Abstract
Background
HIF-1 activates various genes in cancer progression and metastasis. HIF-1α 1772 C/T and 1790 G/A polymorphisms are reportedly associated with cancer risk; however, the results are inconclusive.
Methodology/Principal Findings
A meta-analysis of 34 studies that involved 7522 cases and 9847 controls for 1772 C/T and 24 studies that involved 4884 cases and 8154 controls for 1790 G/A was conducted to identify the association of C/T and G/A polymorphisms with cancer risk. Odds ratio (OR) and 95% confidence intervals (95% CI) were used to assess the strength of association.
HIF-1α 1772 C/T and 1790 G/A polymorphisms were associated with higher cancer risk in homozygote comparison (1772C/T: TT vs. CC: OR = 2.45, 95% CI: 1.52, 3.96; P heterogeneity = 0.028; 1790G/A: AA vs. GG: OR=4.74, 95% CI: 1.78, 12.6; P heterogeneity < 0.01), dominant model (1772C/T: TT/CT vs. CC: OR = 1.27, 95% CI: 1.04, 1.55; P heterogeneity < 0.01, 1790G/A: AA/GA vs. GG: OR = 1.65, 95% CI: 1.05, 2.60; P heterogeneity < 0.01), T allele versus C allele (T vs. C: OR = 1.42, 95% CI: 1.18, 1.70; P heterogeneity < 0.01), and A allele versus G allele (A vs. G: OR = 1.83, 95% CI: 1.13, 2.96; P heterogeneity < 0.01). On a subgroup analysis, the 1772 C/T polymorphism was significantly linked to higher risks for breast cancer, lung cancer, prostate cancer, and cervical cancer, whereas the 1790 G/A polymorphism was significantly linked to higher risks for lung cancer and prostate cancer. A significantly increased cancer risk was found in both Asians and Caucasians for 1772C/T polymorphism, whereas a significantly increased cancer risk was found in Caucasians in the heterozygote comparison and recessive model for 1790G/A polymorphism.
Conclusions
HIF-1α 1772 C/T and 1790 G/A polymorphisms are significantly associated with higher cancer risk.
INTRODUCTION
Cancer, which results from complex interactions between genetic and environmental factors, has become a challenging health problem. An increasing number of studies have been performed in the past few years to assess the relationship between genetic variation and cancer risk [1].
Oxygen (O2) concentration in tumor tissues is significantly lower than that in the surrounding normal tissues. Many studies have focused on hypoxia because of its function in maintaining tumor microenvironments [2]. Hypoxic tumor microenvironment initiates multiple cellular responses, such as proliferation and angiogenesis, triggering the development and progression of cancer. In general, hypoxia may regulate tumor cell phenotypes by altering genes that are sensitive to O2 pressure [3]. Studies have demonstrated that HIF-1 has an important function in the development and progression of cancer by activating various genes associated with angiogenesis, cell adhesion, erythropoiesis, and glucose transportation [4]. HIF-1 is a heterodimer consisting of an oxygen-sensitive subunit HIF-1α and a constitutively expressed subunit HIF-1β; it is degraded rapidly through the von Hippel–Lindau-mediated ubiquitin–proteasome pathway under normoxia conditions [5]. Recent studies have shown that HIF-1α is overexpressed in many human cancers with advanced tumor grade, suggesting that HIF-1α acts as an independent factor of cancer prognosis [6].
The human HIF-1α gene, which is located at chromosome 14q21–24, is composed of 15 exons. It codes for a 3919 bp cDNA and produces an 826 amino acid protein. Single nucleotide polymorphisms (SNPs) in coding regions can mediate amino acid changes and affect the structure and biological activity of the translated protein [7]. The most widely studied HIF-1α polymorphisms are 1772 C/T (Pro582Ser, rs11549465) and 1790 G/A (Ala588Thr, G1790A, rs11549467), which induce proline-to-serine and alanine-to-threonine amino acid substitutions, respectively. Both polymorphic variants can significantly elevate transcriptional activity than the wild type under both hypoxic and normoxic conditions in in vitro studies [8]. Moreover, both polymorphisms are associated with increased tumor microvessel density, thereby contributing to the development and progression of cancer [9].
HIF-1α 1772 C/T and 1790 G/A genetic polymorphisms were previously suggested to be responsible for the risk for various types of cancer. However, the results of epidemiological studies are inconsistent [10–12]. Thus, the relationship between HIF-1α polymorphisms and cancers requires further investigation. Accordingly, we performed a meta-analysis on eligible case–control studies to produce a more powerful estimation of the association of HIF-1α 1772 C/T and 1790 G/A polymorphisms with cancer risk.
Methods
Identification and eligibility of relevant studies
All studies published before June 26, 2013 that investigated the association of HIF-1α 1772 C/T and 1790 G/A polymorphisms with cancer risk were considered in this meta-analysis. A systematic search of literature was carried out using PubMed and Embase. The keywords used for the search were “hypoxia-inducible factor-1” or “HIF-1” concatenated with “SNP,” “polymorphism,” “mutation,” or “variant” and “tumor,” “cancer,” “carcinoma,” or “malignancy.” Only studies with complete data on the comparison of HIF-1α 1772 C/T or 1790 G/A polymorphisms between cancer patients and controls were selected. Case reports, animal studies, review articles, editorials, abstracts, reports with incomplete data, and studies based on pedigree data were excluded.
Data extraction
Two investigators (Yang and Zhu) independently reviewed the articles to exclude irrelevant and overlapping studies. The results were compared, and disagreements were resolved by discussion and consensus. We only included the publication that reported the most extensive information when overlapping articles were found. The following data were extracted for each study: first author, year of publication, country, ethnicity, control source, cancer type, Hardy–Weinberg equilibrium, and the number of cases and controls for each genotype.
Statistical analysis
STATA (version 11.0; StataCorp, College Station, Texas, USA) was used for the meta-analysis. All genotype models for the two HIF-1α polymorphisms were evaluated. We also conducted subgroup analyses by cancer type, ethnicity, and source of control. For cancer type subgroups, we included the subgroup that contained more than three studies.
The existence of heterogeneity between studies was ascertained by Q-statistic. The pooled odds ratio (OR) was estimated with models based on fixed-effects or random-effects assumptions. A random-effects model was used when the significant Q statistic (P < 0.1) indicated the presence of heterogeneity in the studies. Otherwise, a fixed-effects model was selected. The 95% confidence interval (CI) of OR was also calculated. The distribution of genotypes in the controls was checked for Hardy–Weinberg equilibrium. Studies with controls not in the Hardy–Weinberg equilibrium were subjected to sensitivity analysis.
The publication bias among the studies was assayed. Funnel plots of the HIF-1α 1772 C/T polymorphism for T allele versus C allele and the HIF-1α 1790 G/A polymorphism for A allele versus G allele were built to search for any evidence of publication bias. An assymetric funnel plot is indicative of publication bias, whereas a symmetric funnel plot implies the absence of publication bias. Egger’s test, estimated by MIX 1.7 software (Kitasato Clinical Research Center, Kitasato University, Japan), was performed to measure funnel plot asymmetry.
