Abstract
Cell adhesion is a broad topic in cell biology that involves physical interactions between cells and other cells or the surrounding extracellular matrix, and is implicated in major research areas including cancer, development, tissue engineering, and regenerative medicine. While current methods have contributed significantly to our understanding of cell adhesion, these methods are unsuitable for tackling many biological questions requiring intermediate numbers of cells (102–105), including small animal biopsies, clinical samples, and rare cell isolates. To overcome this fundamental limitation, we developed a new assay to quantify the adhesion of ~102–103 cells at a time on engineered substrates, and examined the adhesion strength and population heterogeneity via distribution-based modeling. We validated the platform by testing adhesion strength of cancer cells from three different cancer types (breast, prostate, and multiple myeloma) on both IL-1β activated and non-activated endothelial monolayers, and observed significantly increased adhesion for each cancer cell type upon endothelial activation, while identifying and quantifying distinct subpopulations of cell-substrate interactions. We then applied the assay to characterize adhesion of primary bone marrow stromal cells to different cardiac fibroblast-derived matrix substrates to demonstrate the ability to study limited cell populations in the context of cardiac cell-based therapies. Overall, these results demonstrate the sensitivity and robustness of the assay as well as its ability to enable extraction of high content, functional data from limited and potentially rare primary samples. We anticipate this method will enable a new class of biological studies with potential impact in basic and translational research.
1 Introduction
Biological cells physically interact with and adhere to different materials and elements in their tissue microenvironments. These interactions play important roles in maintaining normal cell behavior, and are implicated in many different pathologies. For example, adhesion is involved in the normal mechanoregulation of vascular and lymphatic endothelium1,2, the differentiation of mesenchymal stem cells (MSCs) on extracellular matrices (ECMs) of various mechanical stiffnesses3, and the attachment of circulating tumor cells (CTCs) at ectopic locations of the vasculature during metastasis.4 Furthermore, the advancement of tissue engineering relies critically on the ability of cells to attach, grow, and remain viable on engineered scaffolds and other biocompatible materials.5,6 Thus, cell adhesion is central to myriad important questions in modern biology and biomedicine, including those related to development, physiology, pathophysiology, tissue regeneration and cell-based therapies.
Many techniques and systems have been developed to measure and characterize adhesion properties of cells.7 The most common approach involves the use of population-based shear flow systems that rely on laminar flow to apply controllable shear rates on cultured cells, and quantify the fractions of adherent (and non-adherent) cells in the entire circulated population. Among these systems, parallel plate flow chambers and cone-and-plate viscometers utilize either increasing or decreasing shear rate protocols to cause detachment or attachment of cells, respectively8,9, whereas variable width or height flow chambers and radial flow systems rely on geometry to generate variable shears at different spatial locations using a single flow rate.10–13 A second major class of adhesion measurement techniques is single-cell manipulation methods where individual cells are subjected to controlled force application using atomic force microscopy or micropipette aspiration to detach cells from their adhered surfaces.7,14–16
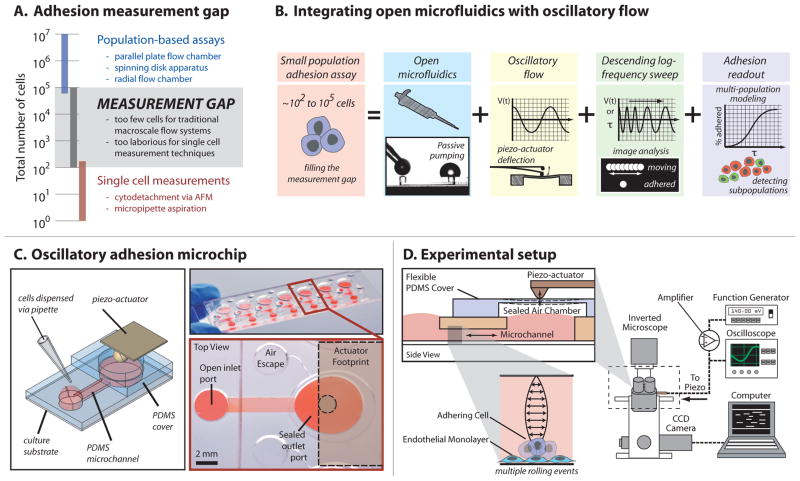
These two traditional classes of cell adhesion assays lie on opposite ends of the population-size spectrum (Fig. 1A). While single-cell techniques can provide detailed information on adhesion properties of individual cells, they are laborious and require expensive, delicate equipment, and as such are typically used to study tens or (at most) hundreds of cells. In contrast, while the majority of population-based shear flow systems can test more than 105 cells in a single experiment, they are limited to average readouts that inherently mask single-cell information, which may reveal important insights on population heterogeneity. Furthermore, these macroscale flow systems often require a minimum of ~105 cells to yield detectable endpoints for each assay, limiting the range of possible biological questions that can be tackled. Thus, a technical gap exists for intermediate cell samples between ~102 to 105 cells, including, for example, primary samples from humans or animal models, and cellular subpopulations isolated via cell sorting. While microfluidic systems have received attention for their ability to increase throughput, reduce reagent volumes and cell sample requirements, and enable new biological studies, these systems have yet to fill this measurement gap for adhesion assays.17–20
Fig. 1.
