocrinology, edited by Davey K. G., Peter R. E., Tobe S. S. National Research Council of Canada, Ottawa. [Google Scholar]
- Schultheis C., Zhou Q., Froschauer A., Nanda I., Selz Y., et al. , 2006. Molecular analysis of the sex-determining region of the platyfish Xiphophorus maculatus. Zebrafish 3: 299–309. [DOI] [PubMed] [Google Scholar]
- Schultheis C., Böhne A., Schartl M., Volff J.-N., Galiana-Arnoux D., 2009. Sex determination diversity and sex chromosome evolution in poeciliid fish. Sex Dev. 3: 68–77. [DOI] [PubMed] [Google Scholar]
- Sebag J. A., Zhang C., Hinkle P. M., Bradshaw A. M., Cone R. D., 2013. Developmental control of the melanocortin-4 receptor by MRAP2 proteins in zebrafish. Science 341: 278–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selz Y., Braasch I., Hoffmann C., Schmidt C., Schultheis C., et al. , 2007. Evolution of melanocortin receptors in teleost fish: the melanocortin type 1 receptor. Gene 401: 114–122. [DOI] [PubMed] [Google Scholar]
- Shibatani T., David L. L., McCormack A. L., Frueh K., Skach W. R., 2005. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry 44: 5982–5992. [DOI] [PubMed] [Google Scholar]
- Song Y., Cone R. D., 2007. Creation of a genetic model of obesity in a teleost. FASEB J. 21: 2042–2049. [DOI] [PubMed] [Google Scholar]
- Spencer J. D., Schallreuter K. U., 2009. Regulation of pigmentation in human epidermal melanocytes by functional high-affinity beta-melanocyte-stimulating hormone/melanocortin-4 receptor signaling. Endocrinology 150: 1250–1258. [DOI] [PubMed] [Google Scholar]
- Stäubert C., Tarnow P., Brumm H., Pitra C., Gudermann T., et al. , 2007. Evolutionary aspects in evaluating mutations in the melanocortin 4 receptor. Endocrinology 148: 4642–4648. [DOI] [PubMed] [Google Scholar]
- Takahashi A., Kawauchi H., 2006. Evolution of melanocortin systems in fish. Gen. Comp. Endocrinol. 148: 85–94. [DOI] [PubMed] [Google Scholar]
- Tao Y. X., 2010. The melanocortin-4 receptor: physiology, pharmacology, and pathophysiology. Endocr. Rev. 31: 506–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G., 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ploeg L. H., Martin W. J., Howard A. D., Nargund R. P., Austin C. P., et al. , 2002. A role for the melanocortin 4 receptor in sexual function. Proc. Natl. Acad. Sci. USA 99: 11381–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff J.-N., Schartl M., 2002. Sex determination and sex chromosome evolution in the medaka, Oryzias latipes, and the platyfish, Xiphophorus maculatus. Cytogenet. Genome Res. 99: 170–177. [DOI] [PubMed] [Google Scholar]
- Volff J.-N., Körting C., Froschauer A., Zhou Q., Wilde B., et al. , 2003. The Xmrk oncogene can escape nonfunctionalization in a highly unstable subtelomeric region of the genome of the fish Xiphophorus. Genomics 82: 470–479. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y., 1981. Establishment of a cell line from the platyfish-swordtail hybrid melanoma. Cancer Res. 41: 679–680. [PubMed] [Google Scholar]
- Wan Y., Zhang Y., Ji P., Li Y., Xu P., et al. , 2012. Molecular characterization of CART, AgRP, and MC4R genes and their expression with fasting and re-feeding in common carp (Cyprinus carpio). Mol. Biol. Rep. 39: 2215–2223. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J., Adam D., Malitschek B., Mäueler W., Raulf F., et al. , 1989. Novel putative receptor tyrosine kinase encoded by the melanoma-inducing Tu locus in Xiphophorus. Nature 341: 415–421. [DOI] [PubMed] [Google Scholar]
- Yang Y., Chen M., Loux T. J., Georgeson K. E., Harmon C. M., 2005. Molecular mechanism of the intracellular segments of the melanocortin-4 receptor for NDP-MSH signaling. Biochemistry 44: 6971–6979. [DOI] [PubMed] [Google Scholar]
- Yang Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Zhang C., Forlano P. M., Cone R. D., 2012. AgRP and POMC neurons are hypophysiotropic and coordinately regulate multiple endocrine axes in a larval teleost. Cell Metab. 15: 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerer E. J., 1982. Size-related courtship strategies in the pygmy swordtail, Xiphophorus nigrensis. Am. Zool. 22: 910. [Google Scholar]