Results
Characteristics of eligible studies
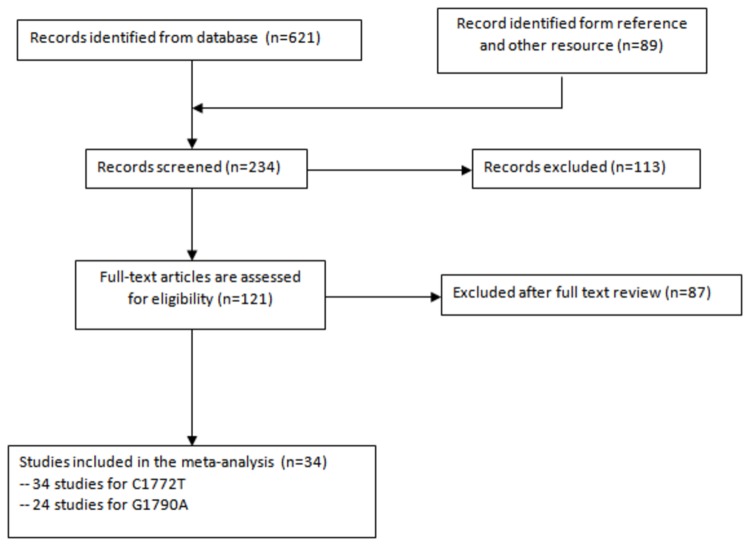
The flow diagram illustrates the main reasons for study exclusion (Figure 1). The selected study characteristics are summarized in Tables 1 and 2. Thirty-four relevant case–control studies concerning the 1772 C/T and 1790 G/A polymorphisms and cancer risk were included in the meta-analysis. Of the 34 studies, 5 concentrated on prostate cancer [13–17], 5 on breast cancer [18–22], 4 on colorectal cancer [23–26], 3 on oral cancer [27–29], 3 on lung cancer [30–32], 2 on pancreatic cancer [33,34], 2 on renal cell carcinoma [35,36], 2 on cervical cancer [37,38], 1 on ovarian cancer, endometrial cancer, and cervical cancer [39], and 7 concentrated separately on esophageal squamous cell carcinoma [40], endometrial cancer [9], liver cancer [41], gastric cancer [42], glioma [43], bladder cancer [44], and head and neck squamous cell carcinoma [8]. Among the eligible studies, 34 presented data on 1772 C/T polymorphism and 24 presented data on 1790 G/A polymorphism. For the 1772 C/T polymorphism, the distribution of the genotypes in the control groups in 5 studies was not in Hardy–Weinberg equilibrium [9,13,28,35,36]. For the 1790 G/A polymorphism, the distribution of the genotypes in the control groups in one study was not in Hardy–Weinberg equilibrium [36]. Among the eligible studies, 1 study provided data on three types of cancer (endometrial cancer, ovarian cancer, and cervical cancer) for both polymorphisms [39].
Figure 1. Reference search and selection of studies in the meta-analysis.
Table 1. Characteristics of eligible studies for the association between the 1772 C/T polymorphism and cancer risk.
| First Author (Reference) | Year | Country | Ethnicity | Control Source | Cancer Type | Cases |
Controls |
HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | CC | CT | TT | |||||||
| Tanimoto | 2003 | Japan | Asian | PB | Head and neck squamous cell carcinoma | 45 | 10 | 0 | 98 | 12 | 0 | 0.545 |
| Kuwai | 2004 | Japan | Asian | PB | Colorectal cancer | 100 | 0 | 0 | 89 | 11 | 0 | 0.561 |
| Ling | 2005 | China | Asian | PB | Esophogeal sqaumous cell carcinoma | 84 | 11 | 0 | 93 | 11 | 0 | 0.569 |
| Kim | 2008 | Korea | Asian | HB | Breast cancer | 81 | 8 | 1 | 93 | 9 | 0 | 0.641 |
| Lee | 2008 | Korea | Asian | PB | Breast cancer | 1207 | 119 | 6 | 1245 | 123 | 1 | 0.25 |
| Nadaoka | 2008 | Japan | Asian | PB | Transitional cell carcinoma of bladder | 197 | 22 | 419 | 42 | 0.35 | ||
| Chen | 2009 | China | Asian | PB | Oral cancer | 163 | 10 | 1 | 334 | 13 | 0 | 0.722 |
| Li | 2009 | China | Asian | PB | Gastric cancer | 83 | 4 | 0 | 93 | 13 | 0 | 0.501 |
| Naidu | 2009 | Malaysia | Asian | PB | Breast cancer | 294 | 100 | 16 | 222 | 50 | 3 | 0.922 |
| Chai | 2010 | China | Asian | HB | Cervical cancer | 65 | 25 | 7 | 94 | 21 | 2 | 0.52 |
| Hsiao | 2010 | China | Asian | HB | Hepatocellular carcinoma | 94 | 8 | 0 | 334 | 13 | 0 | 0.722 |
| Kang | 2011 | Korea | Asian | PB | Colorectal cancer | 38 | 12 | 46 | 4 | |||
| Kim | 2011 | Korea | Asian | HB | Cervical cancer | 177 | 22 | 0 | 187 | 27 | 0 | 0.325 |
| Putra | 2011 | Japan | Asian | HB | Lung cancer | 74 | 9 | 0 | 98 | 12 | 0 | 0.545 |
| Wang | 2011 | China | Asian | HB | Pancreatic cancer | 209, (198) | 54 | 0 | 242 | 29 | 0 | 0.352 |
| Xu | 2011 | China | Asian | HB | Glioma | 121 | 27 | 2 | 135 | 14 | 1 | 0.354 |
| Li | 2012 | China | Asian | HB | Prostate cancer | 612 | 48 | 2 | 659 | 57 | 0 | 0.267 |
| Clifford | 2001 | UK | Caucasian | PB | Renal cell carcinoma | 30 | 5 | 0 | 110 | 27 | 6 | 0.018 |
| Ollerenshaw | 2004 | UK | Caucasian | PB | Renal cell carcinoma | 16 | 54 | 90 | 1 | 90 | 71 | <0.001, |
| Fransen | 2006 | Sweden | Caucasian | PB | Colorectal cancer | 167 | 28 | 3 | 213 | 43 | 2 | 0.916 |
| Konac | 2007 | Turkey | Caucasian | HB | Endometrial, ovarian, and cervical cancer | 48 | 40 | 14 | 68 | 37 | 2 | 0.229 |
| Orr-Urtreger | 2007 | Israel | Caucasian | PB | Prostate cancer | 287 | 99 | 16 | 217 | 80 | 3 | 0.137 |
| Horre´e | 2008 | Netherlands | Caucasian | PB | Endometrial cancer | 50 | 5 | 3 | 463 | 84 | 12 | 0.001 |
| Apaydin | 2008 | Turkey | Caucasian | PB | Breast cancer | 79 | 21 | 2 | 68 | 29 | 5 | 0.415 |
| Foley | 2009 | Ireland | Caucasian | PB | Prostate cancer | 65 | 30 | 0 | 175 | 13 | 0 | 0.623 |
| Muñoz-Guerra | 2009 | Spain | Caucasian | PB | Oral cancer | 57, | 6 | 7 | 113 | 27 | 8 | 0.001 |
| Konac | 2009 | Turkey | Caucasian | HB | Lung cancer | 110, | 31 | 0 | 111 | 43 | 2 | 0.335 |
| Knechtel | 2010 | Austrila | Caucasian | HB | Colorectal cancer | 291 | 77 | 1773 | 383 | >0.05 | ||
| Ruiz-Tovar | 2012 | Spian | Caucasian | PB | Pancreatic cancer | 47 | 1 | 11 | 116 | 28 | 8 | 0.002 |
| Kuo | 2012 | China | Caucasian | HB | Lung cancer | 153 | 94 | 38 | 216 | 73 | 11 | 0.132 |
| Alves | 2012 | Brazil | Caucasian | PB | Oral cancer | 0 | 1 | 39 | 0 | 85 | 3 | <0.001 |
| Zagouri | 2012 | Greece | Caucasian | HB | Breast cancer | 98 | 15 | 0 | 107 | 17 | 0 | 0.413 |
| Chau | 2005 | USA | Mixed | PB | Prostate cancer | 161 | 29 | 6 | 179 | 14 | 3 | <0.001 |
| Li | 2007 | USA | Mixed | PB | Prostate cancer | 818 | 209 | 14 | 175 | 13 | 0 | 0.623 |
Table 2. Characteristics of eligible studies for the association between the 1790 A/G polymorphism and cancer risk.