(A) Open microfluidic oscillatory flow assay addresses the current adhesion measurement gap. (B) A small population adhesion assay capable of handling 102–105 cells was developed by integrating open microfluidics via passive pumping, oscillatory flow via piezo-cantilever deflection, and a descending logarithmic frequency sweep to generate information-rich readouts. (C) The microchip consists of seven PDMS microchannels on a 3″ × 1″ slide that serves as the culture substrate. The culture substrate may be glass (shown, top right), polystyrene (used in reported experiments), or other material. Each microchannel consists of a straight microchannel with a small circular inlet port and large ovoid outlet port, and a flexible diaphragm made of PDMS for sealing the outlet during actuation. (D) The microchip can be positioned on the stage of an inverted microscope. As the piezo-actuator bends, deflections in the diaphragm generate pressures in the sealed air chamber that result in oscillatory flow within the microchannel where cells are cultured. A signal from a function generator is sent through a piezo amplifier, monitored using an oscilloscope, and sent to the piezo-cantilever for actuation. A computer is used to control image-stream acquisition and subsequent processing.
To address this unmet need, we developed a new microfluidic system for conducting cell adhesion assays on limited cell samples consisting of only hundreds to thousands of cells (Fig. 1B). The system enables pipette-based loading of cells and reagents, is amenable to complex microscale geometries and microenvironments, and offers flexibility in custom fluid flow waveforms for various applications. These capabilities, coupled with an information-rich readout, allow this system to be applied to a range of biological experiments. Here we describe the method and demonstrate the ability to observe physiologically relevant changes in adhesion in the context of both cancer biology and cardiology. We describe a method for identifying and quantifying distinct subpopulations of cell-substrate interactions, and also employ cell-derived matrix substrates, demonstrating the ability to integrate engineered microenvironments for developing more biologically relevant in vitro studies.
2 Results
2.1 Design and Operation
The designed microscale system integrates four key features to create an adhesion assay intended to fill the gap in measurement techniques and enable new experiments: (1) open microfluidics via passive pumping, (2) oscillatory flow, (3) a custom data acquisition and image analysis protocol, and (4) a multi-population modeling approach for quantifying adhesion (Fig 1B). Passive pumping, which relies on surface tension to pump fluid from small droplets to larger droplets based on Laplace’s law (Fig 1B),21 reduces dead volumes associated with tubing and media reservoirs common to flow-through systems, and can be used with different channel geometries and substrates to incorporate advanced functionality and micropipette-based operation.22–24 In the current system, a simple straight microchannel (7.5 mm × 1.5 mm × 200 μm) connects a small input port to a large output port to facilitate micropipette delivery of cell suspensions, media, and reagents via passive pumping (Fig. 1C).
Oscillatory flow was generated using a flexible diaphragm fabricated from polydimethylsiloxane (PDMS) positioned over the large output port, and reversibly sealed to the top of the PDMS microchannel (Fig. 1C–D). The diaphragm was actuated using a piezo-cantilever, which bends along its length in response to a voltage signal produced using a function generator and signal amplifier. Deflections of the diaphragm induced pressure changes within the reversibly sealed air chamber to induce oscillatory flow in the microchannel while the ports served as low-volume reservoirs to accommodate volume fluctuations. Although, device geometry and operating frequencies and amplitudes used in the cell-based studies ensure a linear relationship between frequency and shear stress (maximum Womersley number < 1), characterizations of linear and non-linear system behavior were also performed for completeness and potential use in other applications (ESI).
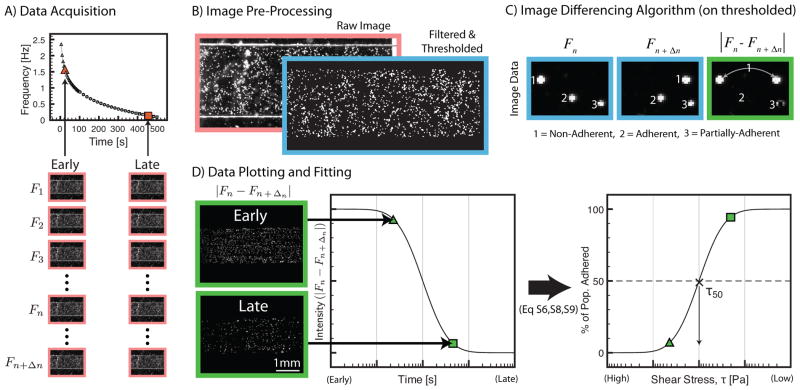
To perform the assay, the piezo-cantilever was set to a frequency of 2 Hz to initiate oscillatory flow and cells were seeded via the input port while the fluid was oscillating. Seeding the cells in this manner prevented undesired cell adhesion before data acquisition began. After cells settled for ~45 s (sufficient for the cells to reach the bottom), data acquisition was initiated and the frequency of oscillation was decreased logarithmically from 2 Hz to 100 mHz over 500 s, i.e., a descending logarithmic frequency sweep (Fig. 1B, 2A, & ESI). As shear stress decreased, an increasing number of cells were allowed to adhere over time. An automated image analysis algorithm was developed to quantify the percent of adhered cells over the 20-fold range in shear-stress for each of the 46 image streams using automated image analysis (Fig. 2).
