Abstract
Background
Deoxynivalenol (DON), a mycotoxin produced by Fusarium species, is one of the most prevalent mycotoxins present in cereal crops worldwide. Due to its toxic properties, high stability and prevalence, the presence of DON in the food chain represents a health risk for both humans and animals. The gastrointestinal microbiota represents potentially the first target for these food contaminants. Thus, the effects of mycotoxins on the human gut microbiota is clearly an issue that needs to be addressed in further detail. Using a human microbiota-associated rat model, the aim of the present study was to evaluate the impact of a chronic exposure of DON on the composition of human gut microbiota.
Methodology/Principal Findings
Four groups of 5 germ free male rats each, housed in 4 sterile isolators, were inoculated with a different fresh human fecal flora. Rats were then fed daily by gavage with a solution of DON at 100 µg/kg bw for 4 weeks. Fecal samples were collected at day 0 before the beginning of the treatment; days 7, 16, 21, and 27 during the treatment; and 10 days after the end of the treatment at day 37. DON effect was assessed by real-time PCR quantification of dominant and subdominant bacterial groups in feces. Despite a different intestinal microbiota in each isolator, similar trends were generally observed. During oral DON exposure, a significant increase of 0.5 log10 was observed for the Bacteroides/Prevotella group during the first 3 weeks of administration. Concentration levels for Escherichia coli decreased at day 27. This significant decrease (0.9 log10 CFU/g) remained stable until the end of the experiment.
Conclusions/Significance
We have demonstrated an impact of oral DON exposure on the human gut microbiota composition. These findings can serve as a template for risk assessment studies of food contaminants on the human gut microbiota.
Introduction
Deoxynivalenol (DON) is a mycotoxin of the trichothecene family which is a fungal secondary metabolite. Produced by the species Fusarium, it is one of the most prevalent mycotoxins present in cereal crops worldwide, and the most frequently occurring type B trichothecene in Europe. A large scale data survey indicated that DON is present in 57% of food samples collected in the European Union [1]. Moreover, the Joint Expert Committee on Food Additives (JECFA) estimates that European cereal consumers have an estimated intake of DON of 1.4 µg/kg body weight (bw) per day [2]. Exposure to DON could also be estimated using biomarkers. Assessment of mycotoxin and its metabolites in urine provide individual data that may establish the prevalence and range of global DON exposure. Therefore, Turner et al, have confirmed that French farmers and United Kingdom adults have been exposed to an almost ubiquitous amounts of DON [3]–[5]. Due to its toxic properties, high stability and prevalence, the presence of DON in the food chain represents an important threat to food safety and therefore represents a health risk for both humans and animals [6].
Epidemiological studies linking DON exposure to adverse health outcomes in humans have been reported in China, India, Japan and Korea [7]–[9]. Human gastroenteritis with nausea, diarrhea and vomiting are the main symptoms linked to Fusarium-contaminated foods. In addition to the symptoms described in humans, DON toxicity in animals is reflected by feed refusal and consequently growth retardation [10]. At the cellular level, DON has been shown to inhibit protein synthesis and to modulate immune responses [11]. Therefore, a No Observable Adverse Effect Level (NOAEL) has been established at 100 µg/kg of bw based on a decrease body weight gain reported in a 2-year feeding study in mice [12].
Risk assessment approaches to determine a NOAEL are based on physicochemical, pharmacological and toxicological studies. From a microbiological perspective, the ingestion of deoxynivalenol also poses a potential risk to human health since changes in the composition of the human gut microbiota may influence host functions after oral exposure to food contaminants. These changes could, for example, impair colonization resistance that protects the host against pathogen proliferation [13]. Some total diet studies have indicated that dietary exposure through the consumption of food contaminated by mycotoxins is frequent in many populations [14]–[18]. Furthermore, results from Wache et al. clearly demonstrated that low doses of DON, which can typically be found in livestock animal feedstuff, had an impact on the swine fecal flora [19]. Despite the fact that the intestine is the major site of DON absorption and the first target of these toxins [20], and the fact that the gastrointestinal tract and its microbiota represent the first barrier against food contaminants, studies describing the effect of DON on the intestinal microbiota are limited. In fact, several in vitro studies have identified intestinal and soil borne bacteria that promote metabolism, binding or detoxification of DON [21]–[28]. By contrast, limited data on the impact of DON or other members of the trichothecene family on intestinal microbiota have been published [19], [29]–[32]. Data on the detrimental effects of DON on the human gut microbiota are very limited and therefore, the actual health risk from an oral contamination is unknown. Thus, the effects of mycotoxins on the human gut microbiota are clearly an issue that needs to be addressed in further detail.
Using an in vivo approach with a human microbiota-associated rat model (HMA rats), the aim of the present study was to evaluate the impact of a subchronic NOAEL dose exposure of DON on the composition of human gut microbiota. The effect of DON was assessed by monitoring changes in the gut microbiota by real-time PCR (qPCR) quantification of dominant and subdominant bacterial groups.
Materials and Methods
Animals
All animal procedures were carried out in strict accordance with the recommendations of the French Ministry of Agriculture. The protocol was approved by Anses's Committee for Ethical Standards and performed in our approved animal breeding facility (Permit Number: D35-137-26).
Axenic Male Sprague-Dawley rats were obtained from the breeding facility of Charles River laboratories (Saint Germain sur l'Arbresle, France). Sterile pelleted feed (SAFE, Scientific Animal Food and Engineering, Augy, France) free of mycotoxin-contamination and sterile water were provided ad libitum. The 20 rats (8 weeks old, 120–150 g bw) were housed individually in polycarbonate cages in 4 sterile isolators. Animals were acclimatized for one week.
Human donors
The procedure of feces sampling in humans does not require unusual and invasive procedures for monitoring and diagnostic, and consequently no ethical permission was mandatory according to the legislation applicable in France in the article of Act L. 1121-1 of 2012. The volunteers signed a consent form for sample utilization and data publication.
The fecal inocula were obtained from 4 healthy adult individuals. These volunteers consumed an unrestricted western-type diet and were not under antibiotic treatment or taking any other drugs known to influence the fecal microbiota composition for at least three months prior to sampling. All subjects were free of known metabolic or gastrointestinal diseases.