| First Author (Reference) | Year | Country | Ethnicity | Control Source | Cancer Type | Cases |
Controls |
HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | GG | GA | AA | |||||||
| Tanimoto | 2003 | Japan | Asian | PB | Head and neck squamous cell carcinoma | 51 | 4 | 0 | 101 | 9 | 0 | 0.655 |
| Kim | 2008 | Korea | Asian | HB | Breast cancer | 87 | 3 | 0 | 94 | 7 | 1 | 0.06 |
| Nadaoka | 2008 | Japan | Asian | PB | Transitional cell carcinoma of bladder | 204 | 15 | 421 | 40 | 0.25 | ||
| Chen | 2009 | China | Asian | PB | Oral cancer | 333 | 14 | 0 | 153 | 20 | 1 | 0.697 |
| Li | 2009 | China | Asian | PB | Gastric cancer | 74 | 13 | 0 | 100 | 6 | 0 | 0.764 |
| Naidu | 2009 | Malaysia | Asian | PB | Breast cancer | 332 | 72 | 6 | 232 | 41 | 2 | 0.898 |
| Hsiao | 2010 | China | Asian | HB | Hepatocellular carcinoma | 27 | 8 | 0 | 200 | 7 | 0 | 0.805 |
| Kim | 2011 | Korea | Asian | HB | Cervical cancer | 187 | 12 | 0 | 200 | 13 | 1 | 0.136 |
| Putra | 2011 | Japan | Asian | HB | Lung cancer | 72 | 9 | 2 | 101 | 9 | 0 | 0.655 |
| Wang | 2011 | China | Asian | HB | Pancreatic cancer | 198 | 64 | 1 | 249 | 22 | 0 | 0.486 |
| Li | 2012 | China | Asian | HB | Prostate cancer | 614 | 47 | 1 | 685 | 31 | 0 | 0.554 |
| Clifford | 2001 | UK | Caucasian | PB | Renal cell carcinoma | 35 | 0 | 0 | 140 | 4 | 0 | 0.866 |
| Ollerenshaw | 2004 | UK | Caucasian | PB | Renal cell carcinoma | 65 | 67 | 14 | 239 | 39 | 10 | <0.001 |
| Fransen | 2006 | Sweden | Caucasian | PB | Colorectal cancer | 189 | 89 | 0 | 247 | 9 | 0 | 0.775 |
| Konac | 2007 | Turkey | Caucasian | HB | Endometrial, ovarian, and cervical cancer | 100 | 2 | 0 | 107 | 0 | 0 | 1 |
| Orr-Urtreger | 2007 | Israel | Caucasian | PB | Prostate cancer | 198 | 2 | 0 | 298 | 2 | 0 | 0.954 |
| Apaydin | 2008 | Turkey | Caucasian | PB | Breast cancer | 102 | 0 | 0 | 94 | 4 | 0 | 0.837 |
| Muñoz-Guerra | 2009 | Spain | Caucasian | PB | Oral cancer | 40 | 21 | 3 | 130 | 9 | 0 | 0.693 |
| Konac | 2009 | Turkey | Caucasian | HB | Lung cancer | 140 | 1 | 0 | 152 | 2 | 0 | 0.936 |
| Knechtel | 2010 | Austrila | Caucasian | HB | Colorectal cancer | 356 | 11 | 2080 | 76 | >0.05 | ||
| Ruiz-Tovar | 2012 | Spian | Caucasian | PB | Pancreatic cancer | 54 | 2 | 3 | 142 | 10 | 0 | 0.675 |
| Kuo | 2012 | China | Caucasian | HB | Lung cancer | 150 | 1 | 41 | 215 | 74 | 11 | 0.154 |
| Alves | 2012 | Brazil | Caucasian | PB | Oral cancer | 2 | 1 | 37 | 81 | 7 | 0 | 0.698 |
| Li | 2007 | USA | Mixed | PB | Prostate cancer | 1053 | 13 | 0 | 1247 | 17 | 0 | 0.81 |
Summary statistics
The meta-analysis for the HIF-1α 1772 C/T polymorphism included 7522 cases and 9847 controls. The prevalence of the CC genotype was the highest, allele C was the most frequent, and the prevalence of the TT genotype was the lowest in both case and control groups.
The meta-analysis for the HIF-1α 1790 G/A polymorphism included 4884 cancer cases and 8154 controls. The prevalence of the GG genotype was the highest, allele G was the most frequent, and the prevalence of the AA genotype was the lowest in both case and control groups.
Overall analysis
Upon pooling of all eligible studies, we observed that both 1772 C/T and 1790 G/A polymorphisms were significantly associated with cancer risk in homozygote comparison (1772C/T: TT vs. CC: OR=2.45, 95% CI: 1.52, 3.96; P heterogeneity = 0.028; 1790G/A: AA vs. GG: OR = 4.74, 95% CI: 1.78, 12.6; P heterogeneity < 0.01), dominant model (1772C/T: TT/CT vs. CC: OR=1.27, 95% CI: 1.04, 1.55; P heterogeneity < 0.01, 1790G/A: AA/GA vs. GG: OR = 1.65, 95% CI: 1.05, 2.60; P heterogeneity < 0.01) (Figures 2 and 3), recessive model (1772C/T: TT vs. CC/CT: OR = 3.18, 95% CI: 1.92, 5.29; P heterogeneity < 0.01, 1790G/A: AA vs. GG/GA: OR = 4.39, 95% CI: 1.61,11.9; P heterogeneity < 0.01), T allele versus C allele (T vs. C: OR = 1.42, 95% CI: 1.18, 1.70; P heterogeneity < 0.01), and A allele versus G allele (A vs. G: OR = 1.83, 95% CI: 1.13,2.96; P heterogeneity < 0.01) (Figures 4 and 5). The association strength between HIF-1α polymorphism and cancer risk is shown in Table 3. No significant association was found in heterozygote comparison (1772C/T: CT vs. CC: OR = 1.15, 95% CI: 0.92, 1.45; P heterogeneity < 0.01, 1790G/A: GA vs. GG: OR = 1.35, 95% CI: 0.82, 2.21; P heterogeneity < 0.01). However, the 1772C/T polymorphism was significantly associated with cancer in the heterozygote model (CT vs. CC: OR = 1.29, 95% CI: 1.04, 1.62; Pheterogeneity < 0.01) when studies not in the Hardy–Weinberg equilibrium were excluded.
Figure 2. Forest plot of dominant model for overall comparison (1772 C/T, TT/CT vs. CC).