Fig. 2.
Data acquisition and image analysis. Red borders = raw images; blue borders = filtered and thresholded images; green borders = absolute difference between two filtered and thresholded images. (A) 46 different image streams, each consisting of multiple frames, are acquired during a descending logarithmic frequency sweep. Data points show times at which image streams are taken and frequency at which the system oscillated during that image stream. Different numbers of frames are required to capture 1 to 2 cycles of cell motion for different frequencies. Triangles consist of 25 frames each, circles 85 frames each, and squares 170 frames each stream. Each frame is represented mathematically as F where the subscript denotes the index of the frame in the image stream. (B) Comparison of raw image with filtered and thresholded images. Filtering and thresholding enhances the seeded cells to be above background artifacts arising from the endothelial cell monolayer or ECM substrate. (C) Illustration of the image differencing algorithm. Cells in motion appear twice in the image (1) while stationary cells disappear (2). Partially adherent cells that exhibit a rocking motion result in a partial signal (3). (D) A difference image is shown that correlates with a difference-signal that is transformed into percent of population adhered (see ESI for further details on the algorithm and definitions of Eq S6, S8, and S9).
Analysis was limited to the center 80% of the channel width to ensure data was taken within a region of uniform shear stress.25 Percentage of adherent cells in this region was plotted as a function of shear stress, τ, and fitted to a sigmoidal curve based on the log-normal cumulative distribution function, FLN, to extract values for τ50 and σ, thereby modeling the data as a single population of cell-substrate interactions (ESI). Physically, τ50 represents the critical shear stress at which 50% of cells are adhered (i.e., the median shear stress), and σ represents the standard deviation of the shear stress on a logarithmic scale, which reveals population heterogeneity of adhesion interactions. The data was also fitted with the linear combination of two FLN functions to model the data as two populations. If the dual-population model provided a sufficient increase in the goodness of fit measure, R2, the dual-population model was used instead of the single-population model to determine a τ50 and σ for each of the two subpopulations. The results of the subpopulations are then combined using a weighted average to estimate overall values for τ50 and σ (ESI).
2.2 Assay Validation: Cancer Cell Adhesion
As validation that our method was sufficiently robust and sensitive for characterizing physiologically relevant changes in adhesion, we tested adhesion of three established cell lines derived from distinct types of cancer, including prostate (PC3-MM2), breast (MDA-MB231), and multiple myeloma (RPMI8226), on cultured monolayers of activated (IL-1β treated) and non-activated human umbilical vein endothelial cells (HUVECs). Studies on the adhesiveness, invasiveness, and metastatic potential of these cell lines have shown that PC3 cells are particularly adhesive and invasive.26,27 MDA-MB231 cells are also highly invasive among breast cancer cell lines28,29, but are known to adhere to activated endothelium only in the absence of shear-stress > 0.05 Pa.28 In contrast to prostate and breast cancers, the adhesiveness and invasiveness of cells of multiple myeloma (MM) origin has been studied to a much lesser extent. Reports of MM cell adhesion that are available suggest that adhesion to (bone marrow) endothelium is mediated by various molecules other than E-selectin.30–32 Thus, we expected that PC3-MM2 and MDA-MB231 cells would exhibit more adhesive phenotypes, while RPMI8226 cells would likely show less adhesion to the endothelial monolayers.
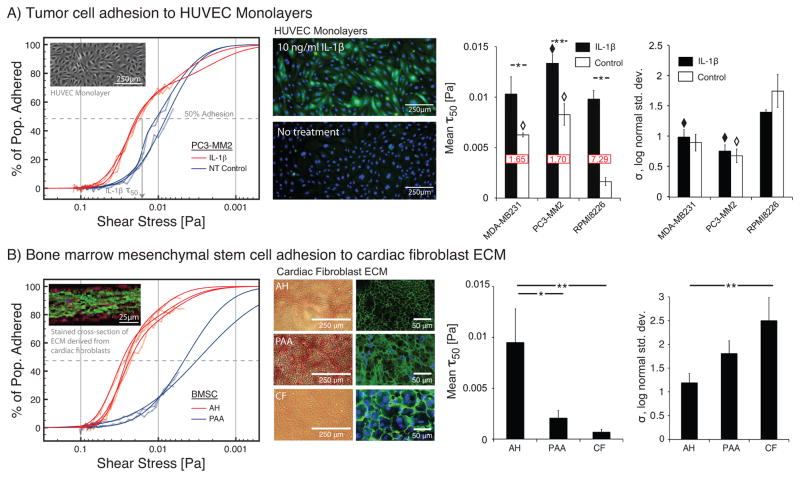
The adhesion data curves were obtained using ~2,500 tumor cells per microchannel. Thus, the total of 50 microchannels that were needed to perform the 6 different sets of experiments and acquire 2,300 adhesion-shear data points required ~125,000 tumor cells total, which is about the number of cells typically needed for a single data point in using traditional methods. Population modeling and the associated τ50 and σ values for each combination of cancer cell type and endothelial surface were analyzed (Fig. 3A). We observed statistically significant increases in τ50 between activated and non-activated endothelial surfaces for all cell types, including RPMI8226 cells. For non-activated endothelium conditions, both MDA-MB231 and PC3-MM2 cells exhibited significantly higher values of τ50 than RPMI8226 cells. However upon activation, RPMI8226 cells exhibited a significantly higher fold-increase than the others, largely due to the very low initial value. Thus, MDA-MB231 and PC3-MM2 cells were likely more adhesive than RPMI8226 cells initially because of intrinsic differences in cell surface properties, which were then amplified for all cell types in the presence of IL-1β induced E-selectin. This was consistent with the literature cited above.