Study design
Transfer of human flora into germ-free rats
Four groups of 5 rats were inoculated with a different fresh human fecal flora. The human fecal specimens were collected and immediately placed in an anaerobic atmosphere (GasPak EZ Anaerobe, BD Diagnostic Systems). In the laboratory, samples were transferred to the Whitley A35 Anaerobic Workstation (AES CHEMUNEX, Bruz, France) for microbiological preparation. Fecal samples from the donors were diluted 1/99 (weight/volume) in prereduced Thioglycollate broth with Resazurine and then given orally to the rats in a volume of 1 ml per rat.
Treatment with deoxynivalenol
After allowing two weeks for microbiota stabilization, 100 µg/kg bw of DON was administrated daily by gavage to the 4 groups of rats for 4 weeks. Fecal samples were collected at day 0 before the beginning of the treatment; days 7, 16, 21, and 27 during the treatment; and 10 days after the end of the treatment at day 37. After collection, they were stored at −80°C and at −20°C until molecular and physicochemical analyses respectively.
Chemical reagents
Deoxynivalenol was purchased from Sigma-Aldrich (Saint-Quentin Fallavier, France) and dissolved in acetonitrile (Sigma-Aldrich) at 1 mg/ml. This solution was stored for a maximum of 1 year at −18°C. The working solutions were diluted in physiological saline solution (B. Braun Avitum, Gradignan, France), stored at room temperature and renewed weekly. Their stability was verified, prior to start the animal experiments, by dosing a working DON solution stored in isolator for 2 weeks. DON concentrations were assessed one time, each week, by an in-house developed and validated High Performance Liquid Chromatography with Ultraviolet detection (HPLC–UV) method.
Bacterial strains and growth
Bacterial type strains used for standard genomic DNA were obtained from the Biological Resource Center of the Institut Pasteur (CRBIP, Paris, France) or Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany) and are presented in table 1.
Table 1. Target organisms, type strains, oligonucleotide primers and probes used in this assay.
| Target organism | Type strain | Primer and probe | Sequence 5′ - 3′ | References |
| All bacteria | Escherichia coli CIP 54.8 | F_Bact 1369 | CGGTGAATACGTTCCCGG | [33] |
| R_Prok 1492 | TACGGCTACCTTGTTACGACTT | |||
| P_TM1389F | 6FAM-CTTGTACACACCGCCCGTC | |||
| Bacteroides/Prevotella group | Bacteroides fragilis CIP 77.16 | F_Bacter 11 | CCTWCGATGGATAGGGGTT | [33] |
| F_Bacter 08 | CACGCTACTTGGCTGGTTCAG | |||
| P_Bac303 | VIC-AAGGTCCCCCACATTG | |||
| Clostridium coccoides group | Blautia coccoides DSM-935 | F_Ccoc 07 | GACGCCGCGTGAAGGA | [33] |
| R_Ccoc 14 | AGCCCCAGCCTTTCACATC | |||
| P_Erec482 | VIC-CGGTACCTGACTAAGAAG | |||
| Clostridium leptum group | Clostridium leptum DSM-753 | F_Clept 09 | CCTTCCGTGCCGSAGTTA | [33] |
| R_Clept 08 | GAATTAAACCACATACTCCACTGCTT | |||
| P_Clep 01 | 6FAM-CACAATAAGTAATCCACC | |||
| Genus Bifidobacterium | Bifidobacterium adolescentis CIP 64.59 | F_Bifid 09c | CGGGTGAGTAATGCGTGACC | [33] |
| R_Bifid 06 | TGATAGGACGCGACCCCA | |||
| P_Bifid | 6FAM-CTCCTGGAAACGGGTG | |||
| Genus Enterococcus | Enterococcus faecium CIP 103014 | F_Entero | CCCTTATTGTTAGTTGCCATCATT | [61] |
| R_Entero | ACTCGTTGTACTTCCCATTGT | |||
| Escherichia coli | Escherichia coli CIP 54.8 | E.coli F | CATGCCGCGTGTATGAAGAA | [62] |
| E.coli R | CGGGTAACGTCAATGAGCAAA | |||
| Lactobacillus/Leuconostoc/Pediococcus group | Lactobacillus acidophilus DSM-20079 | F_Lacto 05 | AGCAGTAGGGAATCTTCCA | [33] |
| R_Lacto 04 | CGCCACTGGTGTTCYTCCATATA |
All strains were inoculated into Tryptic Soy broth, MRS broth and Thioglycollate broth with Resazurine for aerobic, Lactobacillus acidophilus and other anaerobic bacteria respectively. Pure cultures were incubated at 37°C in an aerobic or anaerobic atmosphere (10% H2, 10% CO2, 80% N2). The total number of Colony Forming Units (CFU) of each culture was determined by plating 100 µl of the appropriate 10-fold dilution series on Trypticase Soy Agar with 10% Sheep Blood, on MRS Agar and on Schaedler Agar with Vitamin K1 and 5% Sheep Blood (BD Diagnostic Systems, Le Pont de Claix, France) for aerobic, Lactobacillus acidophilus and anaerobic bacteria respectively.
DNA extraction
Genomic DNA from bacterial cultures was extracted using Wizard Genomic DNA Purification Kit (Promega, Charbonnières Les Bains, France) according to the manufacturer's instructions. Extracted DNA was quantified using a BioSpec-nano (Shimadzu Scientific Instruments, Columbia, U.S.A.).
Fecal DNA was extracted from the 200 mg aliquots of feces as described previously [33].
Oligonucleotide primers and probes
Primers and probes used in this study are presented in table 1. TaqMan qPCR was adapted to quantify the All bacteria system, the Bacteroides/Prevotella, Clostridium coccoides, Clostridium leptum groups and the genus Bifidobacterium. Real-time qPCR using SYBR Green was performed for Escherichia coli, the genus Enterococcus and the Lactobacillus/Leuconostoc/Pediococcus group. Primer and probe specificities were previously assessed by Furet et al. [33]. The TaqMan probes were synthesised by Life Technologies (Saint Aubin, France). The primers were purchased from Sigma Aldrich.