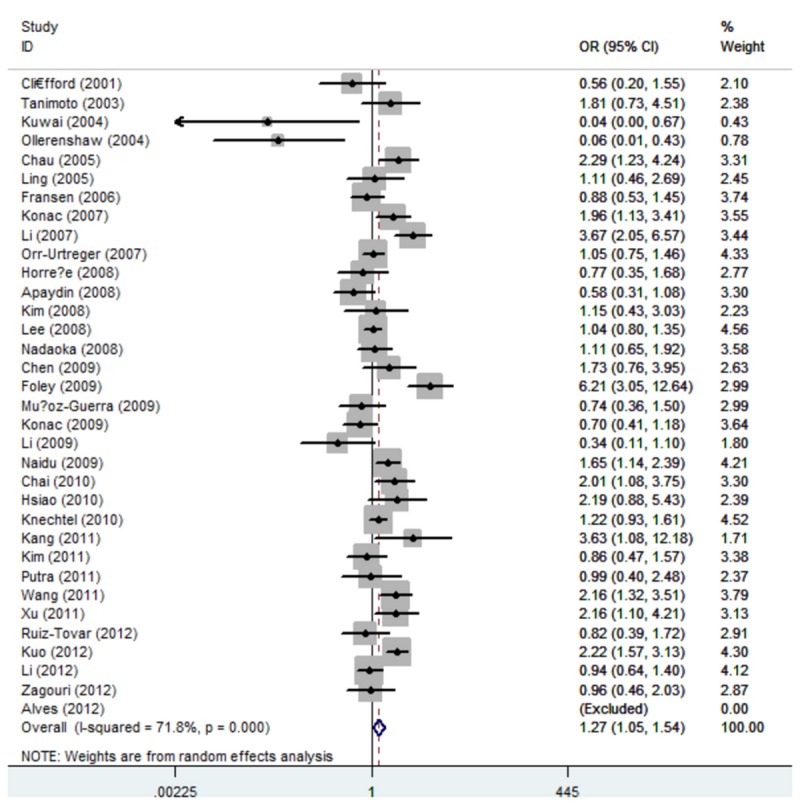
Figure 3. Forest plot of dominant model for overall comparison (1790 G/A, AA/GA vs. GG).
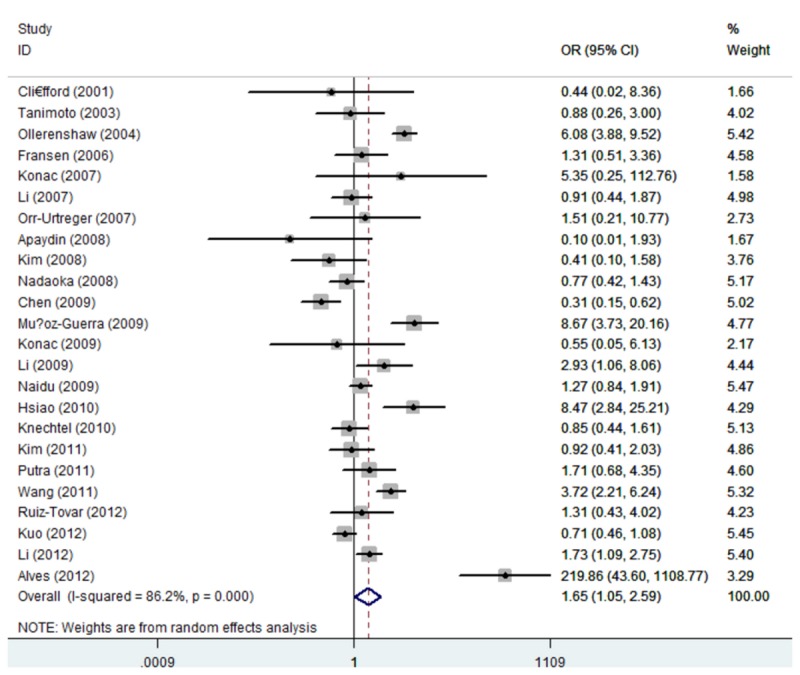
Figure 4. Forest plot of overall comparison (1772 C/T, T allele vs. C allele).
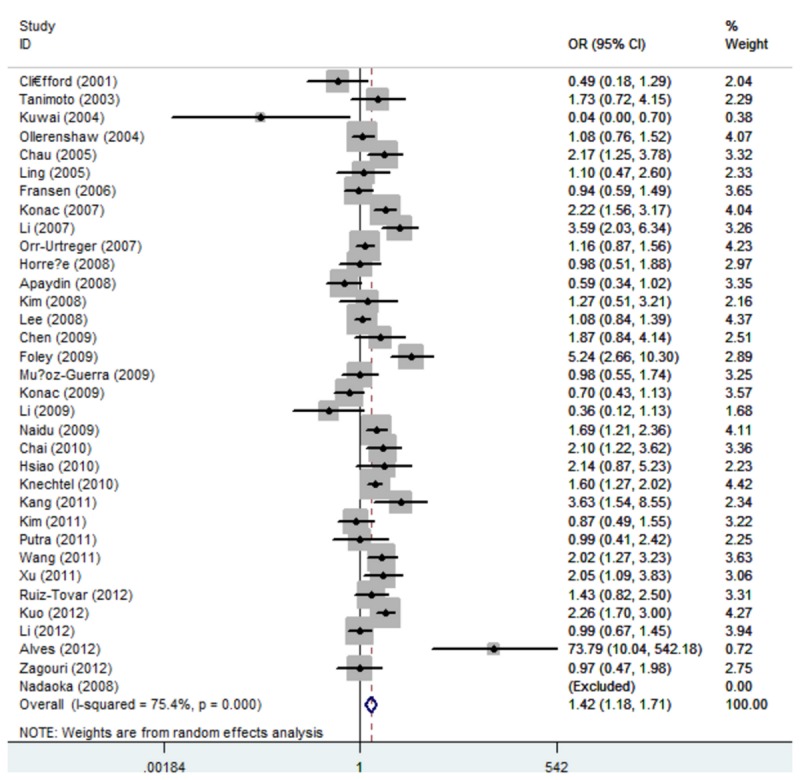
Figure 5. Forest plot of overall comparison (1790 G/A, A allele vs. G allele).
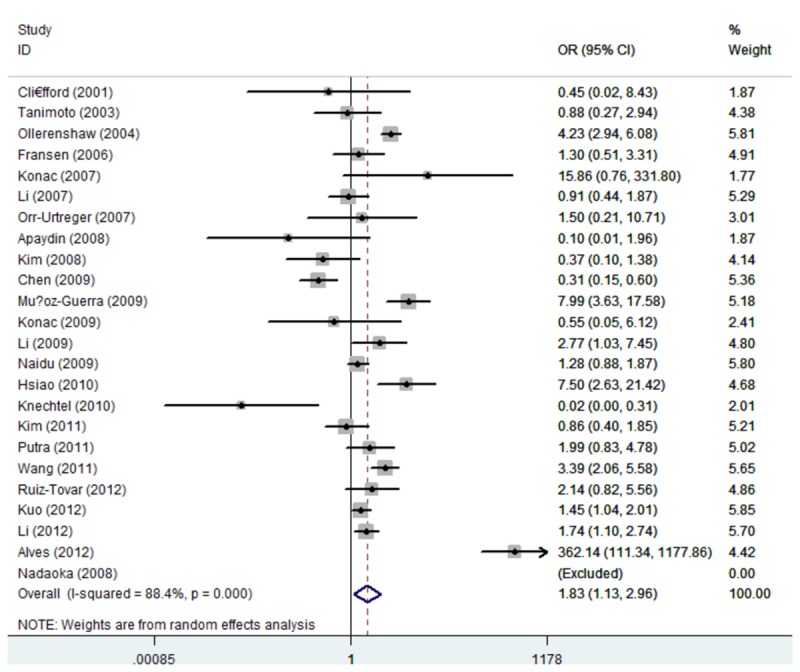
Table 3. Main results of the meta-analysis for the association of HIF1A gene 1772 C/T and 1790 G/A polymorphisms with cancer risk.