Fig. 3.
Adhesion assay results for two different applications. (left) Raw adhesion curves (semi-transparent lines) indicate percent of cell population adhered vs. shear stress. Multi-population modeling results are plotted as solid lines. Population models yield two measures: (i) critical shear stress at which 50% of the population is adhered (τ50), and (ii) the log-normal standard deviation (σ). (left-middle) Images of substrate conditions. (right) Average values of τ50 (fold change of activated vs. non-activated shown in red box) and σ. (A) Tumor cell adhesion to IL-1β-activated and non-activated endothelial monolayers for three different tumor cell lines representing breast cancer (MDA-MB231), prostate cancer (PC3-MM2), and multiple myeloma (RPMI8226). Immunostaining shows E-selectin (green) upregulation via 10 ng/ml IL-1β compared to no treatment. Nuclei stained with Hoechst (blue). (B) Primary BMSC adhesion to 3D-ECM produced by primary cardiac fibroblasts treated via three different methods. Images of ECM conditions show eosin staining and immunostaining. Immunostaining images, including the graph inset image of a cross section of the ECM, are false colored to show fibronectin as green, collagen as red, and nuclei as blue (enlarged versions in ESI). (A,B) Error bars = SE for n ≥ 3. Horizontal bars: “*” = p < 0.05, “**” = p < 0.01. “◆” = p < 0.05 compared to RPMI8226s on IL-1β-activated endothelium. “◇” = p < 0.05 compared to RPMI8226s on non-activated endothelium.
Values of σ were also compared between non-activated and activated endothelium groups and across cell types. We observed that within each cell type, there was no statistical difference in σ between activated and non-activated endothelium. However, σ for RPMI8226 cells was significantly higher than σ for PC3-MM2 and MDA-MB231 cells on activated endothelial monolayers, and significantly higher than σ for PC3-MM2 on non-activated endothelial monolayers. These data suggested that σ, or the population heterogeneity in the strength of cell-substrate interactions, was not influenced by endothelial activation, but only by intrinsic differences between cells.
The change in R2 between the single- and dual-population model fits ( ) was significantly larger in the case of the prostate (p = 0.0002) and multiple myeloma (p = 0.023) cells on activated endothelium compared to the the non-activated cases. This was not observed for the breast cancer cell line (p = 0.55). Thus, although values of σ did not suggest an overall change in heterogeneity upon activation for any cell type, population modeling suggested the emergence of a subpopulation of cell-substrate interactions upon activation for PC3-MM2 and RPMI8226 cells. Notably, in the case of RPMI8226 cells on activated endothelium, on average ~13% of the cell population adhered at a shear stress roughly 30-fold higher than that of the remaining 87% of the population. Thus, while the general trends related to cell adhesion propensity for different cancer cell types agreed with existing literature and validated our platform, the analysis was able to provide additional insights including the overall heterogeneity of the cell-substrate interactions (σ) and the identification and quantification of subpopulations for more detailed examinations of cell attachment and functional response.
The data was further mined in order to assess the potential range of cell sample sizes that could be addressed using this method. Three channels were chosen that exhibited a significant change in the adhesion signal and analyzed using successively smaller ROIs that contained ~500 cells, down to as few as 25 cells. As expected, below ~100 cells the sample became significantly less representative of the larger population, making it difficult to use population modeling to extract parameters, establishing a minimum sample size limit of 102 (Fig. S7 & S8, ESI).
2.3 Assay Application: Primary Bone Marrow Stromal Cell Adhesion to a Primary Cardiac Fibroblast Cell Produced 3-Dimensional Extracellular Matrix
Next, we applied the assay in the field of cardioregenerative medicine to further illustrate the potential for new applications and insights enabled by this technology. Adhesion of therapeutic cells to injured myocardium is a crucial yet understudied component of cell-based cardioregenerative therapy. Current estimates of therapeutic cell retention vary with cell type, mode of delivery and elapsed time from administration, but generally less than 10% of the cells transplanted adhere in the myocardium.33–35 Despite the poor retention rates, small improvements in cardiac function and reduction in ventricular scarring have been reported, providing evidence of potential therapeutic utility.36,37 Such in vivo studies can be challenging and expensive, making it difficult to iteratively test and develop new strategies for improved efficacy. Another common challenge of cell-based therapies is the often-limited source and quantity of the therapeutic cells, which can preclude the use of many conventional adhesion assays, whether in vivo or in vitro. Our adhesion assay addresses these challenges by providing an in vitro approach that enables study of potentially limited populations of therapeutic cells.