Real-time PCR conditions
PCR was performed in 25 µl PCR volumes containing 15 µl Power SYBR® Green PCR Master Mix 2X (Life Technologies) or TaqMan Universal PCR 2 (Life Technologies), 0.20 µM of each primer, 0.25 µM of each probe and 10 µl of template DNA at the appropriate dilution (inhibition testing section). Natural Multiplate™ Low-Profile 96-Well Unskirted PCR Plates and a Chromo4 LightCycler were used (Bio-rad, Marnes-La-Coquette, France). The cycling program included a 10 min incubation at 95°C followed by 40 cycles consisting of 95°C for 30 s, 60°C for 1 min. For SYBR-Green® amplifications, to improve amplification specificity, a melting curve analysis of the PCR products was performed by ramping the temperature to 95°C for 10 s and back to 50°C for 15 s followed by incremental increases of 0.5°C/s up to 95°C. The cycle threshold (Ct), i.e. the number of PCR cycles necessary to reach the threshold fluorescence level, was manually determined for each run by the user. All the samples were analysed in duplicate.
PCR Setup Controls
Multiple Non Template Controls (NTC) were included in every assay and amplification of all NTC wells invalidated the entire qPCR run, leading to a repeat run.
Generation of standard curves
Genomic DNA from the different type strains was used to prepare ten-fold dilution series from 0.1 log10 to 8 log10 CFU equivalent. Sterile water (15 µl) was used as a negative control. A standard curve for each type strain was generated by plotting the Ct against the logarithm of bacterial quantity (log10 CFU equivalent) for each run.
Inhibition Testing
The TaqMan® Exogenous Internal Positive Control Reagents kit (Life Technologies) was used as an exogenous amplification control to check the appropriate dilution used in real-time PCR [34]. This IPC inhibition assay comprises a control qPCR assay in which the IPC DNA is the only amplifiable target performed in the presence of water. This generates a reference Ct value for the IPC amplicon, characteristic of an uninhibited assay. If the water is substituted with DNA from a sample, a shift of greater than one cycle, to a higher Ct and reduced amplification efficiency indicates the presence of PCR inhibitors in the sample. Each qPCR reaction comprised 1X TaqMan Universal PCR 2, 1X Exo IPC Mix, and 5 µl of diluted sample extract or water. Thermal cycling conditions were 1 cycle of 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, 60°C for 1 min.
Determination of DON concentrations in feces
Concentrations of DON and its main metabolite deepoxy-deoxynivalenol (DOM-1) in feces from donors and rats were analysed by an adapted method from Sørensen and Elbæk (2005) using a Liquid Chromatography coupled with tandem Mass Spectrometry (LC-MS/MS) (Laboratory LDA 22, Ploufragan, France) [35].
Data normalization
To overcome the fact that fecal samples may contain more or less water, normalization was done by subtracting the log10 CFU/g obtained for the “all bacteria” group from the log10 CFU/g for the other bacterial groups [33], [36]. The data are presented as the mean of normalized log10 value ± standard deviation of colony-forming unit equivalent per gram of fresh feces (log10 CFU/g).
Statistics
Statistical analysis was performed with SYSTAT V.13 (Systat Software, Chicago, USA). Bacterial levels were compared using the analysis of variance (ANOVA) followed by Dunnett's Multiple Comparison test to compare each sampling time during and after the treatment to the sampling time before the treatment (control). DON concentrations in feces were compared using the analysis of variance (ANOVA). P values <0.05 were considered statistically significant.
Results
Evaluation of the qPCR performances
PCR efficiencies were calculated from the slopes of the log-linear portion of the standard curves that were run in each plate. The efficiency range was 82–111% corresponding to good amplification efficiency. The dynamic range covered at least 5 log10 concentrations of magnitude, and included the expected interval for the target nucleic acids to be quantified.
We assessed the presence of inhibitors in the DNA samples that could interfere with the following qPCR method. One third of the DNA extracts from fecal samples were included in the inhibition assay and results indicated that no inhibition was present in these samples diluted at 10−4 and 10−5. In contrast, results from samples diluted at 10−3 showed shifts of more than 1 Ct with respect to the reference Ct value, indicative of the presence of PCR inhibitors in these samples. DNA extracts were therefore used at the appropriate dilution of 10−4.
The limit of quantification was 6 log10 CFU/g, corresponding to the lowest value quantified in the fecal samples with good repeatability (<0.5 Ct).
Establishment of the human gut microbiota in human microbiota-associated rats
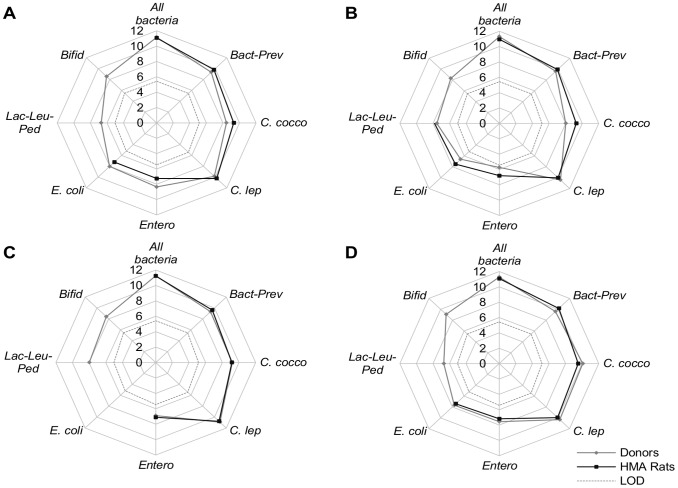
Figure 1 presents the fecal concentration levels for dominant and subdominant bacterial groups in the HMA rats after 2 weeks of stabilization and the corresponding human donors. With the exception of bifidobacteria and the Lactobacillus/Leuconostoc/Pediococcus group, all other groups showed concentrations in rat feces close to their concentrations in human feces. These bacteria from human donors established very well in the rat gut tract, in contrast to bifidobacteria and the Lactobacillus/Leuconostoc/Pediococcus group which were below the limit of detection of 5.46 and 5.03 log10 CFU/g respectively. Our results indicate that these bacterial groups established in rats at lower levels than the inoculating bacterial community from the human donors.
Figure 1. Fecal concentrations of each target bacterial group of the HMA rats and in donors.
(A) Isolator I; (B) Isolator II; (C) Isolator III; (D) Isolator IV; Results obtained by qPCR were expressed as the mean of the log10 value (for rats n = 5 except for isolator IV where n = 4; for humans n = 2 repetitions) of CFU/g. Bact-Prev: Bacteroides-Prevotella group; C. cocco: Clostridium coccoides group; C. lep: Clostridium leptum group; Bifid: Genus Bifidobacterium; Entero: Genus Enterococcus; E. coli: Escherichia coli; Lac-Leu-Ped: Lactobacillus-Leuconostoc-Pediococcus group; LOD: limit of detection.