| 1772 C/T polymorphisms (rs11549465) |
TT VS CC |
CT VS CC |
TT/CT VS CC |
TT VS CT/CC
|
T allele VS C allele |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Sample size | ORa | P b | ORa | P b | ORa | P b | ORa | P b | ORa | P b | |
| Total | 34 | 7522 | 2.45 (1.52–3.96) | 0.028 | 1.15 (0.92–1.45) | <0.001 | 1.27 (1.05–1.55) | <0.001 | 3.18 (1.92–5.29) | <0.001 | 1.42 (1.18–1.70) | <0.001 |
| Total in HWE | 25 | 6575 | 3.65 (2.47–5.40) | 0.318 | 1.29 (1.04–1.62) | <0.001 | 1.35 (1.10–1.65) | <0.001 | 3.38 (2.29–5.00) | 0.476 | 1.40 (1.15–1.71) | <0.001 |
| Cancer types | ||||||||||||
| Breast cancer | 5 | 2047 | 2.30 (1.08–4.91) | 0.084 | 1.07 (0.88–1.29) | 0.188 | 1.12 (0.92–1.35) | 0.711 | 2.27 (1.06–4.87) | 0.120 | 1.09 (0.76–1.55) | 0.022 |
| Lung cancer | 3 | 509 | 1.41 (0.07–30.4) | 0.044 | 1.13 (0.59–2.19) | 0.018 | 1.50 (1.15–1.96) | 0.688 | 3.27 (1.73–6.17) | 0.065 | 1.19 (0.50–2.86) | <0.001 |
| Oral cancer | 3 | 284 | 2.01 (0.75–5.41) | 0.463 | 0.85 (0.24–2.97) | 0.047 | 1.04 (0.61–1.78) | 0.823 | 22.8 (0.28–1888) | <0.001 | 3.93 (0.61–25.4) | <0.001 |
| Colorectal cancer | 4 | 627 | 1.91 (0.32–11.6) | 0.24 (0.01–5.51) | 0.027 | 1.10 (0.87–1.38) | 0.744 | 1.97 (0.33–11.9) | 1.36 (0.68–2.70) | 0.002 | ||
| Prostate cancer | 5 | 2396 | 3.68 (1.58–8.55) | 0.871 | 2.02 (1.01–4.07) | <0.001 | 2.10 (1.08–4.09) | 0.028 | 3.52 (1.52–8.16) | 0.847 | 2.06 (1.15–3.68) | <0.001 |
| Cervical cancer | 3 | 328 | 10.1 (3.12–32.6) | 0.153 | 1.37 (0.92–2.02) | 0.099 | 1.63 (1.12–2.37) | 0.158 | 8.26 (2.64–25.9) | 0.236 | 1.89 (0.84–4.26) | 0.002 |
| Others | 13 | 1331 | 1.68 (0.42–6.80) | <0.001 | 0.97 (0.56–1.68) | <0.001 | 1.20 (0.98–1.47) | 0.512 | 1.99 (1.40–2.84) | 0.100 | 1.37 (0.96–1.97) | <0.001 |
| Ethnicities | ||||||||||||
| Caucasian | 15 | 2151 | 1.70 (0.81–3.55) | 0.001 | 0.86 (0.57–1.31) | <0.001 | 1.05 (0.76–1.46) | <0.001 | 2.97 (1.44–6.14) | <0.001 | 1.32 (0.99–1.75) | <0.001 |
| Asian | 17 | 4134 | 4.42 (2.07–9.43) | 0.997 | 1.25 (0.98–1.60) | 0.010 | 1.33 (1.06–1.68) | 0.006 | 4.12 (1.93–8.77) | 0.955 | 1.40 (1.11–1.78) | 0.002 |
| Mixed | 2 | 1237 | 3.13 (0.90–10.8) | 0.500 | 2.98 (1.92–4.63) | 0.372 | 3.05 (2.00–4.66) | 0.269 | 2.77 (0.80–9.54) | 0.646 | 2.91 (1.96–4.32) | 0.208 |
| Source of control | ||||||||||||
| PB | 21 | 4944 | 1.92 (1.05–3.50) | 0.037 | 0.99 (0.69–1.41) | <0.001 | 1.17 (0.87–1.57) | <0.001 | 3.14 (1.60–6.16) | <0.001 | 1.40 (1.06–1.84) | <0.001 |
| HB | 13 | 2578 | 4.38 (2.64–7.47) | 0.486 | 1.32 (1.13–1.57) | 0.023 | 1.39 (1.09–1.77) | 0.002 | 3.88 (2.32–6.51) | 0.569 | 1.46 (1.16–1.85) | <0.001 |
| 1790 G/A polymorphisms (rs11549465) | AA VS GG | GA VS GG | AA/GA VS GG | AA VS GA/GG | A allele VS G allele | |||||||
| N | Sample size | ORa | P b | ORa | P b | ORa | P b | ORa | P b | ORa | P b | |
| Total | 24 | 5136 | 4.74 (1.78–12.6) | 0.002 | 1.35 (0.82–2.21) | <0.001 | 1.65 (1.05–2.60) | <0.001 | 4.39 (1.61–11.9) | 0.001 | 1.83 (1.13–2.96) | <0.001 |
| Total in HWE | 23 | 5090 | 4.68 (1.34–16.3) | 0.001 | 1.23 (0.77–1.98) | <0.001 | 1.53 (0.99–2.36) | <0.001 | 4.65 (1.35–16.0) | 0.001 | 1.83 (1.13–2.96) | <0.001 |
| Cancer types | ||||||||||||
| Breast cancer | 3 | 521 | 1.44 (0.38–5.44) | 0.336 | 1.03 (0.70–1.52) | 0.115 | 1.05 (0.72–1.53) | 0.077 | 1.41 (0.37-5.37) | 0.356 | 1.07 (0.75-1.52) | 0.055 |
| Lung cancer | 3 | 362 | 5.42 (2.75–10.7) | 0.866 | 0.26 (0.01–7.10) | <0.001 | 0.82 (0.56–1.19) | 0.226 | 7.11 (3.61–14.0) | 0.975 | 1.48 (1.09-2.00) | 0.575 |
| Oral cancer | 3 | 375 | 20.7(0.10–4519) | <0.001 | 2.21 (0.18–26.9) | <0.001 | 7.81 (0.27–224) | <0.001 | 17.5 (0.10–3257) | <0.001 | 9.34 (0.23-388) | <0.001 |
| Prostate cancer | 3 | 1865 | 3.35 (0.14–82.3) | 1.41 (0.97–2.07) | 0.365 | 1.44 (0.98–2.10) | 0.340 | 3.25 (0.13–79.9) | 1.45 (1.00-2.11) | 0.330 | ||
| Others | 14 | 1542 | 4.81 (2.34–9.87) | 0.460 | 1.70 (0.99–2.90) | <0.001 | 1.80 (0.99–3.26) | <0.001 | 3.01 (1.47–6.21) | 0.367 | 1.91 (1.01-3.58) | <0.001 |
| Ethnicities | ||||||||||||
| Caucasian | 12 | 1635 | 17.4 (4.01-75.3) | 0.001 | 1.09 (0.33–3.58) | <0.001 | 2.19 (0.90–5.34) | <0.001 | 15.8(3.42–72.9) | <0.001 | 2.27 (0.92-5.58) | <0.001 |
| Asian | 11 | 2435 | 1.44 (0.60-3.46) | 0.522 | 1.45 (0.85–2.46) | <0.001 | 1.36 (0.83–2.24) | <0.001 | 1.41 (0.58–3.39) | 0.508 | 1.42 (0.84-2.40) | <0.001 |
| Source of control | ||||||||||||
| PB | 14 | 3013 | 9.69 (1.41-66.7) | <0.001 | 1.40 (0.71–2.74) | <0.001 | 1.80 (0.89–3.64) | <0.001 | 8.08 (1.12–58.1) | <0.001 | 2.10 (0.95-4.68) | <0.001 |
| HB | 10 | 2123 | 4.08 (2.26-7.37) | 0.401 | 1.23 (0.53–2.86) | <0.001 | 1.47 (0.85–2.55) | <0.001 | 5.02 (2.79–9.02) | 0.278 | 1.50 (0.86-2.62) | <0.001 |
a Random-effects model was used when the P value for the heterogeneity test was < 0.05; otherwise, fixed-effects model was used.
b P Value of Q-Test for the Heterogeneity Test
N: number of studies included; OR: odds ratio; PB: population-based; HB: hospital-based; HWE= Hardy–Weinberg equilibrium.