In this proof-of-principle study, we quantify adhesion of bone marrow mesenchymal stromal cells (BMSCs), a potential adult therapeutic cell population of interest in cardioregenerative medicine. The BMSCs are harvested from a single rat for a set of experiments to reduce rat-to-rat experimental noise. Thus, as often occurs in small animal primary cell work, the cell quantities are limited. We characterize their adhesion to different preparations of ECM derived from primary rat cardiac fibroblasts (CFs) that are considered relevant for downstream development. Although the flexibility of the assay allows integration of cell-derived matrix and oscillatory flow to approximate some aspects of the therapeutic site for potential improvements in relevance (e.g., the fibroblastic nature of cardiac scar tissue, a cell-derived mixture of matrix proteins and other cellular factors, as well as periodic motion during therapeutic cell attachment), the focus of this study is to provide a relevant demonstration of the ability to efficiently leverage precious cell resources to enable development of new strategies to improve therapeutic cell retention.
Briefly, a confluent monolayer of primary rat CFs were seeded into the microchannels and cultured for 10 ± 3 days to produce the 3D cell derived ECM. Each microchannel was either left untreated, or was decellularized using ammonium hydroxide (AH) or peracetic acid (PAA) protocols (see ESI for methods). The AH treatment left lower levels of cell debris compared to the PAA treatment and required an additional collagen coating on the polystyrene prior to seeding to help anchor the matrix. The CF condition represents untreated cardiac fibroblasts without collagen pretreatment (Fig. 3B, images). These particular preparations were chosen to obtain preliminary results that would help direct future investigations, but also demonstrate the ability to generate various matrices on the channel surfaces for integration with the assay.
Notably, ~1% of the BMSCs from a single rat was needed each day to test multiple conditions with replicates. Population modeling results show that BMSCs adhered to the AH-treated ECM at shear stress levels ~10-fold higher than that achieved in the case of PAA or the untreated cardiac fibroblast cell sheet, CF (Fig. 3B). The AH condition also exhibited a significantly lower heterogeneity (σ) compared to the CF case, suggesting the possible presence of more homogeneous and stronger adhesion interactions. The AH condition also exhibited more dual-population behavior than for CF ( , p = 0.045) but was not significantly different from PAA (p = 1). More importantly, the study shows the ability to leverage a limited cell source for assessing a range of adhesion conditions, thus, enabling a more efficient approach to iteratively developing new hypotheses and strategies for modulating adhesion. Further, it demonstrates the ability to integrate engineered microenvironments to enable improvements in biological relevance.
3 Discussion
We have described a robust, sensitive, and quantitative microscale adhesion assay that fills a gap in the current repertoire of adhesion measurement methods. The system can efficiently utilize as few as 102 cells to characterize the distribution of adhesion interactions at 46 different shear stresses spanning a 20-fold range, significantly broadening the current range of potential cell samples that can be studied using shear flow. Central to the design of the system and assay are four main elements that, when integrated, provide new and previously unachievable functionality.
First, passive pumping enables an open microfluidics design that facilitates loading of minute cell samples with minimal cell loss and dead volume typical of tube-based systems and maintains complete separation of samples to ensure independence of each data point acquired from the array. While passive pumping devices have been widely applied for practical cell-based assays for numerous studies in the past decade22,24,38–40, the current application is the first to achieve tunable, continuous, dynamic fluid flow in such a device over the entire time-course of the experiment. Further, the modular nature of the design allows the microchannel cell culture device and oscillatory flow components to remain uncoupled from one another until the time of the adhesion assay. This modularity simplifies preparation of cell monolayers and surface treatments via pipette-based operations and streamlines logistics during experimental procedures to facilitate integration of engineered microenvironments.
Second, oscillatory fluid flow facilitates monitoring a fixed population of cells. While oscillatory flow is more typically employed as a mechanical stimulus to model physiological conditions where oscillatory flow is relevant,41,42 its use here enables observation and quantification of the same group of cells rocking back-and-forth continuously over time. This is fundamentally different from continuous flow systems where new cells are continually introduced to the adherent substrate in a manner that presents each cell with a limited opportunity to attach. While recirculation can be implemented with continuous flow systems to reduce the total number of cells required for an assay, there remain significant dead volumes associated with tubing and reservoirs.
Third, use of a descending logarithmic frequency sweep and automated image analysis enables quantification of adhesion data at 46 different time-points spanning a 20-fold change in shear stress. This large amount of data would have traditionally required 46 independent experiments each requiring large numbers of cells to obtain the same information. By performing a frequency sweep, our assay efficiently yields a multitude of adhesion characteristics for the fixed cell population and removes the need for time-consuming shear stress titration studies to optimize conditions for each combination of cell type and substrate. The concept of increasing throughput and extracting more data from microscale adhesion systems has been gaining popularity18,20,43,44, and our current work is an extension of this trend. However, in contrast to the majority of such assays which provide useful insights into detachment events, we have instead applied the approach to the study attachment events.