DON concentrations in feces
DON concentrations in donors' feces were below the limit of detection (3 µg/kg of feces).
In rats, fecal samples from each rat housed in the same isolator were pooled at days 16 and 27. Fecal DON concentrations for each isolator, and for each sampling time, ranged from 240 to 360 µg/kg of feces. Differences in DON concentrations between isolators or sampling time were not significant. Therefore, an overall mean of DON was estimated at 284.4±45.8 µg/kg of feces between day 16 and 27.
DOM-1 was not detected in fecal samples of rats (limit of detection: 3 µg/kg of feces).
Changes of the gut microbiota in response to DON treatment
Table 2 shows a general overview of the evolution over time of the bacterial groups targeted for each isolator. Despite a different intestinal microbiota in each isolator, similar trends were generally observed for the dominant and subdominant groups targeted in the feces of the humanized rats.
Table 2. Time course evolution of bacteria levels in the 4 isolators during the experimental period.
| Treatment (days) | |||||||
| Before | During | After | |||||
| Isolator | Target organism | 0 | 7 | 16 | 21 | 27 | 37 |
| ISO I | All bacteria | 11.07±0.14 | 10.77±0.18 | 10.82±0.09 | 10.77±0.26 | 11.18±0.08 | 11.10±0.17 |
| Bact-Prev | −1.28±0.15 | −0.91±0.11** | −0.79±0.04*** | −0.88±0.24** | −1.24±0.12 | −1.25±0.19 | |
| C. cocco | −1.76±0.30 | −1.46±0.33 | −1.39±0.44 | −2.04±0.69 | −2.03±0.36 | −1.89±0.78 | |
| C. lep | −0.82±0.05 | −0.68±0.17 | −0.69±0.04 | −0.87±0.26 | −0.94±0.09 | −1.02±0.07 | |
| Bifid | < LODa | < LODa | < LODa | < LODa | < LODa | < LODa | |
| Entero | −3.81±0.79 | −4.14±0.24 | −4.15±0.55 | −4.72±0.80 | −4.89±0.36 | −4.85±0.88 | |
| E. coli | −3.84±0.30 | −3.60±0.10 | −3.74±0.27 | −3.97±0.32 | −4.94±0.15*** | −4.57±0.31*** | |
| Lac-Leu-Ped | < LODb | < LODb | < LODb | < LODb | < LODb | < LODb | |
| ISO II | All bacteria | 10.94±0.10 | 10.75±0.10 | 10.74±0.12 | 10.75±0.07 | 11.09±0.05 | 11.10±0.13 |
| Bact-Prev | −1.01±0.18 | −0.74±0.11** | −0.69±0.09** | −0.84±0.13 | −1.09±0.08 | −1.22±0.12* | |
| C. cocco | −1.65±0.15 | −1.56±0.25 | −1.48±0.32 | −1.71±0.42 | −1.97±0.25 | −2.18±0.13* | |
| C. lep | −0.90±0.21 | −0.85±0.11 | −0.98±0.06 | −1.02±0.09 | −0.95±0.11 | −0.96±0.14 | |
| Bifid | < LODa | < LODa | < LODa | < LODa | < LODa | < LODa | |
| Entero | −4.13±0.88 | −4.22±0.60 | −4.81±0.50 | −4.79±87 | −4.83±0.71 | −5.01±0.44 | |
| E. coli | −3.44±0.48 | −3.26±0.16 | −3.58±0.16 | −3.69±0.40 | −4.35±0.41*** | −4.38±0.10*** | |
| Lac-Leu-Ped | −3.13±1.20 | −3.32±1.12 | −3.45±1.31 | −3.16±1.67 | −3.56±1.23 | −3.65±1.22 | |
| ISO III | All bacteria | 11.22±0.17 | 10.68±0.20 | 10.74±0.15 | 10.66±0.13 | 11.30±0.15 | 11.28±0.11 |
| Bact-Prev | −1.61±0.20 | −1.08±0.25*** | −1.09±0.11*** | −1.08±0.14*** | −1.44±0.20 | −1.51±0.16 | |
| C. cocco | −2.06±0.39 | −2.01±0.30 | −1.82±0.55 | −1.81±0.50 | −1.91±0.55 | −2.41±0.23 | |
| C. lep | −0.45±0.16 | −0.21±0.15 | −0.15±0.10* | −0.18±0.12* | −0.42±0.20 | −0.35±0.11 | |
| Bifid | < LODa | < LODa | < LODa | < LODa | < LODa | < LODa | |
| Entero | −4.12±0.48 | −4.27±0.62 | −4.32±0.72 | −4.14±0.77 | −4.00±0.38 | −4.71±0.27 | |
| E. coli | < LOD | < LOD | < LOD | < LOD | < LOD | < LOD | |
| Lac-Leu-Ped | < LODb | < LODb | < LODb | < LODb | < LODb | < LODb | |
| ISO IV | All bacteria | 11.07±0.14 | 10.63±0.06 | 10.63±0.15 | 10.65±0.11 | 11.10±0.13 | 11.20±0.08 |
| Bact-Prev | −0.90±0.09 | −0.30±0.16*** | −0.30±0.18*** | −0.20±0.11*** | −0.72±0.16 | −0.83±0.14 | |
| C. cocco | −1.54±0.23 | −0.77±0.47 | −1.19±0.63 | −1.20±0.64 | −1.56±0.44 | −1.27±0.36 | |
| C. lep | −1.12±0.18 | −0.67±0.24 | −0.75±0.36 | −0.74±0.25 | −1.05±0.28 | −1.12±0.14 | |
| Bifid | < LODa | < LODa | < LODa | < LODa | < LODa | < LODa | |
| Entero | −3.87±0.60 | −3.86±0.29 | −4.05±0.50 | −3.36±0.20 | −4.25±0.71 | −3.79±0.47 | |
| E. coli | −3.66±0.54 | −3.31±0.28 | −3.59±0.24 | −3.46±0.24 | −4.03±0.49 | −3.93±0.15 | |
| Lac-Leu-Ped | < LODb | < LODb | < LODb | < LODb | < LODb | < LODb | |
Results obtained by qPCR were expressed for all bacteria as the mean of the log10 value (n = 5 except for isolator IV where n = 4) ± standard deviation of CFU/g. Normalization was done by subtracting the log10 CFU/g obtained for the “all bacteria” group from the log10 CFU/g for the other bacterial groups. Results were expressed for the different bacterial groups as the mean of normalized log10 value (n = 5 except for isolator IV where n = 4) ± standard deviation of CFU/g.