One study contained detailed data on ovarian cancer, endometrial cancer, and cervical cancer. We used the combined data for the overall analysis and the separate data for the subgroup analysis by cancer type.
Subgroup analyses
Subgroup analyses were performed to investigate the effect of cancer type, ethnicity, and source of control. For cancer type, the 1772C/T polymorphism demonstrated an increased risk for breast cancer, lung cancer, prostate cancer, cervical cancer, and other cancers in various models. In the subgroup analyses of “oral cancer” and “colorectal cancer,” we did not find any significant association between the 1772C/T polymorphism and cancer risk. The 1790G/A polymorphism exhibited an increased cancer risk for lung cancer in the homozygote and recessive models (AA vs. GG: OR = 5.42, 95% CI: 2.75, 10.7; P heterogeneity = 0.866; AA vs. GG/GA: OR = 7.11, 95% CI: 3.61, 14.0; P heterogeneity = 0.975; A vs. G: OR = 1.48, 95% CI: 1.09, 2.00; P heterogeneity = 0.575) and for prostate cancer (A vs. G: OR = 1.45, 95% CI: 1.00, 2.11; P heterogeneity = 0.330). We found a significant association between 1772C/T and 1790G/A polymorphisms and cancer risk in both population-based and hospital-based studies.
However, ethnicity significantly affected cancer susceptibility. For the 1772C/T polymorphism, a significantly increased cancer risk was found in both Asians and Caucasians. For the 1790G/A polymorphism, a significantly increased cancer risk was found in Caucasians in the heterozygote comparison (AA vs. GG: OR = 17.4, 95% CI: 4.01, 75.3; P heterogeneity < 0.01) and recessive model (GG/GA: OR = 15.8, 95% CI: 3.42, 72.9; P heterogeneity < 0.01). However, no significant association between these polymorphisms and cancer risk was found in Asians. These results revealed that the effect of HIF-1α polymorphisms on cancer was associated with ethnicity.
Sensitivity analysis
Sensitivity analysis was performed to explore the influence of an individual study on the pooled results by deleting a single study each time from the pooled analysis. The results showed that no individual study significantly affected the pooled OR because no substantial change was found (figure not shown).
Publication bias
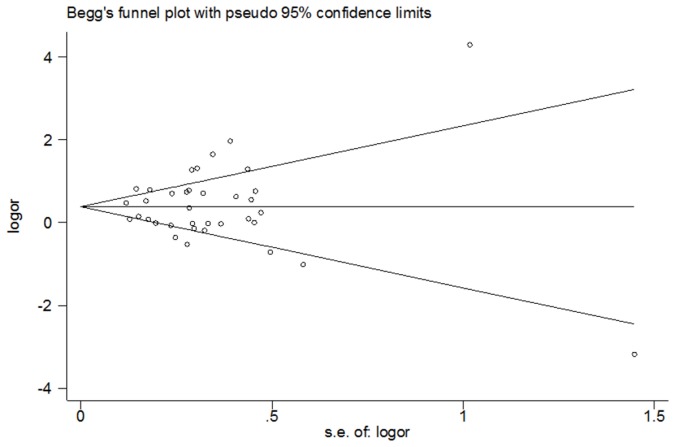
Publication bias was assessed by Begg’s funnel plot and Egger’s test. Begg’s funnel plot for the 1772 C/T polymorphism is shown in Figure 6 (P = 0.589 for T allele vs. C allele). Egger’s test was performed for statistical analysis, and no publication bias was detected (P =0.481 for T allele vs. C allele). The results of Begg’s and Egger’s tests for the 1790 G/A polymorphism were P = 0.785 and P = 0.870, respectively, for A allele versus G allele (Figure 7). Overall, no publication bias was detected in the data.
Figure 6. Funnel plot of heterozygote comparison (1772 C/T, T allele vs. C allele).
Figure 7. Funnel plot of heterozygote comparison (1790 G/A, A allele vs. G allele).
Discussion
HIF-1 has an important function in cancer progression and metastasis by activating various genes that are linked to the regulation of angiogenesis, cell survival, and energy metabolism [3]. The presence of T and A variant alleles of HIF-1α 1772, namely, C/T and 1790 G/A polymorphisms, are associated with high transcriptional abilities and protein synthesis in vitro [8]. In vivo studies related these genetic variations to many aggressive clinical features of cancer, such as the ulcerative growth pattern in colorectal tumors, suggesting that HIF-1α polymorphism is associated with cancer [45]. However, studies on the association of HIF-1α 1772 C/T and 1790 G/A polymorphisms with cancer are conflicting. In 2009, Zhao [10] conducted a meta-analysis using 16 case–control studies and concluded that 1772 C/T is significantly associated with higher cancer risk and that 1790 G/A is only significantly associated with breast cancer. Liu [12] performed a similar meta-analysis from 22 case–control studies, including 5552 cases and 8044 controls for 1772 C/T and 3381 cases and 5830 controls for 1790 G/A, and one study evaluated cancer prognosis by polymorphism [45]. This previous study concluded that the 1790 G/A polymorphism and not the 1772 C/T polymorphism is significantly associated with cancer risk. In the present study, we performed an updated meta-analysis from 34 case–control studies that involved 7522 cases and 9847 controls for 1772 C/T polymorphism and 4884 cases and 8154 controls for 1790 G/A polymorphism.
In the present meta-analysis, we investigated the association of HIF-1α 1772 C/T and 1790 G/A polymorphisms with cancer risk. Subgroup analyses by cancer type and ethnicity were also performed. Our analyses showed that both 1772C/T and 1790G/A polymorphisms were significantly associated with cancer risk. In the subgroup study, various types of cancers, such as breast cancer, lung cancer, prostate cancer, and cervical cancer, were associated with 1772C/T, whereas only lung cancer was linked with 1790G/A. However, the odds ratio values in some of the subgroup analyses were large and lacked statistical power because of the significant heterogeneity. Ethnicity may also significantly affect cancer susceptibility. For the 1790G/A polymorphism, we did not find any association between the 1790G/A polymorphism and cancer risk in Asians. This finding can be explained by the difference in genetic background, environmental exposure, and risk factors relating to lifestyle between Asian and Caucasian populations.
Some limitations of this meta-analysis should be addressed. First, the lack of the the detailed information about environment risk factors for cancer risk from included studies limited our further evaluation of potential gene–gene and gene–environment interactions. Second, the P value of the Hardy–Weinberg equilibrium of three included studies was less than 0.05, suggesting that these study populations were not representative of the broader target population. Despite these limitations, our meta-analysis had some strong advantages. This meta-analysis shed light on the association between HIF-1α polymorphisms and increased risk for various cancers. In addition, the quality of the included studies was satisfactory and met our inclusion criterion. Moreover, substantial numbers of cases and controls were pooled from different studies, which significantly increased the statistical power of the analysis. No publication bias was also found in the collected data.
In summary, this meta-analysis provided insights into the association of HIF-1α 1772 C/T and 1790 G/A gene polymorphisms with cancer risk, supporting the hypothesis that HIF-1α polymorphisms are a susceptibility marker of cancer. However, large sample studies are warranted to validate our findings, especially in some types of cancer, such as breast cancer and cervical cancer. More studies on gene–gene and gene–environment interactions should also be considered in the future to obtain a more comprehensive understanding of the association between HIF-1α polymorphisms and cancer risk.
Supporting Information
PRISMA checklist.