Fourth, multi-population modeling provides the ability to identify and quantify changes in subpopulations that may otherwise be hidden if treated as a single population. This has important implications for many emerging biological questions related to population heterogeneity and the role of specific cellular subpopulations to normal and pathologic development, as well as to regenerative medicine.
Taken together, the first three elements enable a large amount of data to be acquired from as few as 102 cells per microchannel, providing new opportunities to study limited cell sources and significantly advancing previous uses of image processing and oscillatory flow for the study of adhesion.45 The range and resolution of the data obtained from each sample is what enables the fourth component of multi-population modeling, which has broad potential to provide new biological insights for applications with and without cell-source limitations.
To demonstrate these enabling capabilities as well as the broad applicability of the assay, studies were performed in two major areas of biology research, cancer and cardiology. The sensitivity and repeatability of the method was validated by confirming an increase in adhesion of three different tumor cell lines representing three different cancers to a HU-VEC monolayer upon activation of the monolayer using IL-1β. New insight into population heterogeneity of cancer cell lines was enabled by the use of oscillatory flow including the detection and quantification of subpopulations. The system was then used for a novel application in cardioregenerative medicine, where our findings demonstrated the utility of the device for small-animal primary cell adhesion studies where cell quantities are limited. More specifically, given that samples from different individual primary cell donors can vary significantly in terms of biology and preparation, the ability to test multiple experimental conditions on each sample greatly increases the statistical power of a study.
To our knowledge, this is the first demonstration of an adhesion assay that enables extensive insight into population distributions of adhesion strength with such small sample sizes. Further, the modularity of the assay allows integration of engineered substrates and microenvironments to enable a wide variety of applications. Thus, this flexible, high-content, information-rich functional assay has potential to provide new and significant biological insight.
In order to gain some perspective on the flow conditions used in this study, we can compare them to various conditions measured in vivo. As mentioned, the assay used purely oscillatory flow to enable highly efficient use of cell samples and acquisition of high-content adhesion data, while the shear stress range of the current embodiment (~0.001–0.1 Pa) is based on previous studies of tumor cell adhesion to facilitate data comparison and validation of the assay. We note that the assay design can be modified to explore other ranges of flow conditions depending on the amplitudes and frequencies appropriate for a particular area of study (ESI). In the normal vasculature, flow is generally pulsatile with both oscillatory and steady components resulting in a non-zero net flow rate. Measurements of shear stress in human capillaries and post-capillary venules range from ~0.3–9.5 Pa, with an average of ~1.5 Pa, while average wall shear stress in the human common carotid artery is ~1.2 Pa, with peaks of ~3 Pa.46,47 However, arterial shear stress below ~0.4 Pa can occur and has been linked to atherogenesis.46 Flow in the lymphatics, on the other hand, is generally more irregular and staltatory, and exhibits significantly lower shear stress, averaging between ~0.04–0.06 Pa with peaks of ~0.3–1 Pa.48 Thus, generation of flow conditions more appropriate for different biological systems can require modification of the assay device design or input signal.
The current assay has several limitations that can be addressed in future designs to expand the capabilities of the overall methodology. Software to drive the device from the computer audio output would eliminate the need for a separate function generator and provide additional control over input signals. Coordinating the actuation and data acquisition would reduce each image stream down to two images for quantifying cell motion, lowering storage requirements and simplifying data analysis. Currently, the microchannel device is designed for relatively low shear stress applications; thus, applications such as detachment assays will generally require modification to achieve higher shear stress levels. The use of purely oscillatory flow could be limiting for the study of endothelial cell response to shear where a non-zero net flow rate is known to be important for physiologic behavior.49 Additional engineering of the input signal would also be needed to perform sequential descending and ascending sweeps to explore topics such as catch-bond-like adhesion or the hysteresis of different adhesion mechanisms.50 If desired, the multi-population modeling can be quite easily extended to three or more populations, as long as one takes care to avoid over-fitting data with too many parameters, which can lead to false identification of subpopulations (ESI, Data Fitting). In addition, as with other functional adhesion assays, parallel biochemical and genetic strategies are generally needed to help elucidate the molecular mechanisms responsible for observed changes in adhesion. Despite these many opportunities for advancement, we have demonstrated the advantages of the assay for examining characteristics of subpopulations and enabling high-content adhesion studies in areas where cell source limitations can be prohibitive.
4 Materials and Methods
Details regarding device fabrication; system hardware, characterization, and calibration; cell isolation and culture; image acquisition and analysis; and population modeling are provided in the ESI.