*P<0.05;
**P<0.01;
***P<0.001.
(a)Bifid LOD = 5.46 log10 CFU/g;
(b)Lac-Leu-Ped LOD = 5.03 log10 CFU/g; Bact-Prev: Bacteroides-Prevotella group; C. cocco: Clostridium coccoides group; C. lep: Clostridium leptum group; Bifid: Genus Bifidobacterium; Entero: Genus Enterococcus; E. coli: Escherichia coli; Lac-Leu-Ped: Lactobacillus-Leuconostoc-Pediococcus group.
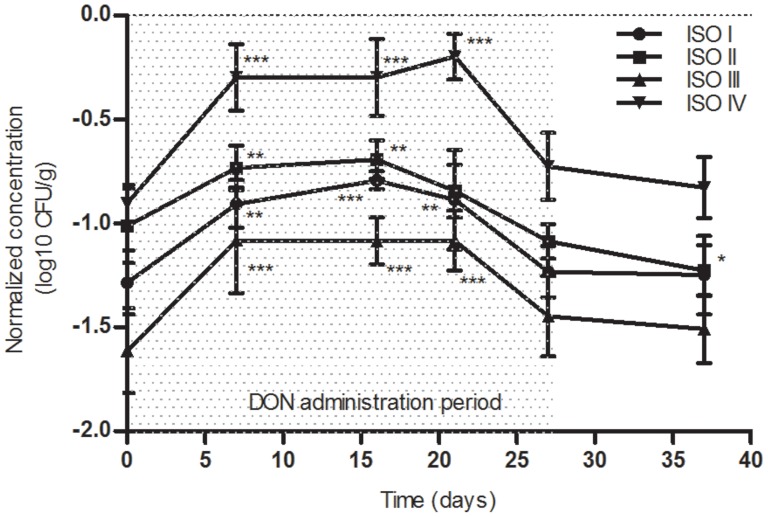
During oral DON exposure, a significant increase of 0.5 log10 was observed for the Bacteroides/Prevotella group during the first 3 weeks of administration in isolators I, III and IV (P<0.01). Concentrations returned to basal levels similar to that of the control at D0 before the end of the treatment, indicating a transient effect (figure 2).
Figure 2. Time course evolution of Bacteroides-Prevotella normalized concentrations in the 4 isolators during the experimental period.
For the different bacterial groups, results were expressed as the mean of normalized log10 value (n = 5 except for isolator IV where n = 4) ± standard deviation of CFU/g.* P<0.05; ** P<0.01; *** P<0.001.
Concentrations of bifidobacteria and the Lactobacillus/Leuconostoc/Pediococcus group remained below the limit of detection of 5.46 and 5.03 log10 CFU/g respectively in all DNA extracts. DON seemed to have no effect on these bacterial groups.
During the experimental period, large interindividual fluctuations with SDs higher than 0.5 log10 CFU/g were observed in the Enterococcus and Clostridium coccoides groups.
The Clostridium leptum group did not seem to be affected by deoxynivalenol, and remained stable in 3 of the 4 isolators. However, in isolator III, a significant increase of 0.3 log10 CFU/g was observed at days 16 and 21 of the treatment (P<0.05).
With the exception of isolator III where Escherichia coli was not detected, concentrations of this species decreased at day 27 i.e. the end of the DON treatment. This decrease (0.9 log10 CFU/g) was significant for isolators I and II (P<0.001). This drop remained stable until the end of the experiment.
Discussion
In this study, we investigated the effect of an oral subchronic exposure of DON at NOAEL on the intestinal microbiota balance. To the best of our knowledge, this study is the first to analyze the impact of DON on the human intestinal microbiota by real-time PCR in HMA rats.
To avoid ethical issues and long monitoring periods, HMA rodents are widely used to evaluate the effect of contaminants or to elucidate whether the gut commensal microbiota is important for human health [13], [37], [38]. In our study, 5 of 7 dominant (Bacteroides/Prevotella, Clostridium coccoides, Clostridium leptum, bifidobacteria) and subdominant (Escherichia coli, Lactobacillus/Leuconostoc/Pediococcus Enterococcus) bacterial groups of human gut microbiota could be successfully established in the germ free rats at levels comparable to the human donors i.e. Bacteroides/Prevotella, Clostridium coccoides and Clostridium leptum groups, Escherichia coli and genus Enterococcus. However, bifidobacteria and the Lactobacillus/Leuconostoc/Pediococcus group were not detected in our experimental conditions. Several studies have previously shown that these bacteria coming from humans decreased in germ free rodents after inoculation, reaching undetectable levels in fecal samples of animals [39]–[42]. Nevertheless, these results demonstrate that the established bacterial community of the feces recipient rats remains similar to the inoculating bacterial community of the human donors. These studies confirm that this animal model is relevant for exploring the effect of food contaminants on the human gut microbiota.
Interactions between bacteria and deoxynivalenol have been demonstrated in several in vitro studies dealing with mycotoxin-transforming microorganisms. Several studies have reported transformation of deoxynivalenol by microorganisms from a variety of environmental samples including field soils, wheat leaves and animal gut contents [27], [28], [43]–[46]. This has been assessed with success for potential applications in detoxifying mycotoxins in contaminated food and feed [47]. For example, lactic acid bacteria strains have the ability to remove DON in vitro by adsorption of the mycotoxin by the cell wall [48], [49]. This specific binding of DON was able to restrict the consequences of DON on Caco-2 as was shown by Turner et al. with the strain Lactobacillus rhamnosus [50]. Eubacterium sp. is also beneficial in counteracting the toxicity of DON in broilers through the deepoxidization of the mycotoxin in contaminated diets [51]. These observations demonstrate clear interactions between DON and bacteria, which could have impacts on both the bacteria and DON levels. DON absorption is incomplete in the intestine with a bioavailability of about 54% in pigs [52] and 50% in conventional rats (personal data), suggesting that a fraction of ingested DON reaches the colon. This suggestion has been confirmed by Nagl et al. in rats, who observed that about 4% of the administered dose of DON were recovered in the feces of animals [53].The quantity of DON that was found in the feces of HMA rats during the treatment suggests that this food contaminant could potentially influence gut microbiota. Indeed, in our study, the significant increase of the Bacteroides/Prevotella group and the significant decrease of Escherichia coli indicate that DON at the NOAEL (100 µg/kg bw) could induce biological effects on the dynamic of the humanized gut microbiota. Our findings are in accordance with Waché et al. who showed changes in the gut microbiota composition of pigs fed with diet naturally contaminated with DON (136 µg/kg bw) [19]. However, these authors reported, using selective media, an increase of aerobic mesophilic bacteria and a decrease of anaerobic sulfite-reducing bacteria during DON treatment [19], which is not consistent with our findings. This difference could be due to the method used to quantify the bacterial population. Indeed, unlike culture-dependent methods, qPCR quantifies all targeted bacteria irrespective of the state (cultivable, viable non-cultivable, non-cultivable and dead bacteria) while conventional microbiological methods only identifies cultivable bacteria (estimated at <30% of the gut microbiota) [54]. In addition, their experimental animal model was the swine where the indigenous gut bacteria are different from those of humans.