(DOC)
Funding Statement
This work was supported by the Natural Science Foundation of China (number 81272504), the Innovation Team (number LJ201123 (EH11)), and Jiangsu Provincial Science and Technology Projects (BK2011854 (DA11)), and “333” Project of Jiangsu Province (BRA2012210 (RS12)), and research grants from Chinese Society of Clinical Oncology (T-H2010-033 (KA10)). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Siegel R, Ward E, Hao Y, Xu J et al. (2009) Cancer statistics, 2009. CA Cancer J Clin 59: 225-249. doi: 10.3322/caac.20006. PubMed: 19474385. [DOI] [PubMed] [Google Scholar]
- 2. Yoshimura M, Itasaka S, Harada H, Hiraoka M (2013) Microenvironment and radiation therapy. Biomed Res Int, 2013: 2013: 685308. PubMed: 23509762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hill RP, Marie-Egyptienne DT, Hedley DW (2009) Cancer stem cells, hypoxia and metastasis. Semin Radiat Oncol 19: 106-111. doi: 10.1016/j.semradonc.2008.12.002. PubMed: 19249648. [DOI] [PubMed] [Google Scholar]
- 4. Tsai YP, Wu KJ (2012) Hypoxia-regulated target genes implicated in tumor metastasis. J Biomed Sci 19: 102. doi: 10.1186/1423-0127-19-102. PubMed: 23241400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kuschel A, Simon P, Tug S (2012) Functional regulation of HIF-1alpha under normoxia--is there more than post-translational regulation? J Cell Physiol 227: 514-524. doi: 10.1002/jcp.22798. PubMed: 21503885. [DOI] [PubMed] [Google Scholar]
- 6. Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA et al. (1999) Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res 59: 5830-5835. PubMed: 10582706. [PubMed] [Google Scholar]
- 7. McMullin MF (2010) HIF pathway mutations and erythrocytosis. Expert. Rev Hematol 3: 93-101. [DOI] [PubMed] [Google Scholar]
- 8. Tanimoto K, Yoshiga K, Eguchi H, Kaneyasu M, Ukon K et al. (2003) Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis 24: 1779-1783. doi: 10.1093/carcin/bgg132. PubMed: 12919954. [DOI] [PubMed] [Google Scholar]
- 9. Horrée N, Groot AJ, van Hattem WA, Heintz AP, Vooijs M et al. (2008) HIF-1A gene mutations associated with higher microvessel density in endometrial carcinomas. Histopathology 52: 637-639. doi: 10.1111/j.1365-2559.2008.02991.x. PubMed: 18370960. [DOI] [PubMed] [Google Scholar]
- 10. Zhao T, Lv J, Zhao J, Nzekebaloudou M (2009) Hypoxia-inducible factor-1alpha gene polymorphisms and cancer risk: a meta-analysis. J Exp Clin Cancer Res 28: 159. doi: 10.1186/1756-9966-28-159. PubMed: 20035632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li D, Liu J, Zhang W, Ren J, Yan L et al. (2013) Association between HIF1A P582S and A588T Polymorphisms and the Risk of Urinary Cancers: A Meta-Analysis. PLOS ONE 8: e63445. doi: 10.1371/journal.pone.0063445. PubMed: 23723982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu J, Zhang HX (2013) 1790 G/A polymorphism, but not 1772 C/T polymorphism, is significantly associated with Cancers: An update study. Gene 523: 58-63. doi: 10.1016/j.gene.2013.03.129. PubMed: 23583797. [DOI] [PubMed] [Google Scholar]
- 13. Chau CH, Permenter MG, Steinberg SM, Retter AS, Dahut WL et al. (2005) Polymorphism in the hypoxia-inducible factor 1alpha gene may confer susceptibility to androgen-independent prostate cancer. Cancer Biol Ther 4: 1222-1225. doi: 10.4161/cbt.4.11.2091. PubMed: 16205110. [DOI] [PubMed] [Google Scholar]
- 14. Li H, Bubley GJ, Balk SP, Gaziano JM, Pollak M et al. (2007) Hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms, circulating insulin-like growth factor binding protein (IGFBP)-3 levels and prostate cancer. Prostate 67: 1354-1361. doi: 10.1002/pros.20589. PubMed: 17624927. [DOI] [PubMed] [Google Scholar]
- 15. Orr-Urtreger A, Bar-Shira A, Matzkin H, Mabjeesh NJ (2007) The homozygous P582S mutation in the oxygen-dependent degradation domain of HIF-1 alpha is associated with increased risk for prostate cancer. Prostate 67: 8-13. doi: 10.1002/pros.20433. PubMed: 16998808. [DOI] [PubMed] [Google Scholar]
- 16. Foley R, Marignol L, Thomas AZ, Cullen IM, Perry AS et al. (2009) The HIF-1alpha C1772T polymorphism may be associated with susceptibility to clinically localised prostate cancer but not with elevated expression of hypoxic biomarkers. Cancer Biol Ther 8: 118-124. doi: 10.4161/cbt.8.2.7086. PubMed: 19106642. [DOI] [PubMed] [Google Scholar]
- 17. Li P, Cao Q, Shao PF, Cai HZ, Zhou H et al. (2012) Genetic polymorphisms in HIF1A are associated with prostate cancer risk in a Chinese population. Asian J Androl 14: 864-869. doi: 10.1038/aja.2012.101. PubMed: 23042446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Apaydin I, Konac E, Onen HI, Akbaba M, Tekin E et al. (2008) Single nucleotide polymorphisms in the hypoxia-inducible factor-1alpha (HIF-1alpha) gene in human sporadic breast cancer. Arch Med Res 39: 338-345. doi: 10.1016/j.arcmed.2007.11.012. PubMed: 18279708. [DOI] [PubMed] [Google Scholar]
- 19. Kim HO, Jo YH, Lee J, Lee SS, Yoon KS (2008) The C1772T genetic polymorphism in human HIF-1alpha gene associates with expression of HIF-1alpha protein in breast cancer. Oncol Rep 20: 1181-1187. PubMed: 18949419. [PubMed] [Google Scholar]
- 20. Lee JY, Choi JY, Lee KM, Park SK, Han SH et al. (2008) Rare variant of hypoxia-inducible factor-1alpha (HIF-1A) and breast cancer risk in Korean women. Clin Chim Acta 389: 167-170. doi: 10.1016/j.cca.2007.12.005. PubMed: 18160046. [DOI] [PubMed] [Google Scholar]
- 21. Naidu R, Har YC, Taib NA (2009) Associations between hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms and risk of developing breast cancer. Neoplasma 56: 441-447. doi: 10.4149/neo_2009_05_441. PubMed: 19580347. [DOI] [PubMed] [Google Scholar]
- 22. Zagouri F, Sergentanis TN, Gazouli M, Tsigginou A, Dimitrakakis C et al. (2012) HSP90, HSPA8, HIF-1 alpha and HSP70-2 polymorphisms in breast cancer: a case-control study. Mol Biol Rep 39: 10873-10879. doi: 10.1007/s11033-012-1984-2. PubMed: 23065205. [DOI] [PubMed] [Google Scholar]
- 23. Kuwai T, Kitadai Y, Tanaka S, Kuroda T, Ochiumi T et al. (2004) Single nucleotide polymorphism in the hypoxia-inducible factor-1alpha gene in colorectal carcinoma. Oncol Rep 12: 1033-1037. PubMed: 15492789. [PubMed] [Google Scholar]