Supplementary Material
Acknowledgments
The research was funded by the following grants: NIH - National Cancer Institue (R33 CA137673); the Innovation & Economic Development Research Program at UW-Madison; the National Library of Medicine (5T15LM007359); the National Heart, Lung, and Blood Institute(1R21HL092477); and the National Institutes of Health, under Ruth L. Kirschstein National Research Service Award (T32 HL 07936) from the National Heart Lung and Blood Institute to the University of Wisconsin-Madison Cardiovascular Research Center. We thank Dr. Erwin Berthier for his contributions to JeXperiment, Rachel Mosher for her assistance during image acquisition.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
References
- 1.Davies P. Physiological Reviews. 1995;75:519–560. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmid-Schoenbein GW. Physiological Reviews. 1990;70:987–1028. doi: 10.1152/physrev.1990.70.4.987. [DOI] [PubMed] [Google Scholar]
- 3.Engler A, Sen S, Sweeney H, Discher D. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Geiger TR, Peeper DS. Biochimica Et Biophysica Acta-Reviews On Cancer. 2009;1796:293–308. doi: 10.1016/j.bbcan.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Nugent H, Edelman E. Circulation Research. 2003;92:1068–1078. doi: 10.1161/01.RES.0000073844.41372.38. [DOI] [PubMed] [Google Scholar]
- 6.Lutolf M, Hubbell J. Nature Biotechnology. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 7.Christ KV, Turner KT. Journal of Adhesion Science and Technology. 2010;24:2027–2058. [Google Scholar]
- 8.Giavazzi R, Foppolo M, Dossi R, Remuzzi A. Journal of Clinical Investigation. 1993;92:3038–3044. doi: 10.1172/JCI116928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jadhav S, Bochner B, Konstantopoulos K. Journal of Immunology. 2001;167:5986–5993. doi: 10.4049/jimmunol.167.10.5986. [DOI] [PubMed] [Google Scholar]
- 10.Heilshorn S, DiZio K, Welsh E, Tirrell D. Biomaterials. 2003;24:4245–4252. doi: 10.1016/s0142-9612(03)00294-1. [DOI] [PubMed] [Google Scholar]
- 11.Usami S, Chen H, Zhao Y, Chien S, Skalak R. Annals of Biomedical Engineering. 1993;21:77–83. doi: 10.1007/BF02368167. [DOI] [PubMed] [Google Scholar]
- 12.Xiao Y, Truskey G. Biophysical Journal. 1996;71:2869–2884. doi: 10.1016/S0006-3495(96)79484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein A, DiMilla P. Journal of Biomedical Materials Research. 2002;59:665–675. doi: 10.1002/jbm.1276. [DOI] [PubMed] [Google Scholar]
- 14.Wang CC, Hsu YC, Su FC, Lu SC, Lee TM. J Biomed Mater Res A. 2009;88:370–83. doi: 10.1002/jbm.a.31604. [DOI] [PubMed] [Google Scholar]
- 15.Sagvolden G, Giaever I, Pettersen EO, Feder J. Proc Natl Acad Sci U S A. 1999;96:471–6. doi: 10.1073/pnas.96.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung KL, Yang L, Whittemore DE, Shi Y, Jin G, Hsieh AH, Akeson WH, Sung LA. Proc Natl Acad Sci U S A. 1996;93:9182–7. doi: 10.1073/pnas.93.17.9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu H, Koo L, Wang W, Lauffenburger D, Griffith L, Jensen K. Analytical Chemistry. 2004;76:5257–5264. doi: 10.1021/ac049837t. [DOI] [PubMed] [Google Scholar]
- 18.Young E, Wheeler A, Simmons C. Lab on a Chip. 2007;7:1759–1766. doi: 10.1039/b712486d. [DOI] [PubMed] [Google Scholar]
- 19.Gutierrez E, Groisman A. Analytical Chemistry. 2007;79:2249–2258. doi: 10.1021/ac061703n. [DOI] [PubMed] [Google Scholar]
- 20.Christophis C, Grunze M, Rosenhahn A. Physical Chemistry Chemical Physics. 2010;12:4498–4504. doi: 10.1039/b924304f. [DOI] [PubMed] [Google Scholar]
- 21.Walker G, Beebe D. Lab on a Chip. 2002;2:131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 22.Meyvantsson I, Warrick J, Hayes S, Skoien A, Beebe D. Lab on a Chip. 2008;8:717–724. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- 23.Puccinelli J, Su X, Beebe D. Journal of the Association for Laboratory Automation. 2010;15:25–32. [Google Scholar]
- 24.Young EWK, Pak C, Kahl BS, Yang DT, Callander NS, Miyamoto S, Beebe DJ. Blood. 2012;119:E76–E85. doi: 10.1182/blood-2011-10-384347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng Y, Lee T, Yu P, Roy P, Low H. Journal of Biomechanical Engineering-Transactions of the Asme. 2006;128:185–193. doi: 10.1115/1.2170118. [DOI] [PubMed] [Google Scholar]