In this study, we observed the significant increase of the Bacteroides/Prevotella group. In Human, this shift may be associated to diseases as reviewed by Clemente et al. [55]. For example, individuals with Crohn's disease or celiac diseases exhibited a higher abundance of Bacteroides than healthy individuals [56], [57].
With the gut microbiota dysbiosis observed in this study, it could be hypothesized that DON may promote the passage of pathogenic micro-organisms present in food and water across the intestinal epithelium. These effects represent a potential health threat as they could contribute to an increase in bacterial infections in animals or humans exposed to DON. This has already been observed by Oswald et al. in piglets treated with fumonisin B1 [31], where mycotoxin treatment was associated with an increased bacterial colonization by pathogenic Escherichia coli in the intestine of animals.
In addition, DON is known to induce proinflammatory responses in experimental animals [58]. In vivo and in vitro studies have also shown that immune cells (including macrophages, B and T lymphocytes and natural killer cells) are very sensitive to DON [59], [60]. DON may therefore contribute to modulate infectious diseases through alterations in immune function.
Although this study and others [19], [30] have shown a potential hazard of mycotoxins on the gut microbiota, the NOAEL of these food contaminants is established only on toxicological data. The International Cooperation on Harmonization of Technical Requirements for Veterinary Medicinal Products (VICH) provides a general approach to establishing a microbiological acceptable daily intake (http://www.vichsec.org) to evaluate the safety of residues of veterinary drugs in animal-derived foods for humans. This approach used for veterinary drugs could be extended to mycotoxins which are prevalent food contaminants.
Conclusions
In conclusion, this study provides data that could help to determine the public health risk of deoxynivalenol on the human gut microbiota.
On one hand, using qPCR we have identified particular genera in human gut microbiota whose concentrations vary after DON exposure at the NOAEL dose. As this effect could have consequences for human health, further investigation would be interesting. Therefore, in order to improve risk assessment in humans, studies on the different functions of the gut microbiota could be performed (barrier effect, enzymatic activities, metabolic profiles) after exposure to DON. Moreover, co-occurrence of mycotoxins is widespread and mycotoxins can be released from their masked forms; therefore, experiments with combinations of mycotoxins and their different forms are necessary and complementary to our study.
On the other hand, we have demonstrated that the NOAEL established for deoxynivalenol based on a toxicological study can have, nonetheless, a microbiologically significant effect by modifying the gut microbiota. Therefore, we suggest that the investigation of the influence of low concentration of mycotoxins on human gut microbiota as a part of the risk assessment process.
Overall, the findings from this investigation could serve as a template for future impact studies of food contaminants on the human gut microbiota.
Funding Statement
This work was funded by the French agency for food, environmental and occupational health and safety (Anses). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schothorst RC, van Egmond HP (2004) Report from SCOOP task 3.2.10 “collection of occurrence data of Fusarium toxins in food and assessment of dietary intake by the population of EU member states”: Subtask: trichothecenes. Toxicology Letters 153: 133–143. [DOI] [PubMed] [Google Scholar]
- 2.Canady R, Coker R, Egan S, Krska R, Kuiper-Goodman T, et al.. (2001) Deoxynivalenol. Safety evaluation of certain mycotoxins in food (pp. 419–555). Joint Expert Committee on Food Additives (JECFA). WHO Food Additives Series 47.
- 3. Turner PC, Burley VJ, Rothwell JA, White KL, Cade JE, et al. (2008) Deoxynivalenol: Rationale for development and application of a urinary biomarker. Food Additives and Contaminants 25: 864–871. [DOI] [PubMed] [Google Scholar]
- 4. Turner PC, Hopton RP, Lecluse Y, White KLM, Fisher J, et al. (2010) Determinants of Urinary Deoxynivalenol and De-epoxy Deoxynivalenol in Male Farmers from Normandy, France. Journal of Agricultural and Food Chemistry 58: 5206–5212. [DOI] [PubMed] [Google Scholar]
- 5. Turner PC, Hopton RP, White KLM, Fisher J, Cade JE, et al. (2011) Assessment of deoxynivalenol metabolite profiles in UK adults. Food and Chemical Toxicology 49: 132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CAST (2003) Mycotoxins: risks in plant animal and human systems. Ames, Iowa: Council for Agricultural Science and Technology. 199.
- 7. Ramakrishna Y, Bhat RV, Ravindranath V (1989) Production of deoxynivalenol by Fusarium isolates from samples of wheat associated with a human mycotoxicosis outbreak and from sorghum cultivars. Applied and Environmental Microbiology 55: 2619–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhat R, Ramakrishna Y, Beedu S, Munshi KL (1989) Outbreak of trichothecene mycotoxicosis associated with consumption of mould-damaged wheat production in Kashmir Valley, India. The Lancet 333: 35–37. [DOI] [PubMed] [Google Scholar]
- 9.Luo X (1988) Outbreaks of moldy cereals poisoning in China. Issues in food safety: 56–63.