- 24. Fransén K, Fenech M, Fredrikson M, Dabrosin C, Söderkvist P (2006) Association between ulcerative growth and hypoxia inducible factor-1alpha polymorphisms in colorectal cancer patients. Mol Carcinog 45: 833-840. doi: 10.1002/mc.20209. PubMed: 16865676. [DOI] [PubMed] [Google Scholar]
- 25. Knechtel G, Szkandera J, Stotz M, Hofmann G, Langsenlehner U et al. (2010) Single nucleotide polymorphisms in the hypoxia-inducible factor-1 gene and colorectal cancer risk. Mol Carcinog 49: 805-809. PubMed: 20572162. [DOI] [PubMed] [Google Scholar]
- 26. Kang MJ, Jung SA, Jung JM, Kim SE, Jung HK et al. (2011) Associations between single nucleotide polymorphisms of MMP2, VEGF, and HIF1A genes and the risk of developing colorectal cancer. Anticancer Res 31: 575-584. PubMed: 21378341. [PubMed] [Google Scholar]
- 27. Chen MK, Chiou HL, Su SC, Chung TT, Tseng HC et al. (2009) The association between hypoxia inducible factor-1alpha gene polymorphisms and increased susceptibility to oral cancer. Oral Oncol 45: e222-e226. doi: 10.1016/j.oraloncology.2009.07.015. PubMed: 19717330. [DOI] [PubMed] [Google Scholar]
- 28. Muñoz-Guerra MF, Fernández-Contreras ME, Moreno AL, Martín ID, Herráez B et al. (2009) Polymorphisms in the hypoxia inducible factor 1-alpha and the impact on the prognosis of early stages of oral cancer. Ann Surg Oncol 16: 2351-2358. doi: 10.1245/s10434-009-0503-8. PubMed: 19449077. [DOI] [PubMed] [Google Scholar]
- 29. Alves LR, Fraga CA, Oliveira MV, Sousa AA, Jorge AS et al. (2012) High HIF-1alpha expression genotypes increase odds ratio of oral cancer. Head Neck Oncol 4: 87 PubMed: 23296286. [Google Scholar]
- 30. Konac E, Dogan I, Onen HI, Yurdakul AS, Ozturk C et al. (2009) Genetic variations in the hypoxia-inducible factor-1alpha gene and lung cancer. Exp Biol Med (Maywood) 234: 1109-1116. doi: 10.3181/0902-RM-49. PubMed: 19546348. [DOI] [PubMed] [Google Scholar]
- 31. Putra AC, Tanimoto K, Arifin M, Hiyama K (2011) Hypoxia-inducible factor-1alpha polymorphisms are associated with genetic aberrations in lung cancer. Respirology 16: 796-802. doi: 10.1111/j.1440-1843.2011.01972.x. PubMed: 21435097. [DOI] [PubMed] [Google Scholar]
- 32. Kuo WH, Shih CM, Lin CW, Cheng WE, Chen SC et al. (2012) Association of hypoxia inducible factor-1alpha polymorphisms with susceptibility to non-small-cell lung cancer. Transl Res 159: 42-50. doi: 10.1016/j.trsl.2011.09.003. PubMed: 22153809. [DOI] [PubMed] [Google Scholar]
- 33. Wang X, Liu Y, Ren H, Yuan Z, Li S et al. (2011) Polymorphisms in the hypoxia-inducible factor-1alpha gene confer susceptibility to pancreatic cancer. Cancer Biol Ther 12: 383-387. doi: 10.4161/cbt.12.5.15982. PubMed: 21709439. [DOI] [PubMed] [Google Scholar]
- 34. Ruiz-Tovar J, Fernandez-Contreras ME, Martín-Perez E, Gamallo C (2012) Association of thymidylate synthase and hypoxia inducible factor-1alpha DNA polymorphisms with pancreatic cancer. Tumori 98: 364-369. PubMed: 22825513. [DOI] [PubMed] [Google Scholar]
- 35. Clifford SC, Astuti D, Hooper L, Maxwell PH, Ratcliffe PJ et al. (2001) The pVHL-associated SCF ubiquitin ligase complex: molecular genetic analysis of elongin B and C, Rbx1 and HIF-1alpha in renal cell carcinoma. Oncogene 20: 5067-5074. doi: 10.1038/sj.onc.1204602. PubMed: 11526493. [DOI] [PubMed] [Google Scholar]
- 36. Ollerenshaw M, Page T, Hammonds J, Demaine A (2004) Polymorphisms in the hypoxia inducible factor-1alpha gene (HIF1A) are associated with the renal cell carcinoma phenotype. Cancer Genet Cytogenet 153: 122-126. doi: 10.1016/j.cancergencyto.2004.01.014. PubMed: 15350301. [DOI] [PubMed] [Google Scholar]
- 37. Chai D, Chen YL, Zheng A, Liu YY, Chu YX et al. (2010) [Relationship between polymorphism of hypoxia inducible factor-1alpha and cervical cancer in Han population in Sichuan Province of China]. Sichuan Xue Xue Bao Yi Xue Ban 41: 674-677. [PubMed] [Google Scholar]
- 38. Kim YH, Park IA, Park WY, Kim JW, Kim SC et al. (2011) Hypoxia-inducible factor 1alpha polymorphisms and early-stage cervical cancer. Int J Gynecol Cancer 21: 2-7. doi: 10.1097/IGC.0b013e318204f6e6. PubMed: 21330825. [DOI] [PubMed] [Google Scholar]
- 39. Konac E, Onen HI, Metindir J, Alp E, Biri AA et al. (2007) An investigation of relationships between hypoxia-inducible factor-1 alpha gene polymorphisms and ovarian, cervical and endometrial cancers. Cancer Detect Prev 31: 102-109. doi: 10.1016/j.cdp.2007.01.001. PubMed: 17418979. [DOI] [PubMed] [Google Scholar]
- 40. Ling TS, Shi RH, Zhang GX, Zhu H, Yu LZ et al. (2005) Common single nucleotide polymorphism of hypoxia-inducible factor-1alpha and its impact on the clinicopathological features of esophageal squamous cell carcinoma. Chin J Dig Dis 6: 155-158. doi: 10.1111/j.1443-9573.2005.00223.x. PubMed: 16246222. [DOI] [PubMed] [Google Scholar]
- 41. Hsiao PC, Chen MK, Su SC, Ueng KC, Chen YC et al. (2010) Hypoxia inducible factor-1alpha gene polymorphism G1790A and its interaction with tobacco and alcohol consumptions increase susceptibility to hepatocellular carcinoma. J Surg Oncol 102: 163-169. doi: 10.1002/jso.21539. PubMed: 20648588. [DOI] [PubMed] [Google Scholar]
- 42. Li K, Zhang Y, Dan Z, Wang Y, Ren ZC (2009) Association of the hypoxia inducible factor-1alpha gene polymorphisms with gastric cancer in Tibetans. Biochem Genet 47: 625-634. doi: 10.1007/s10528-009-9254-2. PubMed: 19504235. [DOI] [PubMed] [Google Scholar]
- 43. Xu G, Wang M, Xie W, Bai X (2011) Hypoxia-inducible factor-1 alpha C1772T gene polymorphism and glioma risk: a hospital-based case-control study from China. Genet Tests Mol Biomarkers 15: 461-464. doi: 10.1089/gtmb.2010.0265. PubMed: 21329466. [DOI] [PubMed] [Google Scholar]
- 44. Nadaoka J, Horikawa Y, Saito M, Kumazawa T, Inoue T et al. (2008) Prognostic significance of HIF-1 alpha polymorphisms in transitional cell carcinoma of the bladder. Int J Cancer 122: 1297-1302. PubMed: 18000826. [DOI] [PubMed] [Google Scholar]
- 45. Szkandera J, Knechtel G, Stotz M, Hofmann G, Langsenlehner U et al. (2010) Association of hypoxia-inducible factor 1-alpha gene polymorphisms and colorectal cancer prognosis. Anticancer Res 30: 2393-2397. PubMed: 20651398. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA checklist.
(DOC)