- 26.Daja M, Niu X, Zhao Z, Brown J, Russell P. Prostate Cancer and Prostatic Diseases. 2003;6:15–26. doi: 10.1038/sj.pcan.4500609. [DOI] [PubMed] [Google Scholar]
- 27.Dimitroff C, Lechpammer M, Long-Woodward D, Kutok J. Cancer Research. 2004;64:5261–5269. doi: 10.1158/0008-5472.CAN-04-0691. [DOI] [PubMed] [Google Scholar]
- 28.Tozeren A, Kleinman H, Grant D, Mercurio MDA, Byers S. International Journal of Cancer. 1995;60:426–431. doi: 10.1002/ijc.2910600326. [DOI] [PubMed] [Google Scholar]
- 29.Lee T, Avraham H, Jiang S, Avraham S. Journal of Biological Chemistry. 2003;278:5277–5284. doi: 10.1074/jbc.M210063200. [DOI] [PubMed] [Google Scholar]
- 30.Okada T, Hawley R. International Journal of Cancer. 1995;63:823–830. doi: 10.1002/ijc.2910630613. [DOI] [PubMed] [Google Scholar]
- 31.Broek I, Vanderkerken K, Camp BV, Riet IV. Clinical & Experimental Metastasis. 2008;25:325–334. doi: 10.1007/s10585-007-9108-4. [DOI] [PubMed] [Google Scholar]
- 32.Katz BZ. Seminars In Cancer Biology. 2010;20:186–195. doi: 10.1016/j.semcancer.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Freyman T, Polin G, Osman H, Crary J, Lu M, Cheng L, Palasis M, Wilensky RL. Eur Heart J. 2006;27:1114–22. doi: 10.1093/eurheartj/ehi818. [DOI] [PubMed] [Google Scholar]
- 34.Hofmann M, Wollert KC, Meyer GP, Menke A, Arseniev L, Hertenstein B, Ganser A, Knapp WH, Drexler H. Circulation. 2005;111:2198–202. doi: 10.1161/01.CIR.0000163546.27639.AA. [DOI] [PubMed] [Google Scholar]
- 35.Forest VF, Tirouvanziam AM, Perigaud C, Fernandes S, Fusellier MS, Desfontis JC, Toquet CS, Heymann MFM, Crochet DP, Lemarchand PF. Stem Cell Res Ther. 2010;1:4. doi: 10.1186/scrt4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Zhang T, Lan X, Du G. Clin Cardiol. 2010;33:296–302. doi: 10.1002/clc.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang W, Ma A, Wang T, Han K, Liu Y, Zhang Y, Zhao X, Dong A, Du Y, Huang X, Wang J, Lei X, Zheng X. Transpl Int. 2006;19:570–80. doi: 10.1111/j.1432-2277.2006.00307.x. [DOI] [PubMed] [Google Scholar]
- 38.Domenech M, Yu H, Warrick J, Badders N, Meyvantsson I, Alexander C, Beebe D. Integrative Biology. 2009;1:267–274. doi: 10.1039/b823059e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berthier E, Surfus J, Verbsky J, Huttenlocher A, Beebe D. Integrative Biology. 2010;2:630–638. doi: 10.1039/c0ib00030b. [DOI] [PubMed] [Google Scholar]
- 40.Sung KE, Yang N, Pehlke C, Keely PJ, Eliceiri KW, Friedl A, Beebe DJ. Integrative Biology. 2011;3:439–450. doi: 10.1039/c0ib00063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chappell D, Varner S, Nerem R, Medford R, Alexander R. Circulation Research. 1998;82:532–539. doi: 10.1161/01.res.82.5.532. [DOI] [PubMed] [Google Scholar]
- 42.Hwang J, Saha A, Boo Y, Sorescu G, McNally J, Holland S, Dikalov S, Giddens D, Griendling K, Harrison D, Jo H. Journal of Biological Chemistry. 2003;278:47291–47298. doi: 10.1074/jbc.M305150200. [DOI] [PubMed] [Google Scholar]
- 43.Young E, Simmons C. Lab on a Chip. 2010;10:143–160. doi: 10.1039/b913390a. [DOI] [PubMed] [Google Scholar]
- 44.Christophis C, Taubert I, Meseck GR, Schubert M, Grunze M, Ho AD, Rosenhahn A. Biophysical Journal. 2011;101:585–593. doi: 10.1016/j.bpj.2011.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Solis-Wever X, Aguas C, Liu Y, Li P, Pappas D. Analytical Chemistry. 2009;81:3334–3343. doi: 10.1021/ac900277y. [DOI] [PubMed] [Google Scholar]
- 46.Reneman RS, Arts T, Hoeks APG. J Vasc Res. 2006;43:251–69. doi: 10.1159/000091648. [DOI] [PubMed] [Google Scholar]
- 47.Koutsiaris AG, Tachmitzi SV, Batis N, Kotoula MG, Karabatsas CH, Tsironi E, Chatzoulis DZ. Biorheology. 2007;44:375–86. [PubMed] [Google Scholar]
- 48.Zawieja DC. Lymphat Res Biol. 2009;7:87–96. doi: 10.1089/lrb.2009.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helmlinger G, Geiger RV, Schreck S, Nerem RM. J Biomech Eng. 1991;113:123–31. doi: 10.1115/1.2891226. [DOI] [PubMed] [Google Scholar]
- 50.Thomas W. Annu Rev Biomed Eng. 2008;10:39–57. doi: 10.1146/annurev.bioeng.10.061807.160427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.