- 10. Pestka JJ (2007) Deoxynivalenol: Toxicity, mechanisms and animal health risks. Animal Feed Science and Technology 137: 283–298. [Google Scholar]
- 11. Pestka J (2010) Deoxynivalenol: mechanisms of action, human exposure, and toxicological relevance. Archives of Toxicology 84: 663–679. [DOI] [PubMed] [Google Scholar]
- 12. Iverson F, Armstrong C, Nera E, Truelove J, Fernie S, et al. (1995) Chronic Feeding Study of deoxynivalenol in B6C3F1 male and female mice. Teratogenesis, Carcinogenesis, and Mutagenesis 15: 283–306. [DOI] [PubMed] [Google Scholar]
- 13. Perrin-Guyomard A, Poul J-M, Laurentie M, Sanders P, Fernández AH, et al. (2006) Impact of ciprofloxacin in the human-flora-associated (HFA) rat model: Comparison with the HFA mouse model. Regulatory Toxicology and Pharmacology 45: 66–78. [DOI] [PubMed] [Google Scholar]
- 14. Sirot V, Fremy J-M, Leblanc J-C (2013) Dietary exposure to mycotoxins and health risk assessment in the second French total diet study. Food and Chemical Toxicology 52: 1–11. [DOI] [PubMed] [Google Scholar]
- 15. Le Bail M, Verger P, Doré T, Fourbet JF, Champeil A, et al. (2005) Simulation of consumer exposure to deoxynivalenol according to wheat crop management and grain segregation: Case studies and methodological considerations. Regulatory Toxicology and Pharmacology 42: 253–259. [DOI] [PubMed] [Google Scholar]
- 16. Leblanc J-C, Malmauret L, Delobel D, Verger P (2002) Simulation of the Exposure to Deoxynivalenol of French Consumers of Organic and Conventional Foodstuffs. Regulatory Toxicology and Pharmacology 36: 149–154. [DOI] [PubMed] [Google Scholar]
- 17. Turner PC, Rothwell JA, White KLM, Gong Y, Cade JE, et al. (2008) Urinary deoxynivalenol is correlated with cereal intake in individuals from the United Kingdom. Environmental Health Perspectives 116: 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Warth B, Sulyok M, Fruhmann P, Berthiller F, Schuhmacher R, et al. (2012) Assessment of human deoxynivalenol exposure using an LC–MS/MS based biomarker method. Toxicology Letters 211: 85–90. [DOI] [PubMed] [Google Scholar]
- 19. Waché Y, Valat C, Postollec G, Bougeard S, Burel C, et al. (2008) Impact of Deoxynivalenol on the Intestinal Microflora of Pigs. International Journal of Molecular Sciences 10: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rotter BA (1996) Toxicology of Deoxynivalenol (Vomitoxin). Journal of Toxicology and Environmental Health 48: 1–34. [DOI] [PubMed] [Google Scholar]
- 21. Gratz SW, Duncan G, Richardson AJ (2013) The human fecal microbiota metabolizes deoxynivalenol and deoxynivalenol-3-glucoside and may be responsible for urinary deepoxy-deoxynivalenol. Applied and Environmental Microbiology 79: 1821–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He P, Young LG, Forsberg C (1992) Microbial transformation of deoxynivalenol (vomitoxin). Appl Environ Microbiol 58: 3857–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ito M, Sato I, Ishizaka M, Yoshida SI, Koitabashi M, et al. (2013) Bacterial cytochrome P450 System catabolizing the Fusarium toxin deoxynivalenol. Applied and Environmental Microbiology 79: 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito M, Sato I, Koitabashi M, Yoshida S, Imai M, et al. (2012) A novel actinomycete derived from wheat heads degrades deoxynivalenol in the grain of wheat and barley affected by Fusarium head blight. Applied Microbiology and Biotechnology: 1–12. [DOI] [PubMed]
- 25. Kollarczik B, Gareis M, Hanelt M (1994) In vitro transformation of the Fusarium mycotoxins deoxynivalenol and zearalenone by the normal gut microflora of pigs. Natural Toxins 2: 105–110. [DOI] [PubMed] [Google Scholar]
- 26. Niderkorn V, Boudra H, Morgavi DP (2006) Binding of Fusarium mycotoxins by fermentative bacteria in vitro . Journal of Applied Microbiology 101: 849–856. [DOI] [PubMed] [Google Scholar]
- 27. Young JC, Zhou T, Yu H, Zhu H, Gong J (2007) Degradation of trichothecene mycotoxins by chicken intestinal microbes. Food and Chemical Toxicology 45: 136–143. [DOI] [PubMed] [Google Scholar]
- 28. Zou ZY, He ZF, Li HJ, Han PF, Meng X, et al. (2012) In vitro removal of deoxynivalenol and T-2 toxin by lactic acid bacteria. Food Science and Biotechnology 21: 1677–1683. [Google Scholar]
- 29. Becker B, Bresch H, Schillinger U, Thiel PG (1997) The effect of fumonisin B1 on the growth of bacteria. World Journal of Microbiology and Biotechnology 13: 539–543. [Google Scholar]
- 30. Burel C, Tanguy M, Guerre P, Boilletot E, Cariolet R, et al. (2013) Effect of Low Dose of Fumonisins on Pig Health: Immune Status, Intestinal Microbiota and Sensitivity to Salmonella . Toxins 5: 841–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oswald IP, Desautels C, Laffitte Jl, Fournout S, Peres SY, et al. (2003) Mycotoxin Fumonisin B1 Increases Intestinal Colonization by Pathogenic Escherichia coli in Pigs. Applied and Environmental Microbiology 69: 5870–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tenk I, Fodor É, Szathmáry C (1982) The effect of pure fusarium toxins (T-2, F-2, DAS) on the microflora of the gut and on plasma glucocorticoid levels in rat and swine. Zentralblatt für Bakteriologie, Mikrobiologie und Hygiene 1 Abt Originale A, Medizinische Mikrobiologie, Infektionskrankheiten und Parasitologie 252: 384–393. [PubMed] [Google Scholar]
- 33. Furet J-P, Firmesse O, Gourmelon M, Bridonneau C, Tap J, et al. (2009) Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiology Ecology 68: 351–362. [DOI] [PubMed] [Google Scholar]
- 34. Johnson GL, Bibby DF, Wong S, Agrawal SG, Bustin SA (2012) A MIQE-Compliant Real-Time PCR Assay for Aspergillus Detection. PLoS ONE 7: e40022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sørensen L, Elbæk T (2005) Determination of mycotoxins in bovine milk by liquid chromatography tandem mass spectrometry. Journal of Chromatography B 820: 183–196. [DOI] [PubMed] [Google Scholar]
- 36. Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, et al. (2009) The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiology 9: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi P, Li L (2011) The germfree murine animal: An important animal model for research on the relationship between gut microbiota and the host. Veterinary Microbiology. [DOI] [PubMed]
- 38. Wos-Oxley ML, Bleich A, Oxley APA, Kahl S, Janus LM, et al. (2012) Comparative evaluation of establishing a human gut microbial community within rodent models. Gut Microbes 3: 234–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, et al. (2011) The Evolution of Host Specialization in the Vertebrate Gut Symbiont Lactobacillus reuteri . PLoS Genet 7: e1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bernbom N, Nørrung B, Saadbye P, Mølbak L, Vogensen FK, et al. (2006) Comparison of methods and animal models commonly used for investigation of fecal microbiota: Effects of time, host and gender. Journal of Microbiological Methods 66: 87–95. [DOI] [PubMed] [Google Scholar]
- 41. Ducluzeau R, Ladiré M, Raibaud P (1984) Effet de l'ingestion de son de blé sur la flore microbienne fécale de donneurs humains et de souris gnotoxéniques receveuses, et sur les effets de barrière exercés par ces flores à l'égard de divers microorganismes potentiellement pathogènes. Annales de l'Institut Pasteur/Microbiologie 135: 303–318. [PubMed] [Google Scholar]
- 42. Hirayama K, Kawamura S, Mitsuoka T (1991) Development and Stability of Human Faecal Flora in the Intestine of Ex-germ-free Mice. Microbial Ecology in Health and Disease 4: 95–99. [Google Scholar]
- 43. Sato I, Ito M, Ishizaka M, Ikunaga Y, Sato Y, et al. (2012) Thirteen novel deoxynivalenol-degrading bacteria are classified within two genera with distinct degradation mechanisms. FEMS Microbiology Letters 327: 110–117. [DOI] [PubMed] [Google Scholar]
- 44. Islam R, Zhou T, Young JC, Goodwin PH, Pauls KP (2012) Aerobic and anaerobic de-epoxydation of mycotoxin deoxynivalenol by bacteria originating from agricultural soil. World Journal of Microbiology and Biotechnology 28: 7–13. [DOI] [PubMed] [Google Scholar]
- 45. Yu H, Zhou T, Gong J, Young C, Su X, et al. (2010) Isolation of deoxynivalenol-transforming bacteria from the chicken intestines using the approach of PCR-DGGE guided microbial selection. BMC Microbiology 10: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guan S, He J, Young JC, Zhu H, Li X-Z, et al. (2009) Transformation of trichothecene mycotoxins by microorganisms from fish digesta. Aquaculture 290: 290–295. [Google Scholar]
- 47. Zhou T, He J, Gong J (2008) Microbial transformation of trichothecene mycotoxins. World Mycotoxin Journal 1: 23–30. [Google Scholar]
- 48. El-Nezami HS, Chrevatidis A, Auriola S, Salminen S, Mykkänen H (2002) Removal of common Fusarium toxins in vitro by strains of Lactobacillus and Propionibacterium . Food Additives and Contaminants 19: 680–686. [DOI] [PubMed] [Google Scholar]
- 49. Franco TS, Garcia S, Hirooka EY, Ono YS, dos Santos JS (2011) Lactic acid bacteria in the inhibition of Fusarium graminearum and deoxynivalenol detoxification. Journal of Applied Microbiology 111: 739–748. [DOI] [PubMed] [Google Scholar]
- 50. Turner P, Wu Q, Piekkola S, Gratz S, Mykkänen H, et al. (2008) Lactobacillus rhamnosus strain GG restores alkaline phosphatase activity in differentiating Caco-2 cells dosed with the potent mycotoxin deoxynivalenol. Food and Chemical Toxicology 46: 2118–2123. [DOI] [PubMed] [Google Scholar]
- 51. Awad W, Bohm J, Razzazi-Fazeli E, Ghareeb K, Zentek J (2006) Effect of addition of a probiotic microorganism to broiler diets contaminated with deoxynivalenol on performance and histological alterations of intestinal villi of broiler chickens. Poultry Science 85: 974–979. [DOI] [PubMed] [Google Scholar]
- 52. Goyarts T, Dänicke S (2006) Bioavailability of the Fusarium toxin deoxynivalenol (DON) from naturally contaminated wheat for the pig. Toxicology Letters 163: 171–182. [DOI] [PubMed] [Google Scholar]
- 53. Nagl V, Schwartz H, Krska R, Moll W-D, Knasmüller S, et al. (2012) Metabolism of the masked mycotoxin deoxynivalenol-3-glucoside in rats. Toxicology Letters 213: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraher MH, O'Toole PW, Quigley EMM (2012) Techniques used to characterize the gut microbiota: a guide for the clinician. Nat Rev Gastroenterol Hepatol advance online publication. [DOI] [PubMed]
- 55. Clemente JC, Ursell LK, Parfrey LW, Knight R (2012) The impact of the gut microbiota on human health: An integrative view. Cell 148: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nadal I, Donant E, Ribes-Koninckx C, Calabuig M, Sanz Y (2007) Imbalance in the composition of the duodenal microbiota of children with coeliac disease. Journal of Medical Microbiology 56: 1669–1674. [DOI] [PubMed] [Google Scholar]
- 57. Dicksved J, Halfvarson J, Rosenquist M, Jarnerot G, Tysk C, et al. (2008) Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J 2: 716–727. [DOI] [PubMed] [Google Scholar]
- 58. Amuzie CJ, Harkema JR, Pestka JJ (2008) Tissue distribution and proinflammatory cytokine induction by the trichothecene deoxynivalenol in the mouse: Comparison of nasal vs. oral exposure. Toxicology 248: 39–44. [DOI] [PubMed] [Google Scholar]
- 59. Atkinson HAC, Miller K (1984) Inhibitory effect of deoxynivalenol, 3-acetyldeoxynivalenol and zearalenone on induction of rat and human lymphocyte proliferation. Toxicology Letters 23: 215–221. [DOI] [PubMed] [Google Scholar]
- 60. Islam MR, Roh YS, Kim J, Lim CW, Kim B (2013) Differential immune modulation by deoxynivalenol (vomitoxin) in mice. Toxicology Letters 221: 152–163. [DOI] [PubMed] [Google Scholar]
- 61. Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A (2004) Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. Journal of Applied Microbiology 97: 1166–1177. [DOI] [PubMed] [Google Scholar]
- 62. Huijsdens XW, Linskens RK, Mak M, Meuwissen SGM, Vandenbroucke-Grauls CMJE, et al. (2002) Quantification of Bacteria Adherent to Gastrointestinal Mucosa by Real-Time PCR. Journal of Clinical Microbiology 40: 4423–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]